Abstract
Protein phosphatase 1 (PP1) is one of the major phosphatases responsible for protein dephosphorylation in eukaryotes. So far, only few specific phosphorylation sites of PP1 regulatory subunit 12A (PPP1R12A) have been shown to regulate the PP1 activity. The effect of insulin on PPP1R12A phosphorylation is largely unknown. Utilizing a mass spectrometry based phosphorylation identification and quantification approach, we identified 21 PPP1R12A phosphorylation sites (7 novel sites, including Ser20, Thr22, Thr453, Ser478, Thr671, Ser678, and Ser680) and quantified 16 of them under basal and insulin stimulated conditions in hamster ovary cells overexpressing the insulin receptor (CHO/IR), an insulin sensitive cell model. Insulin stimulated the phosphorylation of PPP1R12A significantly at Ser477, Ser478, Ser507, Ser668, and Ser695, while simultaneously suppressing the phosphorylation of PPP1R12A at Ser509 (more than 2-fold increase or decrease compared to basal). Our data demonstrate that PPP1R12A undergoes insulin stimulated/suppressed phosphorylation, suggesting that PPP1R12A phosphorylation may play a role in insulin signal transduction. The novel PPP1R12A phosphorylation sites as well as the new insulin-responsive phosphorylation sites of PPP1R12A in CHO/IR cells provides targets for investigation of the regulation of PPP1R12A and the PPP1R12A-PP1cδ complex in insulin action and other signaling pathways in other cell models, animal models, and humans.
Keywords: PPP1R12A, phosphorylation, HPLC-ESI-MS/MS, quantification
1. Introduction
Protein phosphatase 1 (PP1), an abundant serine/threonine phosphatase, is responsible for the majority of protein dephosphorylation in eukaryotes [1–3], regulating diverse biological processes. The interaction between the catalytic subunit of PP1 (PP1c) and the regulatory subunits of PP1 (PP1R) leads to the formation of numerous PP1 complexes with unique substrate specificity, distinct subcellular localization, and various regulatory mechanisms [1–3].
Protein phosphatase 1 regulatory subunit 12A (PPP1R12A), also known as myosin phosphatase target subunit 1 (MYPT1 or MBS), is highly expressed in non-muscle and smooth muscle cells [4] Recently, it has been reported that PPP1R12A is highly expressed in mouse EDL and soleus muscles [5]. One of the well known functions of PPP1R12A is to form a myosin phosphatase holoenzyme with the δ isoform of protein phosphatase 1 catalytic subunit (PP1cδ) in non-muscle and smooth muscle cells, thus modulating the specificity and activity of PP1cδ against phosphorylated myosin, which ultimately regulates subsequent muscle contraction and cell migration [4, 6–8]. In addition, the PPP1R12A/PP1cδ complex has been shown to dephosphorylate polo-like kinase 1 (PLK1), leading to mitotic arrest [9]. Moreover, a number of other proteins involved in different biological processes have been shown to interact with PPP1R12A, such as interleukin-16 and telokin, suggesting that PPP1R12A may be involved in various additional cell functions [4, 6, 8]. Recently, we have identified PPP1R12A as a novel endogenous, insulin stimulated interaction partner of insulin receptor substrate-1 (IRS-1), implying that PPP1R12A might play a role in IRS-1 dephosphorylation and insulin signaling [10].
The domain structure of human PPP1R12A consists of eight ankyrin repeats (residues 39–296 in the human sequence) at the N-terminus and four leucine zipper motifs at the C-terminus, which serve as platforms for protein-protein interactions [6–8, 11]. There is a PP1c-binding motif (K/R-I/V-X-F/W, residues 35–38 in the human sequence) at the N-terminus within PPP1R12A [4], which is required for the binding of PPP1R12A to PP1cδ. In addition, the crystal structure of a PPP1R12A N-terminal fragment (residues 1–299) complexed with PP1cδ indicates that PPP1R12A interacts with PP1cδ through the ankyrin repeats and a myosin phosphatase N-terminal element (“MyPhoNE motif”: RxxQV/I/LK/RxY/W, residues 10–17 in the human PPP1R12A sequence) [12].
It is well known that protein phosphorylation often regulates the parent protein activity in response to various stimuli. Human PPP1R12A has 1,030 amino acid residues, including 120 serines, 90 threonines, and 24 tyrosines. Approximately 40 sites of phosphorylation in PPP1R12A have been identified from different cell lines and various species, and these are listed in four large phosphorylation site databases: www.phosphosite.org, phospho.elm.eu.org, www.uniprot.org, and www.phosida.com. However, only a few of these PPP1R12A phosphorylation sites have been characterized for function. Ser432, Ser473, and Ser601 phosphorylation of PPP1R12A modulates the dephosphorylation of PLK1 and results in mitotic arrest [9]. Phosphorylation of PPP1R12A at Ser445, Ser472 and Ser910 leads to the formation of a complex between PPP1R12A and 14-3-3, and subsequent cell detachment [13]. Phosphorylation of PPP1R12A at Thr696 [14] or Thr853 [15] reduces the binding of PPP1R12A to myosin, leading to inhibition of myosin phosphatase activity and loss of muscle contraction in smooth muscle cells. Nonetheless, phosphorylation status of PPP1R12A in insulin sensitive cells remains to be elucidated.
As a potent anabolic hormone, insulin regulates a wide variety of biological processes including glucose metabolism. Insulin induced protein phosphorylation and dephosphorylation is a key to linking events at the plasma membrane with intracellular machinery. Abnormalities in this process are considered to be one of the main contributing factors to for a large number of disease conditions, such as insulin resistance, the metabolic syndrome, type 2 diabetes, cardiovascular diseases, and cancers. Extensive research has been carried out to study the role of kinases in insulin action. However, a mechanism for serine/threonine phosphatase action in insulin signal transduction is largely unknown. In the present work, multi-segment High Performance Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry (HPLC-ESI-MS/MS) was used to identify and quantify PPP1R12A phosphorylation sites that are regulated by insulin in a widely used insulin sensitive cell model, CHO/IR cells. The peak area of MS2 or MS3 generated fragment ions, an approach developed in our laboratory [16], was utilized to obtain relative quantification information of PPP1R12A phosphorylation upon insulin treatment.
2. Material and methods
2.1. Materials
The following suppliers were used: sequencing-grade trypsin, Sigma (St. Louis, MO); C18 ZipTip, Millipore (Billerica, MA); anti-myc antibody, Santa Cruz (Santa Cruz, CA); anti-phospho-PPP1R12A (Ser507), Cell Signaling Technology (Cell Signaling, MA); anti-PPP1R12A, Millipore (Billerica, MA). Chinese hamster ovary cells overexpressing the insulin receptor (CHO/IR) were a gift from Dr. Feng Liu (University of Texas Health Science Center at San Antonio, TX). The cDNA encoding full-length wild-type human myc-PPP1R12A was a gift from Dr. Masumi Eto [9].
2.2. Cell culture, transfection, immunoprecipitation, and SDS-PAGE
CHO/IR cells were maintained in HAM’s F-12 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Cells were subcultured by trypsinization of subconfluent cultures using 0.25% trypsin with EDTA and were seeded at 2.5 × 106 cells per 10 cm dish. Cells were transfected with 5–10 μg of Myc-tagged PPP1R12A plasmid DNA using Lipofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were serum starved for 4 h at 37°C and treated with or without insulin (100 nM) for 15 min at 37°C. The cells were lysed with 1 mL of lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 20 mM sodium pyrophosphate, 10 mM NaF, 20 mM beta-glycerophosphate, 1% Triton, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin and aprotinin). Bradford Assay (Bio-Rad) was used to estimate protein concentration using bovine serum albumin (BSA) (Sigma, St Louis, MO) as a standard. Cell lysates (1 mg) were diluted in lysis buffer and incubated with 2 μg of anti-myc antibody for PPP1R12A purification. The immunoprecipitates were collected with Protein A agarose beads (Sigma, St. Louis, MO) overnight at 4°C with gentle rotation and were then washed three times with phosphate buffered saline (PBS). Samples were boiled in sodium dodecyl-sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and resolved on 10% 1D-SDS-PAGE. The proteins were then visualized by Coomassie blue staining (Sigma Chemical Co., St. Louis, MO).
2.3. In-gel digestion and Mass spectrometry
All were performed as described previously [16, 17]. Briefly, the gel portions containing PPP1R12A were excised, destained twice with 300 μL of 50% acetonitrile (ACN) in 40 mM NH4HCO3, and dehydrated with 100% ACN for 15 minutes. After removal of ACN by aspiration, the gel pieces were dried in a vacuum centrifuge at 60°C for 30 minutes, and subjected to trypsin digestion overnight. The resulting peptides were desalted and analyzed by on-line HPLC on a Linear Trap Quadrupole-Fourier Transform Ion Cyclotron Resonance (LTQ-FT). Please see the supplemental word file for details.
“Top-10” data-dependent MS/MS analysis was performed to identify PPP1R12A peptides and to obtain their HPLC retention times, as described before [18]. For quantification, the following multi-segment strategy was employed: one survey scan, followed by one parent-list Collision-Induced Dissociation (CID) scan and six targeted CID scans. Included in the parent list were the +2 or +3 charge states of the eight representative PPP1R12A peptides selected from the prominent ions reproducibly observed in the top-10 data-dependent tandem-MS analysis. These peptides were used as internal standards for PPP1R12A protein content (see below). They were selected according to the following criteria: (1) detected by HPLC-ESI-MS with high intensity among PPP1R12A peptides; (2) no or few MS/MS spectra assigned to the respective peptides with missed cleavage; (3) no methionine in the sequence to avoid variability due to methionine oxidization; (4) distributed throughout the PPP1R12A sequence. The 2+ ions of the phosphopeptides of interest were placed on the target list. In order to assess the relative quantities of a large number of phosphopeptides in each experiment and yet still maintain acceptable mass analysis cycle times, the targeted m/z values were grouped into segments based on their expected HPLC retention times. One main reason for a targeted approach is that it offers the capability to quantify co-eluting isobaric phosphopeptides using unique fragment ions as described before [16].
Tandem mass spectra were extracted from Xcalibur ‘RAW’ files and searched against the SwissProt database (Version SwissProt_2011_05, taxonomy, human, 20239 entries) using Mascot (Matrix Science, London, UK; version 2.1). Cross-correlation of Mascot search results with X! Tandem was accomplished with Scaffold (Proteome Software, Portland, OR). The search variables that were used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Probability assessment of peptide assignments and protein identifications were made through the use of Scaffold (version Scaffold_3.0.9.1; Proteome Software, Portland, OR, USA). Only peptides with ≥ 95% probability were considered. Phosphorylation sites were assigned using Scaffold PTM (version 1.0.3, Proteome Software, Portland, OR, USA), a program based on the Ascore algorithm [19–21]. Sites with Ascores > 13 have a p-value < 0.05 and > 95% certainty for correct phosphorylation site localization. Sites with Ascores >19 have a p-value <0.01 and > 99% certainty for correct phosphorylation site localization. In this study, sites with Ascores > 13 (P < 0.05) were considered confidently localized and each site was further conformed by manual inspection of the MS/MS spectrum.
Peak areas for each of the 8 representative PPP1R12A peptides were obtained by integration of the appropriate reconstructed ion chromatograms with 10 ppm error tolerance for precursor ion masses acquired using the Fourier Transform Ion Cyclotron Resonance (FTICR), and mean peak area was calculated. Peak areas for each PPP1R12A phosphopeptides were obtained by integration of the appropriate reconstructed ion chromatograms with 0.5D error tolerance for the fragment ions acquired using the LTQ mass analyzer. If a phosphorylation site existed in various miss cleaved isoforms due to incomplete trypsin digestion, the isoform with the largest peak area was selected for quantification of the phosphorylation site. The peak area for a phosphopeptide was divided by the mean peak area for the 8 representative PPP1R12A peptides during the same HPLC-ESI-MS/MS run to obtain the normalized peak-area ratio. Relative quantification of each phosphopeptide was then obtained by comparing normalized peak-area ratios for control and insulin treated samples [16–18]. A greater than two-fold increase or decrease is considered as a significant change.
2.4 Western blot analysis
CHO/IR cells or CHO/IR cells transfected with PPP1R12A plasmid was serum starved for 4 h at 37°C and treated with or without insulin (100 nM) for 15 min at 37°C. The cells were lysed and protein concentration was estimated using the Bradford assay. Samples were boiled in SDS-PAGE sample buffer and resolved on 10% 1D-SDS-PAGE, transferred onto nitrocellulose membranes (Bio-Rad), and analyzed by Western blotting (WB) with the appropriate antibodies, and the immune complex was detected by chemiluminescence
3. Results and discussion
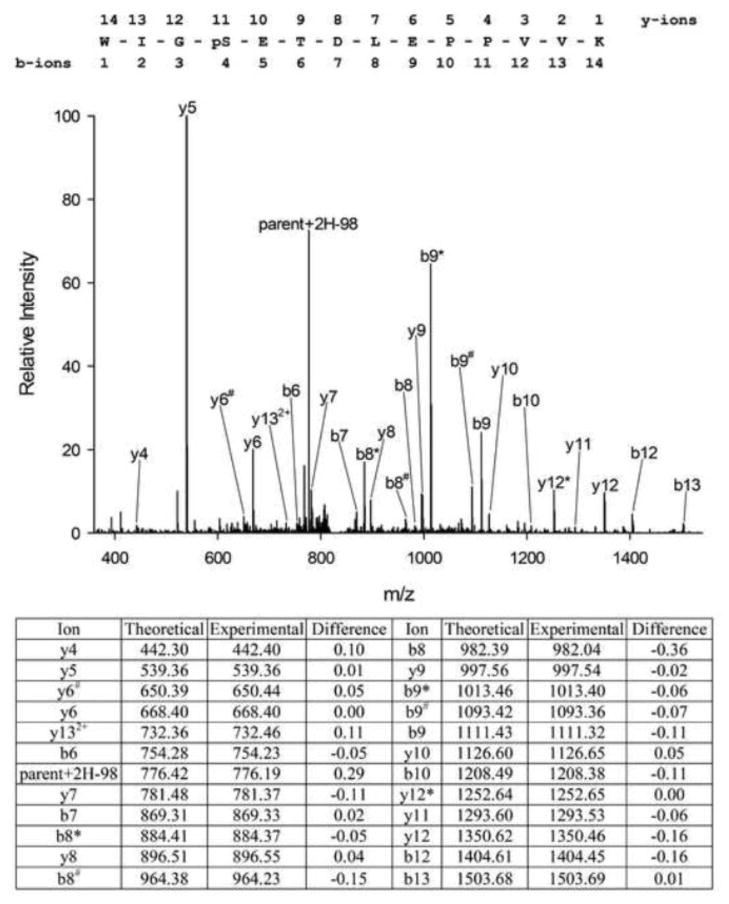
We hypothesized that insulin would regulate phosphorylation of PPP1R12A in CHO/IR cells, a widely used cell model for insulin signaling, and therefore set out to identify PPP1R12A phosphorylation sites and assess how they respond to insulin. Towards this end, myc-tagged PPP1R12A was isolated from CHO/IR cells by immunoprecipitation, and mass spectrometry analysis was performed as described in the Experimental section. The spectra obtained by HPLC-ESI-MS/MS confirmed the presence of PPP1R12A with 64% sequence coverage (Figure 1). Table 1 lists the PPP1R12A phosphopeptides detected and their respective predominant phosphorylation sites. If a phosphorylation site existed in various miscleaved isoforms due to incomplete trypsin digestion, the isoform with the highest Ascore was selected for the phosphorylation site. In all, 21 phosphorylation sites were detected, 7 of which were not listed in the four large phosphorylation site databases (http://www.phosphosite.org; http://phospho.elm.eu.org; http://www.uniprot.org; http://www.phosida.de), and thus appear to be novel. These previously unknown phosphorylation sites include: Ser20, Thr22, Thr453, Ser478, Thr671, Ser678, and Ser680. The MS/MS spectrum for the peptide containing phosphor-Ser20, a novel site, was shown in Figure 2. After the manuscript is accepted for publication, we will post the Scaffold file on our website so that readers can access all MS/MS spectra after installation of the Scaffold viewer, which is freely available on http://www.proteomesoftware.com.
Figure 1.
Combined coverage map of peptides detected in tryptic digests of PPP1R12A expressed in CHO/IR cells using HPLC-ESI-MS/MS analysis. 64% PPP1R12A sequence coverage was obtained. Detected peptides are highlighted yellow, methionine oxidation sites are in lowercase, serine/threonine phosphorylation sites are highlighted green, and novel phosphorylation sites are in bold.
Table 1.
PPPR12A Phosphopeptides identified by HPLC-ESI-MS/MS.
| Start | Stop | Peptide Sequence | Phosphorylation Site | Best Ascore |
|---|---|---|---|---|
| 17 | 30 | WIGpSETDLEPPVVK | S20# | 45.01 |
| 17 | 30 | WIGSEpTDLEPPVVK | T22# | 14.04 |
| 296 | 315 | DKKpSPLIESTANMDNNQSQK | S299 | 100.87 |
| 296 | 315 | DKKSPLIEpSTANMDNNQSQK | S304 | 25.66 |
| 421 | 428 | IpSPKEEER | S422 | 1000.00 |
| 443 | 456 | pTGSYGALAEITASK | T443 | 17.01 |
| 443 | 456 | TGpSYGALAEITASK | S445 | 26.02 |
| 442 | 456 | KTGSYGALAEIpTASK | T453# | 17.32 |
| 476 | 483 | LpSSSLDNK | S477 | 40.00 |
| 476 | 483 | LSpSSLDNK | S478# | 17.01 |
| 505 | 514 | LApSTSDIEEK | S507 | 33.98 |
| 504 | 514 | RLASpTSDIEEKENR | T508 | 16.90 |
| 505 | 514 | LASTpSDIEEK | S509 | 27.96 |
| 504 | 514 | RLApSTpSDIEEK | S507, S509 | 21.94,14.02 |
| 668 | 682 | pSYLTPVRDEESESQR | S668 | 21.98 |
| 668 | 682 | SYLpTPVRDEESESQR | T671# | 76.78 |
| 668 | 682 | SYLTPVRDEEpSESQR | S678# | 54.01 |
| 668 | 682 | SYLTPVRDEESEpSQR | S680# | 51.51 |
| 695 | 709 | pSTQGVTLTDLQEAEK | S695 | 16.99 |
| 852 | 880 | STGVSFWTQDSDENEQEQQpSDTEEGSNKK | S871 | 19.86 |
| 908 | 917 | SGpSYSYLEER | S910 | 30.46 |
| 993 | 1002 | RIpSEMEEELK | S995 | 1000.00 |
The preceding p indicates phosphorylated amino acid;
Novel phosphorylation sites
Figure 2.
Tandem mass spectrum of a novel site of phosphorylation, pSer20, in PPP1R12A from a tryptic digest of PPP1R12A as well as the theoretical and experimental m/z values for detected fragment ions. *Loss of H3PO4 (98 units) from the indicated fragment. #Loss of H2O (18 units).
Among the sites identified, Ser20 and Thr22 are at the N-terminus and in close proximity to the PP1c binding motif (K/R-I/V-X-F/W, residues 35–38 in human PPP1R12A) and the MyPhoNE motif (RxxQV/I/LK/RxY/W, residues 10–17 in the human sequence) [6]. In addition, Ser299 and Thr304 are adjacent to the ankyrin repeats (residues 39–295 in human PPP1R12A). Moreover, Ser422, Thr443, Ser445, Thr453, Ser477, Ser478, Ser507, Thr508, Ser509 are within the middle region of PPP1R12A, which may interact with PP1cδ [22], and seven-in-absentia-homolog 2 (SIAH2, binding sites residues 445–632 in human PPP1R12A) [23]. Ser695 precedes Thr696, a well known inhibitory site of PPP1R12A in smooth muscle cells. Furthermore, three phosphorylation sites were identified in the C-terminus, Ser871, Ser910, and Ser995. Ser995 is within the leucine zipper motif (residues 979–1030 in human PPP1R12A).
To assess the effect of insulin on PPP1R12A phosphorylation, serum starved, CHO/IR cells overexpressing myc-tagged PPP1R12A were either left untreated or treated with insulin. myc-tagged PPP1R12A was immunoprecipitated and resolved by 10% SDS-PAGE. Coomassie blue staining was used for protein visualization, and the gel area corresponding to PPP1R12A was excised and subject to trypsin digestion. Relative phosphorylation quantification by HPLC-ESI-MS/MS was performed as described in the Experimental section. Six independent biological replicates (six basal and six insulin treated, total 12 samples) were utilized to increase the confidence of our findings. The basal and insulin-stimulated samples that were harvested on the same day, resolved on the same gel, and analyzed by HPLC-ESI-MS/MS during the same period of time were paired to minimize day-to-day variations. Eight non-phosphorylated PPP1R12A peptides were chosen to serve as endogenous internal standards as a measure of total PPP1R12A present per sample (supplemental Table 1) based on the criteria described in the Experimental section.
Analysis of PPP1R12A phosphorylation revealed that several PPP1R12A phosphopeptides contain multiple phosphorylation sites (Table 1). Retention time analysis indicated some of the phosphorylation isoforms were co-eluting. Quantification for co-eluting isobaric phosphorylated peptides was obtained by generating MS2 or MS3 fragment ions and using the peak areas of the fragment b and y ions that were unique to each phosphorylation site within each phosphopeptide as described in Langlais et al. [16]. Quantification for other phosphorylated peptides was also obtained by generating MS2 or MS3 fragment ions derived from individual phosphopeptide. Among the 21 phosphorylation sites identified, we were able to obtain quantitative information for 16 of them (Table 2). The detailed quantification data were included in Supplemental Table 2 and 3. Each PPP1R12A phosphorylation site was normalized by the average value of the respective basal sample and then expressed as a fold-change over basal ± SEM (n=6). Phosphorylation of PPP1R12A at Ser20, Ser299, Ser304, Ser422, Thr443, Ser445, Thr671, Ser678, Ser910, and Ser995 was not significantly affected by insulin (Table 2 and Supplemental Table 3). In contrast, more than 2-fold insulin stimulation was observed for the phosphorylation of PPP1R12A at Ser477, Ser478, Ser507, Ser668, and Ser695 (Table 2 and Supplemental Table 2). On the other hand, Ser509 phosphorylation decreased 2-fold upon the treatment of insulin. The increased or decreased phosphorylation of PPP1R12A upon insulin treatment has not been previously reported for these sites. The global phosphorylation profile of PPP1R12A upon insulin stimulation implies the activation or deactivation of various kinases and/or phosphatases.
Table 2.
Summary of the effect of insulin on PPP1R12A phosphorylation
| Start | Stop | Peptide Sequence | Phosphor-Site | Fold Change a | Log2 fold change |
|---|---|---|---|---|---|
| 17 | 30 | WIGpSETDLEPPVVK | Ser20 | 1.88±0.20 | 0.91 |
| 296 | 315 | DKKpSPLIESTANMDNNQSQK | Ser299 | 0.99±0.16 | −0.01 |
| 296 | 315 | DKKSPLIEpSTANMDNNQSQK | Ser304 | 1.00±0.17 | 0.00 |
| 421 | 428 | IpSPKEEER | Ser422 | 0.85±0.17 | −0.23 |
| 443 | 456 | pTGSYGALAEITASK | Thr443 | 0.97±0.16 | −0.04 |
| 443 | 456 | TGpSYGALAEITASK | Ser445 | 1.24±0.06 | 0.31 |
| 476 | 483 | LpSSSLDNK | Ser477 | 10.92±2.97* | 3.45 |
| 476 | 483 | LSpSSLDNK | Ser478 | 4.84±1.39* | 2.28 |
| 505 | 514 | LApSTSDIEEK | Ser507 | 6.53±1.37* | 2.71 |
| 505 | 514 | LASTpSDIEEK | Ser509 | 0.48±0.08* | −1.06 |
| 504 | 514 | RLApSTpSDIEEK | Ser507, Ser509 | 3.49±0.61* | 1.80 |
| 668 | 682 | pSYLTPVRDEESESQR | Ser668 | 2.25±0.38* | 1.17 |
| 668 | 682 | SYLpTPVRDEESESQR | Thr671 | 0.66±0.06 | −0.60 |
| 668 | 682 | SYLTPVRDEEpSESQR | Ser678 | 0.60±0.11 | −0.74 |
| 695 | 709 | pSTQGVTLTDLQEAEK | Ser695 | 2.38±0.18* | 1.25 |
| 908 | 917 | SGpSYSYLEER | Ser910 | 1.19±0.20 | 0.25 |
| 993 | 1002 | RIpSEMEEELK | Ser995 | 1.00±0.11 | 0.00 |
Fold change: obtained by dividing the normalized peak area for a phosphopeptide in each experiment by the average of the normalized peak area under the control condition, n=6.
Indicate insulin stimulated or suppressed phosphorylation at these sites by more than 2-fold.
We performed WB analyses to confirm the insulin stimulated phosphorylation of Ser507 (pS507) in human PPP1R12A overexpressed in CHO/IR cells, and to determine whether insulin has similar effect on pS507 in human PPP1R12A overexpressed in CHO/IR cells and in endogenous PPP1R12A in CHO/IR cells. Ser507 are conserved in human, rat, mouse, as well as many other species including Chinese hamster from which the CHO/IR cell line was derived. As can be seen from Figure 3A, pS507 in human PPP1R12A overexpressed in CHO/IR cells increased upon insulin stimulation, which is consistent with the results obtained by HPLC-ESI-MS/MS. In addition, as shown in Figure 3B, pS507 in endogenous PPP1R12A in CHO/IR cells also increased after insulin treatment. These results indicated that insulin has similar stimulating effect on pS507 in overexpressed PPP1R12A and endogenous PPP1R12A in CHO/IR cells.
Figure 3.
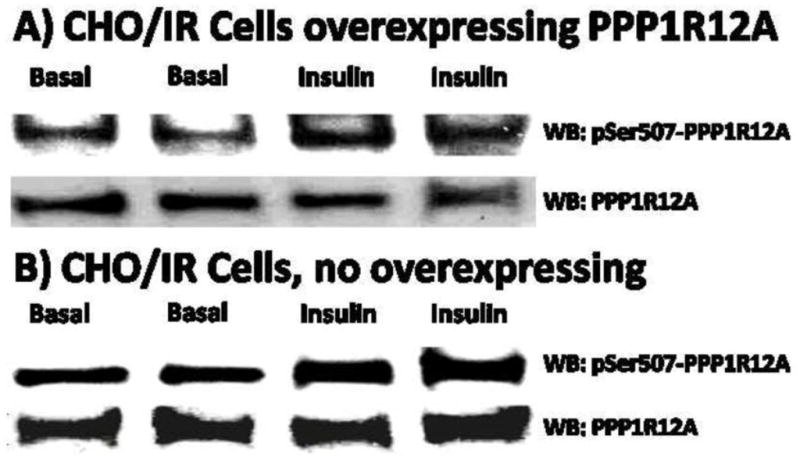
Western blotting analysis of pSer507 in PPP1R12A. A), Western blotting confirmation of insulin stimulated pSer507 in PPP1R12A overexpressed in CHO/IR cells. B), insulin stimulated pSer507 in endogenous PPP1R12A in CHO/IR cells.
As discussed before, Ser20 and Thr22, novel sites identified in this study, are in close proximity to the PP1c binding motif and the MyPhoNE motif [6]. As the crystal structure of the PP1 complex between chicken PP1cδ isoform (also called β isoform) and amino acids 1–299 of PPP1R12A (also called MYPT1) indicates, residues 1–34, which precede the PP1c binding motif in human PPP1R12A (residues 35–38), also interact with PP1cδ [12]. It has been shown that a short peptide (residues 23–38) of PPP1R12A containing the PP1c binding motif but lacking the N terminus, binds to PP1c but has no effect on PP1cδ catalytic activity [22], while a peptide containing residues 1–38 of PPP1R12A interacts with PP1c and increases its phosphatase activity. It is possible that pSer20 may play a role in regulating PP1cδ activity when in a complex with PPP1R12A. Without pSer20, PPP1R12A may still bind to PP1cδ through the PP1c binding motif; however, the resulting complex may not have the full phosphatase activity against its substrates. In addition, Ser20 is also near a nuclear localization signal (residues 27–33) within PPP1R12A. When PPP1R12A does not bind to PP1c or other ligands, it is mainly localized in the nucleus [24]. Insulin stimulated phosphorylation at pSer20, even though less than 2-fold, might be required for the translocation of PPP1R12A from the nucleus to the cytoplasm.
Ser477, Ser478, and Ser507 of PPP1R12A exhibited over a 4-fold increase of phosphorylation after insulin treatment (10.92±2.97, 4.84±1.39, 6.53±1.37, respectively). It was reported that PP1cδ may interact with the PPP1R12A truncation containing residues 304–511 by Surface Plasmon Resonance [22], and it is tempting to speculate that phosphorylation at Ser477, Ser478, and Ser507 may also be involved in the interaction between PPP1R12A and PP1cδ. Furthermore, SIAH2 can interact with PPP1R12A at residues 445–632, resulting in accelerated proteasomal degradation of PPP1R12A [23]. A SIAH-binding motif, RLAYVAP (residues 493–499), is required for the interaction to occur. Thus, phosphorylation at these 3 sites might be involved in the regulation of PPP1R12A protein abundance. The increase in phosphorylation of PPP1R12A at Ser477, Ser478, and Ser507 represents one of the strongest fold changes of any insulin stimulated serine and threonine phosphorylation we have studied to date using this mass spectrometry technique for the quantification of protein phosphorylation [16–18, 25–29]. The strength of insulin stimulated PPP1R12A phosphorylation at Ser477, Ser478, and Ser507 could be interpreted as a major regulatory mechanism responsible for the control of PPP1R12A function in insulin signaling.
Contrary to Ser507 phosphorylation, insulin suppressed Ser509 phosphorylation by more than 2-fold. It has been reported that phosphorylation at adjacent residues may interplay with each other, such as Ser629 and Ser636 of IRS-1. Insulin stimulates phosphorylation of IRS-1 at Ser629, leading to decreased phosphorylation of IRS-1 at Ser636 and enhanced downstream signaling [29]. Whether Ser507 and Ser509 play a similar role warrants further investigation.
It is noted that one double phosphorylation peptide, RLApSTpSDIEEK (phosphor-Ser507 and Ser509), was identified with AScore >13. Quantification by HPLC-ESI-MS/MS indicated a 3.49±0.61 fold increase of the double phosphorylated peptide upon insulin stimulation. As discussed before, insulin enhanced Ser507 phosphorylation while suppressed Ser509 substantially. It is possible that a portion of the decrease observed for Ser509 might due to the monophosphorylated peptides containing Ser509 became double phosphorylated.
It has been shown that phosphorylation of PPP1R12A at Ser695 by cGMP-dependent protein kinase (PKG), which may prevent Thr696 phosphorylation in smooth muscle cells, leads to increased myosin phosphatase activity and causes smooth muscle relaxation [30]. Based on Scaffold PTM analysis, Thr696 was not phosphorylated in CHO/IR cells either in basal or insulin stimulated conditions, indicating that Ser695 phosphorylation might prevent Thr696 phosphorylation in CHO/IR cells. Whether Ser695 phosphorylation can enhance PPP1R12A-PP1cδ complex activity in CHO/IR cells remains to be elucidated.
We used literature searches and NetworKIN 2.0, an online bioinformatics tool, to predict kinases capable of phosphorylating PPP1R12A (Table 3) [31, 32]. The potential kinases for the PPP1R12A phosphorylation sites that underwent insulin stimulation, Ser477, Ser478, Ser507, Ser668, and Ser695, included AMPK-related protein kinase 5 (NUAK1), p21-activated kinases (PAKs), Casein kinase II (CK2), Ribosomal protein S6 kinase 1/2 (RPS6KB1/2), Myotonic dystrophy protein kinase (DMPK), cGMP-dependent protein kinase (PKG), all of which are kinases that have been shown to be activated by insulin [33–37]. In addition, the potential kinases for insulin suppressed site Ser509 included casein kinases. More studies will be needed to better understand the involvement of these kinases in the insulin stimulated or suppressed phosphorylation of PPP1R12A.
Table 3.
Potential kinases for PPP1R12A phosphorylation sites
| Start | Stop | Peptide Sequence | Phosphor-Site | Predicted Kinases |
|---|---|---|---|---|
| 17 | 30 | WIGpSETDLEPPVVK | Ser20 | CSNK2A2, CK2A1 |
| 296 | 315 | DKKpSPLIESTANMDNNQSQK | Ser299 | CDK2, CDK3 |
| 296 | 315 | DKKSPLIEpSTANMDNNQSQK | Ser304 | NEK2, DMPK |
| 421 | 428 | IpSPKEEER | Ser422 | CDK2, CDK3 |
| 443 | 456 | pTGSYGALAEITASK | Thr443 | DMPK, AURKB, NEK2 |
| 443 | 456 | TGpSYGALAEITASK | Ser445 | PRKAA1, PRKAA2, NUAK1 |
| 476 | 483 | LpSSSLDNK | Ser477* | PAK4, PAK6, PAK7 |
| 476 | 483 | LSpSSLDNK | Ser478* | CSNK2A2, CK2A1 |
| 505 | 514 | LApSTSDIEEK | Ser507* | CSNK2A2, CK2A1 |
| 505 | 514 | LASTpSDIEEK | Ser509* | CSNK2A2, CK2A1 |
| 668 | 682 | pSYLTPVRDEESESQR | Ser668* | RPS6KB1, RPS6KB2, PAK4, PAK6, PAK7, DMPK, Pim1/2, CLK1, CLK2 |
| 668 | 682 | SYLpTPVRDEESESQR | Thr671 | CDK2, CDK3 |
| 668 | 682 | SYLTPVRDEEpSESQR | Ser678 | CSNK2A2, CK2A1, TGFBR2, ACVR2A, ACVR2B |
| 695 | 709 | pSTQGVTLTDLQEAEK | Ser695* | PKA, PKG, PIM2 |
| 908 | 917 | SGpSYSYLEER | Ser910 | AKT1, AKT2, AKT3, PAK4, PAK6, PAK7, NUAK1 |
| 993 | 1002 | RIpSEMEEELK | Ser995 | CSNK2A2, CK2A1 |
Insulin stimulated or suppressed phosphorylation at these sites by more than 2-fold.
Using NetworKIN 2.0 Beta (http://networkin.info/version_2_0/newPrediction.php) and additional literature search.
Kinase abbreviations: Activin receptor type IIA/B precursor (ACVR2A/B), RAC serine/threonine-protein kinase (AKT1/2/3), Serine/threonine-protein kinase aurora-B (AURKB), Cell division protein kinase 2/3 (CDK2/3), casein kinase 2, alpha 1 polypeptide (CK2A1), casein kinase 2, alpha prime polypeptide (CSNK2A2 or CK2A2), Dual specificity protein kinase CLK1/2 (CLK1/2), Myotonin-protein kinase (DMPK), Serine/threonine-protein kinase Nek2 (NEK2), AMPK-related protein kinase 5 (NUAK1), Serine/threonine-protein kinase PAK 4/6/7 (PAK4/6/7, also known as p21-activated kinase 4/6/7), cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), Proto-oncogene serine/threonine-protein kinase Pim-1/2 (PIM1/2), 5′-AMP-activated protein kinase, catalytic alpha-1/2 chain (PRKAA1/2 or AMPK alpha-1/2), Ribosomal protein S6 kinase 1/2 (RPS6KB1/2), TGF-beta receptor type II precursor (TGFBR2).
4. Conclusions
The present work provides a global phosphorylation profile of PPP1R12A upon insulin stimulation in CHO/IR cells, an established insulin sensitive cell model, using HPLC-ESI-MS/MS. We are the first to report that insulin stimulated phosphorylation of PPP1R12A at Ser477, Ser478, Ser507, Ser668, and Ser695 by more than 2-fold, while suppressed phosphorylation at Ser509 by more than 2-fold. These results indicate that insulin has a profound effect on PPP1R12A phosphorylation, and activation/deactivation of a variety of kinases or phosphatases may be required to regulate PPP1R12A phosphorylation and downstream signaling. We have also identified seven, previously unreported PPP1R12A phosphorylation sites, including Ser20, Thr22, Thr453, Ser478, Thr671, Ser678, and Ser680. Even though most of these novel sites did not respond to insulin in CHO/IR cells, they provide targets for investigation on the regulation of PPP1R12A and/or PP1cδ in other cells, such as smooth muscle cells or cardiomyocytes, by insulin or other stimuli. A summary of these PPP1R12A phosphorylation findings is provided in Figure 4. This work demonstrates again that the HPLC-ESI-MS/MS peak area based label-free quantification strategy developed in our laboratory have the capability to provide relative quantification of multiple phosphorylation sites simultaneously. Future experiments will aim to increase the sequence coverage and phosphorylation sites discovery using additional enzymes for digestion, such as Glu-c and chymotrypsin, and will concentrate on confirming the effect of insulin on PPP1R12A phosphorylation in both animal and human muscle, while site-specific mutagenesis will be employed to assess the role of PPP1R12A phosphorylation in insulin signaling within in vitro insulin signaling models, such as L6 myotubes. Elucidating the function of PPP1R12A phosphorylation in insulin signaling may provide novel insight into the reduced insulin action that is associated with insulin resistance and the metabolic syndrome.
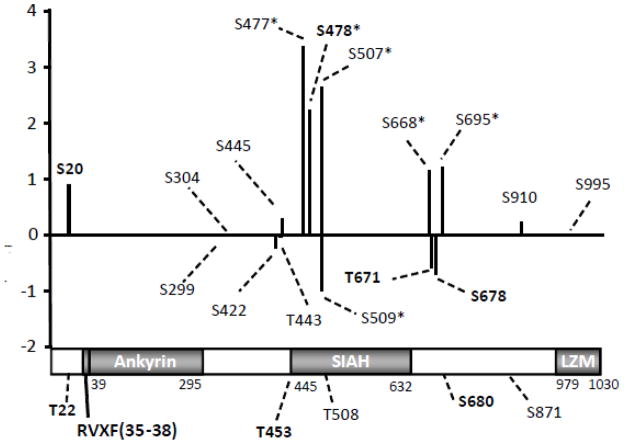
Figure 4.
Summary of PPP1R12A phosphorylation. Insulin stimulated changes relative to basal values are expressed as the base-2 log of the change; thus, a 100% increase corresponds to a value of 1.0, and a 50% decrease corresponds to a value of 1.0. For sites where there was no change in relative phosphorylation, the value is 0. Among the 21 phosphorylation sites identified, we were able to obtain quantitative information for 16 of them. The amino acid residue number for each site of phosphorylation that is quantifiable is given on the x-axis, while the 5 unquantiable sites were indicated in the domain structure of PPP1R12A. The positions of the Ankyrin repeats, SIAH, and LZM domains are shown in gray. The black bar preceding the Ankyrin repeats represents the PP1c binding motif, aa35–38. * indicates sites with more than 2-fold increase or decrease in phosphorylation upon insulin stimulation, n=6. Novel sites are indicated in bold.
Supplementary Material
Highlights.
Identified 21 PPP1R12A phosphorylation sites, 7 of which were novel
Insulin stimulated or suppressed PPP1R12A phosphorylation at multiple sites
Identified a phosphatase regulatory subunit as a new insulin signaling protein
Provide targets for studying phosphatases in insulin action in health and disease
Demonstrated again mass spectrometry can quantify multiple phosphorylation sites
Acknowledgments
This research was supported by funds from the National Institute of Health R01DK081750 (ZY) and an American Diabetes Association Clinical/translational Research Award 7-09-CT-56 (ZY). We thank Thangiah Geetha for help with the sample preparation.
Footnotes
The supplemental table 1, 2 and 3 are available free of charge via the Internet.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–8. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–56. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 3.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–45. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: Many roles in cell function. Biochemical and biophysical research communications. 2008;369:149–56. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryder JW, Lau KS, Kamm KE, Stull JT. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J Biol Chem. 2007;282:20447–54. doi: 10.1074/jbc.M702927200. [DOI] [PubMed] [Google Scholar]
- 6.Grassie ME, Moffat LD, Walsh MP, Macdonald JA. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Arch Biochem Biophys. 2011 doi: 10.1016/j.abb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto R, Kato T, Mizoguchi A, Takahashi N, Nakakuki T, Mizutani H, et al. Characterization and function of MYPT2, a target subunit of myosin phosphatase in heart. Cell Signal. 2006;18:1408–16. doi: 10.1016/j.cellsig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem. 2004;259:197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- 9.Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, et al. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Developmental cell. 2008;14:787–97. doi: 10.1016/j.devcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geetha T, Langlais P, Luo M, Mapes R, Lefort N, Chen SC, et al. Label-free proteomic identification of endogenous, insulin-stimulated interaction partners of insulin receptor substrate-1. J Am Soc Mass Spectrom. 2011;22:457–66. doi: 10.1007/s13361-010-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shichi D, Arimura T, Ishikawa T, Kimura A. Heart-specific small subunit of myosin light chain phosphatase activates rho-associated kinase and regulates phosphorylation of myosin phosphatase target subunit 1. J Biol Chem. 2010;285:33680–90. doi: 10.1074/jbc.M110.122390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–4. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 13.Zagorska A, Deak M, Campbell DG, Banerjee S, Hirano M, Aizawa S, et al. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Science signaling. 2010;3:ra25. doi: 10.1126/scisignal.2000616. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa K, Ito M, Hartshorne DJ. Phosphorylation of the large subunit of myosin phosphatase and inhibition of phosphatase activity. J Biol Chem. 1996;271:4733–40. doi: 10.1074/jbc.271.9.4733. [DOI] [PubMed] [Google Scholar]
- 15.Velasco G, Armstrong C, Morrice N, Frame S, Cohen P. Phosphorylation of the regulatory subunit of smooth muscle protein phosphatase 1M at Thr850 induces its dissociation from myosin. FEBS Lett. 2002;527:101–4. doi: 10.1016/s0014-5793(02)03175-7. [DOI] [PubMed] [Google Scholar]
- 16.Langlais P, Mandarino LJ, Yi Z. Label-free relative quantification of co-eluting isobaric phosphopeptides of insulin receptor substrate-1 by HPLC-ESI-MS/MS. J Am Soc Mass Spectrom. 2010;21:1490–9. doi: 10.1016/j.jasms.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi Z, Langlais P, De Filippis EA, Luo M, Flynn CR, Schroeder S, et al. Global assessment of regulation of phosphorylation of insulin receptor substrate-1 by insulin in vivo in human muscle. Diabetes. 2007;56:1508–16. doi: 10.2337/db06-1355. [DOI] [PubMed] [Google Scholar]
- 18.Yi Z, Luo M, Mandarino LJ, Reyna SM, Carroll CA, Weintraub ST. Quantification of phosphorylation of insulin receptor substrate-1 by HPLC-ESI-MS/MS. J Am Soc Mass Spectrom. 2006;17:562–7. doi: 10.1016/j.jasms.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 20.Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res. 2008;7:1675–82. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth A, Kiss E, Herberg FW, Gergely P, Hartshorne DJ, Erdodi F. Study of the subunit interactions in myosin phosphatase by surface plasmon resonance. Eur J Biochem. 2000;267:1687–97. doi: 10.1046/j.1432-1327.2000.01158.x. [DOI] [PubMed] [Google Scholar]
- 23.Twomey E, Li Y, Lei J, Sodja C, Ribecco-Lutkiewicz M, Smith B, et al. Regulation of MYPT1 stability by the E3 ubiquitin ligase SIAH2. Experimental cell research. 2010;316:68–77. doi: 10.1016/j.yexcr.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Muranyi A, Erdodi F, Hartshorne DJ. Localization of myosin phosphatase target subunit and its mutants. Journal of muscle research and cell motility. 2005;26:123–34. doi: 10.1007/s10974-005-2579-5. [DOI] [PubMed] [Google Scholar]
- 25.Højlund K, Yi Z, Lefort N, Langlais P, Bowen B, Levin K, et al. Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia. 2010;53:541–51. doi: 10.1007/s00125-009-1624-0. [DOI] [PubMed] [Google Scholar]
- 26.Holmes RM, Yi Z, De Filippis E, Berria R, Shahani S, Sathyanarayana P, et al. Increased abundance of the adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain and leucine zipper motif (APPL1) in patients with obesity and type 2 diabetes: evidence for altered adiponectin signalling. Diabetologia. 2011;54:2122–31. doi: 10.1007/s00125-011-2173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langlais P, Yi Z, Finlayson J, Luo M, Mapes R, De Filippis E, et al. Global IRS-1 phosphorylation analysis in insulin resistance. Diabetologia. 2011 doi: 10.1007/s00125-011-2271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langlais P, Yi Z, Mandarino LJ. The Identification of Raptor as a Substrate for p44/42 MAPK. Endocrinology. 2011;152:1264–73. doi: 10.1210/en.2010-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo M, Langlais P, Yi Z, Lefort N, De Filippis EA, Hwang H, et al. Phosphorylation of human insulin receptor substrate-1 at Serine 629 plays a positive role in insulin signaling. Endocrinology. 2007;148:4895–905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wooldridge AA, MacDonald JA, Erdodi F, Ma C, Borman MA, Hartshorne DJ, et al. Smooth muscle phosphatase is regulated in vivo by exclusion of phosphorylation of threonine 696 of MYPT1 by phosphorylation of Serine 695 in response to cyclic nucleotides. J Biol Chem. 2004;279:34496–504. doi: 10.1074/jbc.M405957200. [DOI] [PubMed] [Google Scholar]
- 31.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–26. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–9. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Khalid S, Rozakis-Adcock M, Fantus IG, Jin T. P-21-activated protein kinase-1 functions as a linker between insulin and Wnt signaling pathways in the intestine. Oncogene. 2009;28:3132–44. doi: 10.1038/onc.2009.167. [DOI] [PubMed] [Google Scholar]
- 34.Tanasijevic MJ, Myers MG, Jr, Thoma RS, Crimmins DL, White MF, Sacks DB. Phosphorylation of the insulin receptor substrate IRS-1 by casein kinase II. J Biol Chem. 1993;268:18157–66. [PubMed] [Google Scholar]
- 35.Llagostera E, Catalucci D, Marti L, Liesa M, Camps M, Ciaraldi TP, et al. Role of myotonic dystrophy protein kinase (DMPK) in glucose homeostasis and muscle insulin action. PLoS One. 2007;2:e1134. doi: 10.1371/journal.pone.0001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergandi L, Silvagno F, Russo I, Riganti C, Anfossi G, Aldieri E, et al. Insulin stimulates glucose transport via nitric oxide/cyclic GMP pathway in human vascular smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology. 2003;23:2215–21. doi: 10.1161/01.ATV.0000107028.20478.8e. [DOI] [PubMed] [Google Scholar]
- 37.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circulation research. 2007;100:328–41. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.