Abstract
Background
During the 2009 influenza A(H1N1) pandemic, household transmission studies were implemented to understand better the characteristics of the transmission of the novel virus in a confined setting.
Methods
We conducted a systematic review and meta-analysis to assess and summarize the findings of these studies. We identified 27 articles, around half of which reported studies conducted in May and June 2009.
Results
In 13 of the 27 studies (48%) that collected respiratory specimens from household contacts, point estimates of the risk of secondary infection ranged from 3 to 38%, with substantial heterogeneity. Meta-regression analyses revealed that a part of the heterogeneity reflected varying case ascertainment and study designs. The estimates of symptomatic secondary infection risk, based on 20 studies identifying febrile acute respiratory illness among household contacts, also showed substantial variability, with point estimates ranging from 4% to 37%.
Conclusions
Transmission of the 2009 pandemic virus in households appeared to vary in different countries and settings, with differences in estimates of the secondary infection risk also partly due to differences in study designs.
In 2009 influenza A (H1N1-2009) virus (abbreviated as pH1N1) emerged to cause the first influenza pandemic of the 21st Century.1 Many epidemiologic studies were carried out to characterize the epidemiology of pH1N1 and inform decisions about possible countermeasures. Of particular early interest was the frequency of transmission from confirmed cases to their close contacts. The household, defined as a person or a group of people living in the same residence, provides a strategic setting to track infections among close contacts of cases: one, the denominator is well-defined, and two, household contacts can generally be identified and followed with fewer resources than other types of contacts. It is also strategic to monitor transmission in households where up to 30% of influenza virus transmission is believed to occur.2,3
A substantial fraction of influenza infections are asymptomatic or associated with mild disease that does not require medical attention. Epidemiologic studies of severe cases (e.g. hospitalized cases) can provide detailed information on the risk factors of death and thus help examine the prognosis and clinical effectiveness of treatments among the severe fraction of cases. However, studies in households can observe the full range of illness associated with influenza, as well as examining transmission in a confined setting.4 In a confined setting, one way to quantify the overall risk of infection (which may be used as a relative measurement of the transmissibility of an influenza virus), is to estimate the proportion of susceptible household contacts of an index case who subsequently become infected.5 The conditional risk of infection in households given exposure to an index case can be estimated overall, or compared among subgroups (e.g. children vs adults), or between persons who did or did not receive specific interventions such as antiviral prophylaxis.6,7 The clinical onset serial interval (defined as the time from illness onset of an index case to illness onset of a secondary case infected by the index case8) offers insight into the natural history of influenza and is key to interpretation of the early growth of cases using mathematical modeling techniques.9 Estimates of the mean serial interval in households are often regarded as a proxy for the average time between successive host generations of infection in a population,9 although the applicability of household results to other settings is still subject to clarification.
Timely household studies can be highly informative.10 The household secondary infection risk (SIR), more commonly referred to as the secondary attack rate (SAR), represents an overall risk of infection among household contacts for a defined time period. Throughout this paper we use SIR, because (1) the household outcome of our interest is not necessarily best referred to as an attack in that many influenza infections are mild and (2) rate is a misnomer and the SIR is a measure of risk under certain assumptions5 (see Methods section below). Here we report a systematic review of household transmission studies of pH1N1. Our objectives were to review the design and implementation of household studies during the pandemic, to compare and contrast their findings and summarize the epidemiologic characteristics of pH1N1 in households, and to identify important considerations for future household studies of influenza transmission.
METHODS
This systematic review and meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.11
Search strategy
Studies containing data on household transmission of the pH1N1 virus were retrieved from the Scopus and Medline (PubMed) electronic databases on 29 June 2011. We used the following free-text search terms in “All fields”:
-
#1
“influenza” OR “flu” OR “H1N1*” OR “pH1N1” OR “nH1N1” OR “vH1N1”
-
#2
“family” OR “household” OR “house” OR “home”
-
#3
“transmission” OR “infection” OR “contagion” OR “spread” OR “attack”
-
#4
#1 AND #2 AND #3
We limited the search to studies published after March 2009 (i.e. subsequent to the emergence of pH1N1) through 12 December, 2011. Additional relevant studies identified by the authors were manually retrieved from other databases.
Study selection
All titles identified by the search strategy were independently screened by two authors (L.L.H.L and B.J.C.). Abstracts of potentially relevant titles were then reviewed for eligibility, and articles were selected for closer examination if a description of household transmission was available. Eligible articles reported a household SIR for pH1N1 or sufficient data to retrieve a SIR, and must have reported data based on ≥5 households. Studies that followed persons only in households in which transmission occurred were also excluded. Multiple reports of the same dataset were assessed and the most comprehensive report of a study was included.
Ascertainment of secondary cases
Household SIRs were calculated as the number of identified cases divided by the number of household contacts. Although not always explicitly mentioned, the estimation of SIRs as an overall conditional risk of infection given exposure involves the following assumptions: (1) household contacts are equally susceptible, (2) SIR is the conditional risk given exposure to the index case(s), and is examined for a reasonable length of time following illness onset in the index case, and (3) the SIR as defined here is a mixture of the risks of infection in households as well as in the community, and, thus, is regarded as an overall risk of infection among household contacts.5 Household SIRs could be estimated either through identification of pH1N1 virus by virologic testing, by serologic evidence of infection, or based on clinical diagnosis or self-reported signs and symptoms. Virologic methods to confirm pH1N1 infections included reverse transcription polymerase chain reaction (RT-PCR) or viral culture on specimens collected from the respiratory tract. Serologic methods included analysis of paired serological specimens by hemagglutination inhibition or viral neutralization assays, with a 4-fold or greater rise between baseline and convalescence conventionally used to indicate infection. Many studies assessed incidence of febrile acute respiratory illness among household contacts, where the illness was a febrile upper respiratory tract infection mostly defined as the presence of fever plus cough or sore throat (a common surveillance definition of influenza-like illness). In some studies the definition was extended to include the presence of fever plus one or more of the following symptoms: coryza, shortness of breath, sneezing, rhinorrhea, sore throat, feverishness, arthralgia, myalgia, prostration or headache. The threshold for body temperature used to classify fever varied from 37.5°C to 38.0°C. Some studies also reported the occurrence of acute respiratory illness among contacts where acute respiratory illness was a broad definition of febrile or afebrile upper respiratory tract infection, typically the presence of two or more influenza-related signs or symptoms. To distinguish the estimated SIR by various approaches to ascertainment, we define SIRPCR, SIRFARI, and SIRARI as the SIR ascertained by RT-PCR, febrile acute respiratory illness, and acute respiratory illness, respectively.
Data extraction
The primary data extracted were the total number of household contacts and infected contacts according to laboratory or clinical outcome measures. Whenever available, we extracted SIRs stratified by age group, household size and antiviral use. Children were usually defined as those up to 15 years of age, although the age threshold in some studies differed by one or two years. We also extracted estimated mean or median serial intervals with 95% confidence intervals, mean household sizes, numbers of households, and index case age distributions if they were reported. Infections identified in household contacts could potentially be generalizable to all naturally-acquired pH1N1 infections (in contrast to, for example, only cases presenting for outpatient medical care). Therefore, data on illness profiles associated with pH1N1 infection were extracted from studies that confirmed infection by laboratory testing, regardless of whether household contacts were symptomatic or asymptomatic. All data were extracted onto a standardized form.
Statistical analysis
SIRs were stratified according to method of ascertainment (virologic or clinical), and further divided into adult and child age groups when data permitted. Combined estimates of the SIR were based on proportions transformed by logit transformation12,13 and combined using a DerSimonian-Laird random-effects model.14 We assessed statistical heterogeneity by the I2 statistic, with higher values signifying greater degree of variation.15 Meta-regression analyses were conducted using multivariate mixed effects models,16 and missing data were dealt with using multiple imputation. All analyses were conducted with R version 2.11.1 (R Development Core Team, Vienna, Austria) and the metafor package.17
RESULTS
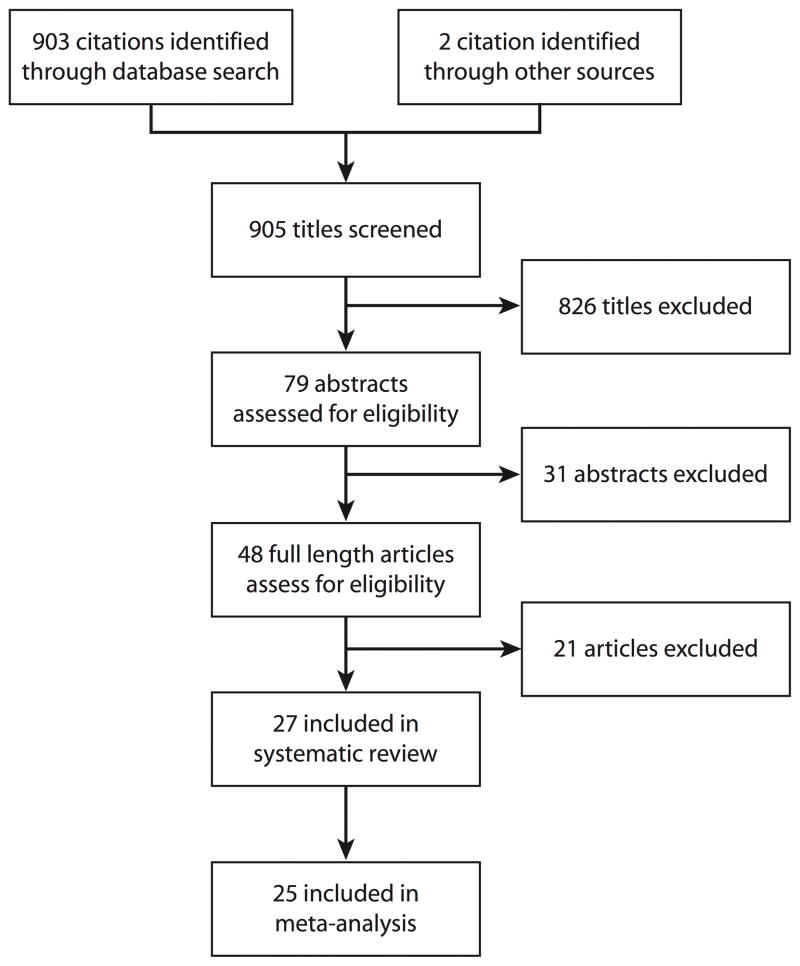
Of the 903 titles initially identified from the database search, 48 full-length articles were assessed for inclusion, of which 21 were excluded (eTable 1 (http://links.lww.com)) and 27 studies were determined to be eligible and included in this systematic review (Figure 1).18–45 We excluded one study26 because it re-analyzed a subset of study subjects that had been recruited in an earlier study that was already included in the analysis24; we counted only the earlier study when we extracted serological testing results from the follow-up study.26 One article46 reported preliminary analyses of data from the “First Few 100” study in the United Kingdom, and the results of this study were subsequently updated in a more comprehensive publication with a slightly larger dataset.36 We therefore excluded the preliminary report from our systematic review and meta-analysis.
Figure 1.
Flow diagram of study selection.
The 27 included articles are summarized in Table 1. Most studies (25/27) identified index cases through various means (e.g. recruitment at primary health care providers, surveying specific exposed populations such as schools and camps, follow-up of cases reported to national public health databases, or through general practice sentinel surveillance networks) and then followed up their households for 1–4 weeks to observe transmission. The other 2 studies recruited subjects from cohorts that had been established prior to observation of household transmission. There was variation in the definition of household and household members or contacts used among the studies that explicitly defined household contacts (eTable 2, http://links.lww.com). Some studies used a traditional definition of household contacts (those who resided with the index), while other studies broadened the definition to include close contacts who spent 1 or more nights in the household, or even any exposure of at least 1 hour to the infectious index case in a household setting.
Table 1.
Summary of household studies of influenza A (H1N1-2009) included in the systematic reviewa
| Location (City, Country) |
Study Period |
No. of Households |
No. of Subjects |
Recruitment Strategyb |
Multiple Index |
Index case ascertainment |
Primary Outcome |
All contacts |
Primary SIR, % | Secondary Outcome(s) |
Secondary SIR(s), % |
Follow-up duration of households |
Serial interval (days) |
Mean Household size | Serology Collected |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calatayud et al 201018 | London, UK | Apr 09 – May 09 | n/a | 47 | Cohort | N | RT-PCR | RT-PCR | N | 17% | n/a | n/a | n/a | n/a | n/a | N |
| Carcione et al 201119 | Western Australia | May 09 – Aug 09 | 595 | 2184 | Case ascertained | N | RT-PCR | RT- PCR/FARI combined | N | 20% | n/a | n/a | 8 | Mean: 3.2, Median: 3 | 3.8 | N |
| Cauchemez et al 200920 | USA | Apr 09 – May 09 | 216 | 816 | Case ascertained | N | RT-PCR | ARI | N | 13% | FARI | 10% | 7 | Mean: 2.9 | n/a | N |
| Chang et al 201121 | Taiwan | Aug 09 – Nov 09 | 87 | 223 | Case ascertained | N | RT-PCR + serology | RT-PCR + serology | Y | 27% | n/a | n/a | 7 | Mean: 2,.3 | n/a | Y |
| Cowling et al 201022 | Hong Kong | Jul 09 – Aug 09 | 105 | 309 | Case ascertained | N | RT-PCR | RT-PCR | Y | 7.8% | ARI, FARI | 26%, 6.1% | 7 | Mean: 3.2 | n/a | Y |
| Doyle et al 201123 | USA | Jun 09 | 53 | 163 | Case ascertained | Y | FARI | FARI | N | 3.4% | n/a | n/a | n/a | Mean: 3 | n/a | N |
| France et al 201024 | New York, USA | Apr 09 | 322 | 1042 | Case ascertained | N | RT- PCR/FARI | FARI | N | 11% | n/a | n/a | n/a | Median: 3 | 4.1 | N |
| Goldstein et al 201025 | Milwaukee, USA | Apr 09 – Jun 09 | 135 | 546 | Case ascertained | N | RT-PCR | FARI | N | 13% | n/a | n/a | 7 | Mean: 3.32 | n/a | N |
| Komiya et al 201027 | Osaka, Japan | May 09 | 124 | 503 | Case ascertained | N | RT-PCR | RT-PCR | N | 3.7% | n/a | n/a | n/a | Median: 3 | 3.1 | N |
| Leung et al 201128 | Hong Kong | Jun 09 | 65 | 270 | Case ascertained | N | RT-PCR | RT-PCR | N | 5.9% | n/a | n/a | 14 | Mean: 2.8 | n/a | N |
| Looker et al 201029 | Victoria, Australia | May 09 – Aug 09 | 132 | 483 | Case ascertained | N | RT-PCR | FARI | N | 33% | n/a | n/a | n/a | n/a | 3.9 | N |
| Loustalot et al 201130 | Texas, USA | May 09 | 668 | 640 | Cohort | N | FARI | FARI | N | 3.7% | n/a | n/a | 10 | n/a | n/a | N |
| Mohamed et al 201131 | Riyadh, Saudi Arabia | Sep 09 – Oct 09 | 69 | 432 | Case ascertained | N | RT-PCR | FARI | N | 17% | n/a | n/a | 12 | n/a | n/a | N |
| Morgan et al 201032 | Texas, USA | Apr 09 – May 09 | 77 | 349 | Case ascertained | N | RT-PCR/FARI | RT-PCR | N | 4.2% | ARI, FARI | 13%, 9.3% | 9 | Median: 4 | n/a | N |
| Nishiura et al 201133 | Japan | May 09 – Feb 10 | 1547 | 6156 | Case ascertained | N | RT- PCR/FARI | FARI | N | 11% | n/a | n/a | 7 | Mean: 3.1, Median: 3 | n/a | N |
| Odaira et al 200934 | Kobe, Japan | May 09 – Jun 09 | 97 | 400 | Case ascertained | N | RT-PCR | RT-PCR | N | 4.1% | ARI | 4.8% | 8 | Mean: 2.9, Median: 3 | n/a | N |
| Papenburg et al 201035 | Quebec, Canada | May 09 – Jul 09 | 42 | 162 | Case ascertained | Y | RT-PCR | RT-PCR + Serology | Y | 45% | RT- PCR, ARI, FARI | 38%, 51%, 29% | 11 | Mean: 3.9, Median: 3 | 3.8 | Y |
| Pebody et al 201136 | UK | Apr 09 – Jun 09 | 259 | 1046 | Case ascertained | Y | RT-PCR | RT-PCR | N | 8.3% | ARI, FARI | 16%,11% | 7 | n/a | 4.3 | N |
| Pedroni et al 201037 | Chile | Jun 09 | 57 | 302 | Case ascertained | N | RT-PCR | FARI | N | 37% | n/a | n/a | 14 | Mean: 3.6, Median: 3 | 3.3 | N |
| Savage et al 201138 | Ontario, Canada | Apr 09 – Jun 09 | 87 | 353 | Case ascertained | N | RT-PCR | ARI | N | 20% | FARI | 10% | 14 | Median: 3 | n/a | N |
| Sikora et al 201039 | Alberta, Canada | Apr 09 – Jun 09 | 87 | 349 | Case ascertained | N | RT-PCR | FARI | N | 30% | n/a | n/a | 14 | n/a | 4.0 | N |
| Simmerman et al 201140 | Bangkok, Thailand | Apr 09 – Aug 09 | 348 | 1233 | Case ascertained | N | RT-PCR | RT-PCR | Y | 21% | FARI | 14% | 7 | Mean: 3.32, Median: 2.94 | n/a | Y |
| Suess et al 201041 | Germany | Apr 09 – Aug 09 | 36 | 119 | Case ascertained | N | RT-PCR | RT-PCR | Y | 18% | n/a | n/a | 8 | Mean: 2.6, Median: 3 | n/a | N |
| Sugimoto et al 201142 | Washington, USA | May 09 – Jun 09 | 41 | 178 | Cohort | Y | RT- PCR/FARI | FARI | N | 5.9% | n/a | n/a | 7 | n/a | n/a | N |
| van Boven et al 201043 | Netherlands | Apr 09 – Jun 09 | 47 | 156 | Case ascertained | N | RT-PCR | RT-PCR | N | 8.3% | n/a | n/a | 7 | n/a | n/a | N |
| van Gemert et al 201144 | Victora, Australia | May 09 – Jun 09 | 36 | 167 | Retrospective cross- sectional | N | RT-PCR | FARI | N | 15% | n/a | n/a | 9 | Median: 2 | 3.1 | N |
One study (Jackson et al26) was not included in this table because it provided only supplementary serologic data of a previous study (France et al24). Data are included in the main text.
Case ascertained means that index cases were identified through symptom screening or laboratory confirmation; Cohort, index cases were identified from specific populations (e.g. camps, schools, etc) and households without index cases were also followed up
Studies in which it would have been possible for more than one person in a household to be identified as index case (i.e. identification of potential co-primary cases).
RT-PCR indicates respiratory specimen positive for influenza by reverse transcription polymerase chain reaction; ARI, acute respiratory illness; FARI, febrile acute respiratory illness; primary SIR, secondary infection risk based on primary outcome; secondary SIR, secondary infection risk based on secondary outcome; Y, yes; N, no; n/a: not applicable.
In 23 of the 27 studies (85%), all index cases were confirmed by RT-PCR, while in the other four studies, index cases were either clinically confirmed or retrospectively identified through symptom surveys. In (63%) 17 of these studies, households were followed up through telephone, postal or internet surveys, while the remaining studies involved one or more home visits. Respiratory specimens were collected for virologic testing from household contacts in 13 of the studies (48%); and 5 collected sera from household contacts.
Four studies reported that they were able to capture household transmission from the earliest local confirmed cases of pH1N1 in their country.18,41,46,47 Three studies utilized active surveillance systems for recruitment,29,41,48 while seven studies recruited index cases and their households following specific local outbreaks in schools or summer camps.18,23,24,28,30,34,42 Most studies were conducted very early in the pandemic; twelve studies (44%) did not recruit cases after June 2009 (eFigure 1, http://links.lww.com). Only 7% (2/27) of studies were published before January 2010.
Secondary Infection Risks
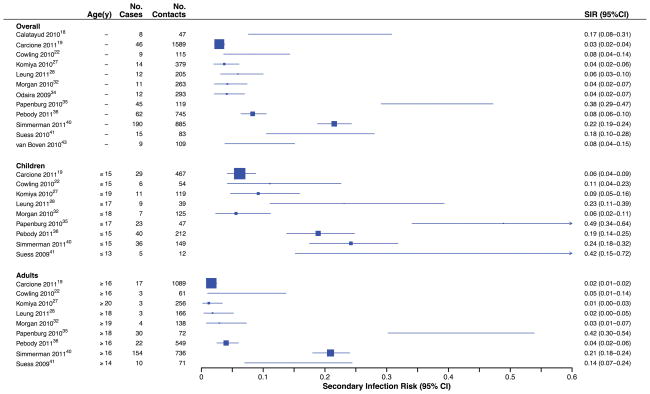
Twelve studies collected respiratory specimens from symptomatic or asymptomatic household contacts (or both) for laboratory confirmation of influenza A (H1N1). These studies reported SIRPCR which ranged from 3 to 38%, with a I2 of 96.5%, indicating substantial heterogeneity (Figure 2). In 9 of the 12 studies, results were stratified by age, yielding a range of SIRPCR from 6% to 49% [I2=91.1%] in children and 1% to 42% (I2=96.8%) in adults. To identify potential moderators of high levels of heterogeneity observed among studies, we conducted meta-regression analysis (Table 2). Studies that collected respiratory specimens from all contacts reported a higher SIRPCR than studies that swabbed only symptomatic contacts. The factors included in the model accounted for 75% of the observed heterogeneity.
Figure 2.
Secondary infection risks (SIRPCR) of laboratory-confirmed infection by RT-PCR among household contacts.
Table 2.
Meta-regression analysis of variables that could influence the secondary infection risk (SIR)
| Confirmed by RT-PCR (n=12)a | Febrile acute respiratory illness (FARI) (n=20)b | |||
|---|---|---|---|---|
| OR | (95% CI) | OR | (95% CI) | |
| Contacts tested by RT-PCR | ||||
| All c | 1.00 | 1.00 | ||
| Only symptomatic | 0.28 | (0.11–0.74)* | 0.99 | (0.44–2.22) |
| No. index cases per householdc,d | ||||
| Multiple | 1.00 | 1.00 | ||
| One | 0.26 | (0.07–1.00) | 0.74 | (0.32–1.70) |
| Duration of follow up | ||||
| ≤8 daysc | 1.00 | 1.00 | ||
| >8 days | 1.75 | (0.48–6.33) | 1.47 | (0.74–2.92) |
| Type of outbreakc | ||||
| General | 1.00 | 1.00 | ||
| Single sourcee | 1.99 | (0.63–6.36) | 0.34 | (0.16–0.74)* |
| Percent of index cases who were childrenc | ||||
| ≤50% | 1.00 | 1.00 | ||
| >50% | 0.49 | (0.10–2.40) | 0.61 | (0.29–1.30) |
| Percent of household contacts who were childrenc | ||||
| ≤30% | 1.00 | 1.00 | ||
| >30% | 0.86 | (0.32–2.35) | 1.35 | (0.65–2.80) |
SIR confirmed by RT-PCR: original τ2: 1.0756, meta-regression τ2: 0.2667, 75.2% of original heterogeneity explained
SIR estimated by FARI: original τ2: 0.2811, meta-regression τ2: 0.1856, 34.0% of original heterogeneity explained
Reference category
Studies in which it would have been possible for more than one person in a household to be identified as index case (i.e. identification of potential co-primary cases).
Single-source outbreak denotes studies which followed a specific outbreak e.g. summer camps or schools.
τ2 is the moment-based estimate of the between-studies variance.
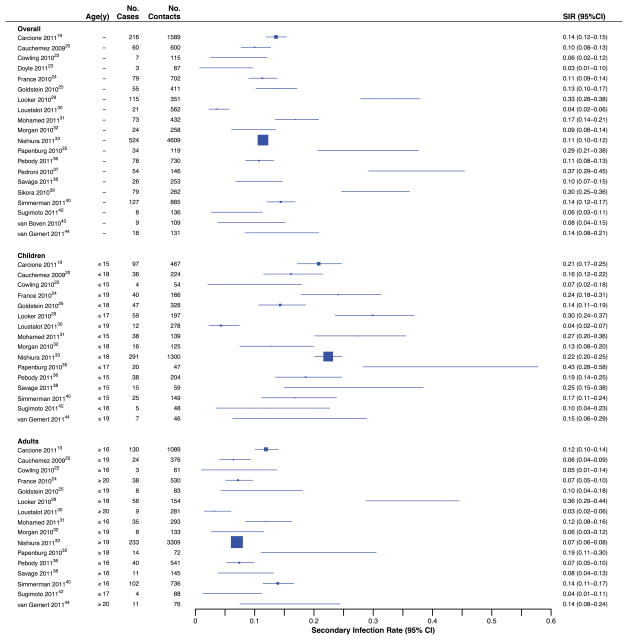
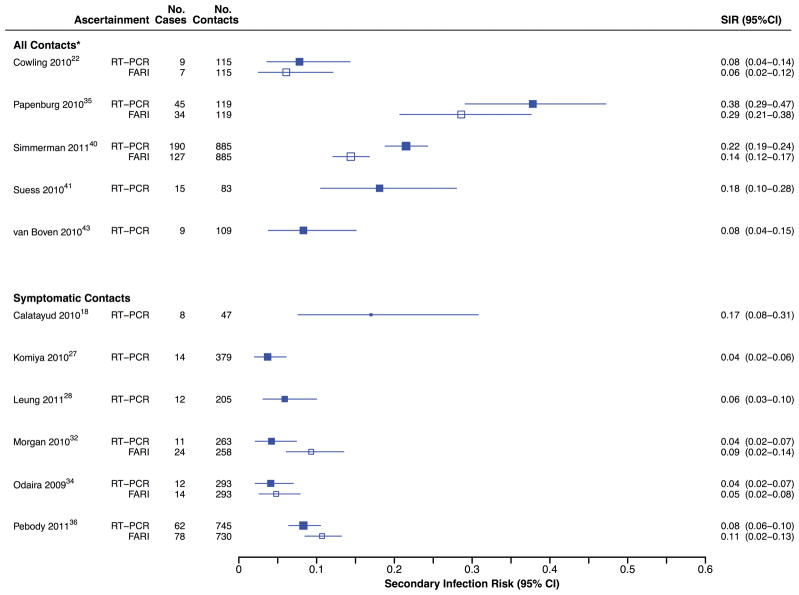
Every study included in this review reported clinical influenza among household contacts. Most (74%; 20/27) reported SIRFARI, when the definition of infection included fever, with point estimates ranging from 3% to 37% [I2=94.7%] (Figure 3). Age-stratified data in 16/20 studies reported SIRFARI with ranges of 4% to 43% [I2=83.7%] and 3% to 36% [I2=92.0%] for children and adult contacts, respectively. Substantial heterogeneity in SIRFARI was observed overall and after stratifying by age. Meta-regression analysis revealed that for studies estimating SIRFARI, only general versus specific outbreaks (i.e. sampling from community-wide major epidemic rather than from specific confined setting such as camp or school clusters) yielded a substantial association with the symptomatic SIR, and the model accounted for 34% of the heterogeneity. Among studies that aimed to collect virologic specimens from all contacts regardless of illness, SIRFARI tended to be lower than SIRPCR estimates for confirmed infection, whereas the reverse was true among studies that aimed to collect virologic specimens only from contacts that reported illness (Figure 4).
Figure 3.
Secondary infection risks (SIRFARI) according to report of febrile acute respiratory infection among household contacts.
Figure 4.
Estimates of the SIRPCR and SIRFARI in 5 studies that aimed to collect respiratory specimens from all household contacts, and 6 studies that aimed to collect respiratory specimens only from ill contacts.
Three studies reported that both SIRPCR and SIRFARI were higher among household contacts of younger index cases,22,25,33 two studies reported that they were lower with younger index cases,19,29 and one study found no difference by index age.41 Four studies that stratified by sex found no difference in SIRs,27,34,40,41 while one study reported female contacts to be at greater risk of infection24 and another that adult women were more likely to transmit pH1N1 to children.36
Eight studies reported SIRARI, with point estimates ranging from 13% to 51% [I2=94.3%] (eFigure 2, http://links.lww.com). Stratified analysis reported ranges of 15% to 55% [I2=86.0%] in children and 10% to 49% [I2=90.6%] in adults.
In addition to the effects of contact and index age on household transmission, some studies analyzed the effects of antiviral treatment and prophylaxis with oseltamivir or zanamivir, vaccination history and household size. Eleven studies recorded infection rates among household contacts that received antiviral prophylaxis,19,23,24,27,28,32–34,36,44,46 but only one study reported the prophylaxis group to be more susceptible to pH1N1 (eFigure 3, http://links.lww.com). One study reported seasonal influenza vaccination history to have no effect on the SIR,45 while three studies24,30,35 reported elevated SIRs among people who had been vaccinated for seasonal influenza. The SIR was variously observed to increase with household size25,30,35,39 or not to be associated with household size.19,24,27,29,33 In one study with a broader definition of household contacts (eTable 2, http://links.lww.com), the SIR decreased in larger households.20
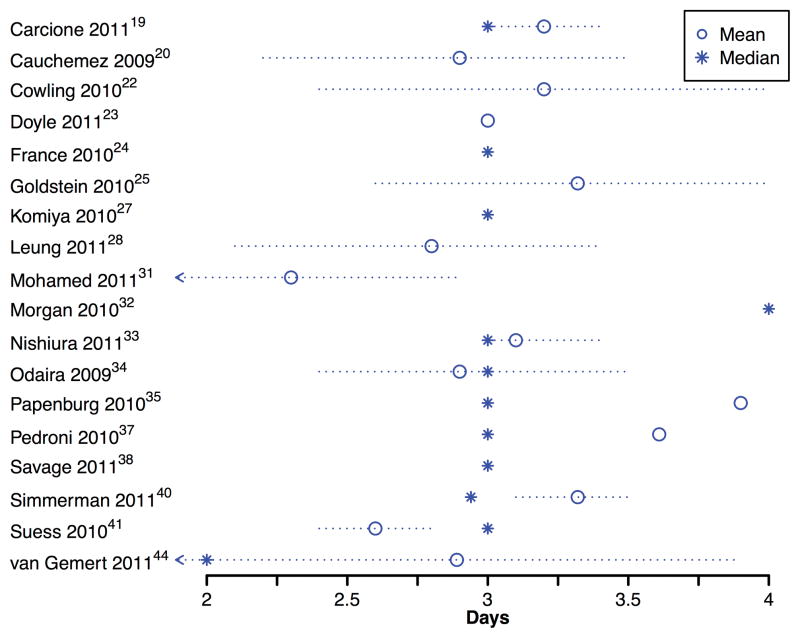
Serial intervals
Among the 27 included studies, 18 (67%) reported either the mean or median household serial interval, including 8 that reported both (Figure 5). Mean serial intervals ranged from 2.6 to 3.9 days while median serial intervals ranged from 2.0 to 4.0 days. Of the 26 point estimates of the serial interval, 20 (77%) fell within the range 2.8–3.5 days.
Figure 5.
Point estimates of the mean and median household serial intervals in 16 studies.
Illness in household contacts with confirmed pH1N1 infection
The clinical signs and symptoms associated with confirmed pH1N1 infection among household contacts are summarized in Table 3 from three studies that collected respiratory specimens from household contacts regardless of reported illness. Cough was the most commonly reported symptom, while fever was reported in approximately 60% of the confirmed cases. Two studies reported asymptomatic fractions among virologically confirmed cases to be 11% and 7%,22,35,35,41 and when cases under antiviral prophylaxis were included, the asymptomatic fraction was 20%.41 One study reported a subclinical fraction (9%),22 and the other study similarly reported that 9% of household contacts with serologic evidence of infection remained asymptomatic.35 One study that reported only serological data found a crude asymptomatic fraction of 25%.26 Two studies reported the proportion of household contacts with various clinical signs, symptoms and syndromes that were confirmed with pH1N1 (eTable 3, http://links.lww.com).
Table 3.
Signs and symptoms reported by household contacts with confirmed influenza in studies where respiratory specimens were collected from household contacts regardless of illness
| Cowling 201022(n=9) No. (%) |
Papenburg 201035(n=45) No. (%) |
Suess 201041(n=15) No. (%) |
|
|---|---|---|---|
| Cough | 8 (88.9) | 38 (84.4) | 10 (66.7) |
| Fever | 5 (55.6) | 30 (66.7) | 8 (53.3) |
| Sore throat | 6 (66.7) | -- | 3 (20.0) |
| Headache | 4 (44.4) | -- | 4 (26.7) |
| Myalgia | 3 (33.3) | -- | 4 (26.7) |
| Diarrhoea | -- | 11 (24.4) | -- |
| Nausea | -- | 7 (15.6) | -- |
| Runny nose | 4 (44.4) | -- | -- |
| Asymptomatic | 1 (11.1) | 3 (6.7) | 3 (20.0) |
Serologic findings
Serologic data on household contacts were available from five studies.21,22,26,35,40 Two studies reported SIRs based solely on serology to be 20% and 27%.21,26 In addition to using of serology to identify asymptomatic infections,35 two studies used serology and RT-PCR results in combination to estimate pH1N1 SIRs.35,40 One study reported no evidence of a protective effect for subjects with elevated baseline antibody titer levels against RT-PCR-confirmed infection.22
DISCUSSION
During the 2009 pandemic, household studies were conducted in many countries to improve understanding of the epidemiologic characteristics of the novel pH1N1 virus in a specific community setting. We described the design of household transmission studies conducted during the pandemic, and we compared the findings of the studies, including the household SIR, the household serial interval, and the symptom profiles and the asymptomatic fraction in household contacts. Among these, the SIR and household serial interval are relatively imprecise in that they are influenced by transmissions both in household and community settings, as well as pre-existing immunity among contacts. We therefore conducted meta-regression analysis to identify potential factors associated with higher or lower household SIRs (Table 2).
There were substantial heterogeneities in estimated SIRs from the various studies, with point estimates of the SIR based on RT-PCR-confirmed secondary cases ranging from 3% to 38% (Figure 2). Estimates were similarly heterogeneous when based on febrile acute respiratory illness (Figure 3) and acute respiratory illness (eFigure 2, http://links.lww.com). The intrinsic transmissibility of the pH1N1 virus is not thought to have varied substantially in different countries; indeed, a recent review identified similarity in the estimates of the reproduction number from a range of studies.9 A review of serologic studies also found similar estimates of cumulative incidence of infection over the first pH1N1 wave in several countries.49
A number of factors may have led to the observed differences in estimated household SIRs. Meta-regression analyses revealed that rigorous case ascertainment with RT-PCR testing for all contacts (including asymptomatic contacts), elevated the SIRPCR, whereas the SIRFARI was unaffected (Table 2, Figure 4). Although rigorous testing undoubtedly increases the cost, our results indicate that studies that tested all household contacts by RT-PCR, regardless of illness, identified more infections. Studies that test only symptomatic contacts will not identify all infected contacts, nor will they achieve a timely collection of specimens within 3 days of onset (when RT-PCR sensitivity is highest50). It might be expected that studies with longer durations of follow-up would pick up not only those transmissions within households but also those from the community; our findings were consistent with increases in SIRPCR and SIRFARI with longer follow-up.
The highest SIRs were observed in Chile,37 Australia,29 and Canada.35,39 Among these, two studies with differing SIRs (estimated SIRPCR of 33% and 15%) were reported from Victoria, Australia at similar times.29,44 The study with the larger SIR estimate recruited subjects by 31 August 2009 while the other ended on 3 June 2009; the proportion of child index cases (aged ≥5 years) was 86% in the study with SIRPCR =15%, compared with 37% in the other study. These are consistent with our findings from meta-regression (Table 2); the smaller SIRPCR in studies with a greater proportion of child index cases may also suggest case ascertainment bias. The Chilean and Canadian studies enrolled households during peak periods of pH1N1 activity, and household contacts were followed up for 2 weeks37,39 and 3–4 weeks,35 potentially increasing the risk of misclassifying infections from the community or household tertiary cases as household secondary cases.
In comparison, studies that report inter-pandemic influenza transmission in households have also reported widely varying SIRs, from 7% to 31%.22,47,51–55 It is likely that factors that led to the significant variation and heterogeneity among the studies in this review of the SIR of pH1N1 also affected the SIR of inter-pandemic influenza. Only three studies directly compared the household transmission of inter-pandemic and pandemic influenza concurrently during a single season in a single population, and each reported comparable SIRs between inter-pandemic and pandemic strains.22,31,40 One study that was excluded from our review also reported similar secondary-infection risks for inter-pandemic and pandemic influenza based on serologic evidence from a cohort study in 2008–2009, explicitly estimating the risks of infections directly caused by household index cases.56 The variations in SIRs for both pH1N1 and inter-pandemic influenza highlight a critical need to formulate guidelines for conducting household studies of influenza so that we can gain more explicit insights into the natural history as well as the transmission within households.
Household serial interval estimates were reported in 18 of the 27 studies (67%) included in our review, with most point estimates falling in the range 2.8–3.5 days (Figure 5). Correction for multiple chains of transmission (e.g. tertiary cases) could reduce serial interval estimates, and shorten the estimated mean to 2.5 days.9 The household serial interval is not a biological constant but instead reflects a combination of the infectivity profile of index cases, contact patterns within households, transmission dynamics in the community, and incubation period -- and these may vary in different settings and by individual characteristics such as age.57–59 Although estimates of the household serial interval have been used to infer the reproduction number from exponential growth rate of cases,60 further studies are needed to estimate the serial interval in various settings. For example, one recent study in the United States estimated that the serial interval of pH1N1 in schools had a mean of just 1.1 days.57
Some household studies permitted estimation of the fraction of virologically confirmed pH1N1 infections with asymptomatic illness at 7–11%, and the profile of signs and symptoms associated with pH1N1 infection (Table 3). Estimation of the asymptomatic fraction was achieved by studies that collected specimens from contacts regardless of the presence or absence of symptoms.22,35,40,41,43 Only 5 of the 27 studies included in this review reported serologic data.21,22,26,35,40 The inclusion of serology can provide additional information on asymptomatic infections, as well as on the degree of protection associated with higher baseline humoral antibody titers. There is currently no consensus on the definition of asymptomatic infections versus subclinical infections. The proportion of pH1N1 infections associated with afebrile illness could be a reasonable definition of the subclinical fraction, with estimates ranging from 33% to 47% (Table 3). Only a small fraction of confirmed infections were completely asymptomatic. 22,26,35,41
Two major technical problems have yet to be solved regarding the analysis of data from household transmission studies. The first is the unobservable nature of infection events. In many studies, household follow-up was truncated at 7–9 days after illness onset of the index case, after which little primary household transmission occurs.61 Although not explicitly mentioned, the cut-off point should be set at a reasonable length to exclude tertiary cases and those acquiring infection in the community (based on the right tail of the serial interval distribution). Except for one study,20 the reviewed studies did not explicitly address decomposition of secondary, tertiary and community infections when estimating the SIR. However, two other studies used viral sequencing to confirm homology between the strains infecting the index cases and corresponding secondary cases in households.35,62 Given that the larger SIRs and longer mean serial interval in some studies35,37 are suggestive of the presence of chains of transmission or community infections and consequent overestimation of the SIR, it is important to try to identify transmissions that occur only within households. Whereas several statistical methods are available to address this point at least partially (using either SIR stratified by household size or observed serial interval distribution59,63), those datasets were unfortunately fairly scarce, and irregular timing of observation during the course of an epidemic has made it difficult to remove the co-primary cases from the observed serial intervals. Furthermore, considering the dependent nature of household and community infection risks,64 we refrained from imposing strong epidemiologic assumptions to build a simplistic statistical model, and also from decomposing the observed data into those attributable to transmissions in the household and community. To address these issues, influence of household study characteristics on the estimate of SIR (including the length of follow-up and the timing of observation) were examined instead, demonstrating that the study design -- most notably case ascertainment -- was an important source of heterogeneity (Table 2). In particular, our meta-analysis has demonstrated that household studies can provide invaluable data on influenza infection; in studies that used febrile illness reports, the resulting estimates of the SIR could underestimate the true SIR (Figure 4) because a substantial fraction of influenza infections are not associated with febrile illness (Table 3).
The second technical problem is the lack of an ideal approach to recruitment and follow-up of households during the course of an epidemic. Studies conducted during the early stages of the pandemic can provide timely estimates of epidemiologic characteristics such as the SIR and serial interval, but the community risk of infection varies throughout the course of the epidemic, and the risk of infection within the household could be influenced by many factors such as changing contact behavior upon diagnosis. In one study, households recruited were quarantined and advised to remain at home; in such situations there could be a higher degree of household contact leading to elevated SIRs.29 However, sampling households only around the peak period of the pandemic could lead to confusion of community infections as secondary cases from within the household, as well as to inclusion of some contacts that have already been infected and are immune. Furthermore, the sources and characteristics of index cases may affect subsequent transmission dynamics as discussed above. All these reservations likely apply equally to household studies of inter-pandemic influenza in households.
Based on our review, we have formulated some recommendations for household transmission study protocols for future studies of pandemic as well as inter-pandemic influenza. First, while many studies were conducted within a few months of the initial World Health Organization global pandemic alert in April 2009, other studies were delayed by requirements for protocol development, ethical approval, and funding. Some of the earliest studies were conducted as part of containment measures or routine public health investigations, and these studies often had the most haphazard approaches to recruitment and follow-up of households. The First Few 100 study conducted in the United Kingdom36 provides an excellent model of a household transmission study that was prepared in advance of the pandemic with a detailed protocol, relevant approvals, and funding in place before the pandemic.
Second, laboratory outcome measures are preferable in community-based studies of influenza because many other co-circulating pathogens are associated with upper respiratory tract infections.65 Febrile acute respiratory infections among contacts provided fairly specific criteria for confirmed pH1N1 infection, and estimates based on febrile acute respiratory infections could be corrected for the fraction of infections developing such illness to provide more reasonable estimates of the SIR. Given the technical issue discussed above regarding the direct interpretation of point estimates of overall SIRs, as well we the heterogeneity reported in our meta-analysis (Figures 2 and 3), the most important information provided by household transmission studies may be on differences in infectivity and susceptibility by age, and the effects of specific interventions such as antiviral use. Household SIRs have been used to provide these estimates in the literature because the exposure of household contacts to a single index case permits fairly straightforward analysis.66
Third, it must be remembered that the household SIR is theoretically defined to reflect the risk of infection among “susceptible” contacts,5 while many older adults are likely to have been partially or fully immune to pH1N1.67 Inclusion of serology in household studies could provide information on humoral immunity. With adequate laboratory capacity, other correlates of protection, such as cell-mediated and mucosal immunity can be assessed through collection of whole blood and nasal washes..
Finally, there was considerable uncertainty in the early stages of the pandemic regarding the fraction of infections that were asymptomatic or subclinical. Along with well-designed prospective symptom diaries, collection of acute and convalescent serology from household contacts in household transmission studies could provide key information on asymptomatic cases, which is essential to interpreting epidemic curves of symptomatic cases and forecasting the course of the pandemic. One potential limiting factor, however, is the availability of validated serologic assays early in the next pandemic.
Household transmission studies can provide important information on influenza epidemiology. However our review suggests that interpretation and comparison of estimates of the SIR from individual studies are substantially affected by differential diagnostic methods and case ascertainment. Furthermore, the unbiased risk of household secondary infection is only approximated by the crude household SIR, and it remains technically challenging to estimate the fraction of secondary cases that were directly infected by the index case. By building a consensus on the appropriate approaches to studying transmission in households (via, for example, common survey protocols), it is likely that household transmission studies could be greatly improved and provide valuable insights into the epidemiology of pandemic and inter-pandemic influenza.
Supplementary Material
Acknowledgments
Source of Funding:
This work received financial support from the Harvard Center for Communicable Disease Dynamics from the National Institute of General Medical Sciences (grant no. U54 GM088558), and the Area of Excellence Scheme of the University Grants Committee of Hong Kong (grant no. AoE/M-12/06). HN is supported by the JST PRESTO program. BJC has received research funding from MedImmune Inc. HAK has received funding from CSL Biotherapies. DKMI has received research funding from Roche.
We thank Mark Simmerman and Dale Carcione for providing detailed breakdowns of the data on pH1N1 transmission from studies in Bangkok and Australia, respectively. We thank Richard Pebody for clarification on the data included in the series of publications based on the First Few 100 study in the United Kingdom. We thank Vicky Fang for technical assistance, and Katie Glass, Geoff Mercer and Peter Horby for helpful discussions.
Footnotes
Conflicts of Interest
The authors report no other potential conflicts of interest.
SDC Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com). This content is not peer-reviewed or copy-edited; it is the sole responsibility of the author.
References
- 1.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360(25):2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.Chao DL, Halloran ME, Obenchain VJ, Longini IM., Jr FluTE, a publicly available stochastic influenza epidemic simulation model. PLoS Comput Biol. 2010;6(1):e1000656. doi: 10.1371/journal.pcbi.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson NM, Cummings DA, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006;442(7101):448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monto AS. Studies of the community and family: acute respiratory illness and infection. Epidemiol Rev. 1994;16(2):351–373. doi: 10.1093/oxfordjournals.epirev.a036158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halloran ME. Secondary Attack Rate. Encyclopedia of Biostatistics. 2005 [Google Scholar]
- 6.Glezen WP. Emerging infections: pandemic influenza. Epidemiol Rev. 1996;18(1):64–76. doi: 10.1093/oxfordjournals.epirev.a017917. [DOI] [PubMed] [Google Scholar]
- 7.Halloran ME, Longini IM, Jr, Struchiner J. Design and Analysis of Vaccine Studies (Statistics for Biology and Health) Springer; 2009. [Google Scholar]
- 8.Fine PE. The interval between successive cases of an infectious disease. Am J Epidemiol. 2003;158(11):1039–1047. doi: 10.1093/aje/kwg251. [DOI] [PubMed] [Google Scholar]
- 9.Boelle PY, Ansart S, Cori A, Valleron AJ. Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza Other Respi Viruses. 2011;5(5):306–316. doi: 10.1111/j.1750-2659.2011.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuchat A, Bell BP, Redd SC. The science behind preparing and responding to pandemic influenza: the lessons and limits of science. Clin Infect Dis. 2011;52 (Suppl 1):S8–12. doi: 10.1093/cid/ciq007. [DOI] [PubMed] [Google Scholar]
- 11.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JJ. The inverse of the Freeman-Tukey double arcsine transformation. Am Stat. 1978;32:138. [Google Scholar]
- 13.Freeman MF, Tukey JW. Transformation related to the angular and the square root. Ann Math Statist. 1950;21:607–611. [Google Scholar]
- 14.Laird NM, Mosteller F. Some statistical methods for combining experimental results. Int J Technol Assess Health Care. 1990;6(1):5–30. doi: 10.1017/s0266462300008916. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 16.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36(3):1–48. [Google Scholar]
- 17.metafor: Meta-analysis Package for R [computer program]. Version 1.6-0 2011. [Google Scholar]
- 18.Calatayud L, Kurkela S, Neave PE, et al. Pandemic (H1N1) 2009 virus outbreak in a school in London, April-May 2009: an observational study. Epidemiol Infect. 2010;138(2):183–191. doi: 10.1017/S0950268809991191. [DOI] [PubMed] [Google Scholar]
- 19.Carcione D, Giele CM, Goggin LS, et al. Secondary attack rate of pandemic influenza A(H1N1) 2009 in Western Australian households, 29 May-7 August 2009. Euro Surveill. 2011;16(3) [PubMed] [Google Scholar]
- 20.Cauchemez S, Donnelly CA, Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med. 2009;361(27):2619–2627. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang LY, Chen WH, Lu CY, et al. Household Transmission of Pandemic (H1N1) 2009 Virus, Taiwan. Emerg Infect Dis. 2011;17(10):1928–1931. doi: 10.3201/eid1710.101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowling BJ, Chan KH, Fang VJ, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010;362(23):2175–2184. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle TJ, Hopkins RS. Low secondary transmission of 2009 pandemic influenza A (H1N1) in households following an outbreak at a summer camp: relationship to timing of exposure. Epidemiol Infect. 2011;139(1):45–51. doi: 10.1017/S095026881000141X. [DOI] [PubMed] [Google Scholar]
- 24.France AM, Jackson M, Schrag S, et al. Household transmission of 2009 influenza A (H1N1) virus after a school-based outbreak in New York City, April-May 2009. J Infect Dis. 2010;201(7):984–992. doi: 10.1086/651145. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein E, Cowling BJ, O’Hagan JJ, et al. Oseltamivir for treatment and prevention of pandemic influenza A/H1N1 virus infection in households, Milwaukee, 2009. BMC Infect Dis. 2010;10:211. doi: 10.1186/1471-2334-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson ML, France AM, Hancock K, et al. Serologically confirmed household transmission of 2009 pandemic influenza A (H1N1) virus during the first pandemic wave--New York City, April-May 2009. Clin Infect Dis. 2011;53(5):455–462. doi: 10.1093/cid/cir437. [DOI] [PubMed] [Google Scholar]
- 27.Komiya N, Gu Y, Kamiya H, et al. Household transmission of pandemic 2009 influenza A (H1N1) virus in Osaka, Japan in May 2009. J Infect. 2010;61(4):284–288. doi: 10.1016/j.jinf.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Leung YH, Li MP, Chuang SK. A school outbreak of pandemic (H1N1) 2009 infection: assessment of secondary household transmission and the protective role of oseltamivir. Epidemiol Infect. 2011;139(1):41–44. doi: 10.1017/S0950268810001445. [DOI] [PubMed] [Google Scholar]
- 29.Looker C, Carville K, Grant K, Kelly H. Influenza A (H1N1) in Victoria, Australia: a community case series and analysis of household transmission. PLoS One. 2010;5(10):e13702. doi: 10.1371/journal.pone.0013702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loustalot F, Silk BJ, Gaither A, et al. Household transmission of 2009 pandemic influenza A (H1N1) and nonpharmaceutical interventions among households of high school students in San Antonio, Texas. Clin Infect Dis. 2011;52 (Suppl 1):S146–153. doi: 10.1093/cid/ciq057. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed AG, Binsaeed AA, Al-Habib H, Al-Saif H. Communicability of H1N1 and seasonal influenza among household contacts of cases in large families. Influenza Other Respi Viruses. 2011 doi: 10.1111/j.1750-2659.2011.00308.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan OW, Parks S, Shim T, et al. Household transmission of pandemic (H1N1) 2009, San Antonio, Texas, USA, April-May 2009. Emerg Infect Dis. 2010;16(4):631–637. doi: 10.3201/eid1604.091658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiura H, Oshitani H. Household transmission of influenza (H1N1-2009) in Japan: age-specificity and reduction of household tranmission risk by zanamivir treatment. J Int Med Res. 2011;39:619–628. doi: 10.1177/147323001103900231. [DOI] [PubMed] [Google Scholar]
- 34.Odaira F, Takahashi H, Toyokawa T, et al. Assessment of secondary attack rate and effectiveness of antiviral prophylaxis among household contacts in an influenza A(H1N1)v outbreak in Kobe, Japan, May-June 2009. Euro Surveill. 2009;14(35) [PubMed] [Google Scholar]
- 35.Papenburg J, Baz M, Hamelin ME, et al. Household transmission of the 2009 pandemic A/H1N1 influenza virus: elevated laboratory-confirmed secondary attack rates and evidence of asymptomatic infections. Clin Infect Dis. 2010;51(9):1033–1041. doi: 10.1086/656582. [DOI] [PubMed] [Google Scholar]
- 36.Pebody RG, Harris R, Kafatos G, et al. Use of Antiviral Drugs to Reduce Household Transmission of Pandemic (H1N1) 2009, United Kingdom. Emerg Infect Dis. 2011;17(6):990–999. doi: 10.3201/eid1706.101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedroni E, Garcia M, Espinola V, et al. Outbreak of 2009 pandemic influenza A(H1N1), Los Lagos, Chile, April-June 2009. Euro Surveill. 2010;15(1) doi: 10.2807/ese.15.01.19456-en. [DOI] [PubMed] [Google Scholar]
- 38.Savage R, Whelan M, Johnson I, et al. Assessing secondary attack rates among household contacts at the beginning of the influenza A (H1N1) pandemic in Ontario, Canada, April-June 2009: A prospective, observational study. BMC Public Health. 2011;11(1):234. doi: 10.1186/1471-2458-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikora C, Fan S, Golonka R, et al. Transmission of pandemic influenza A (H1N1) 2009 within households: Edmonton, Canada. J Clin Virol. 2010;49(2):90–93. doi: 10.1016/j.jcv.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Simmerman JM, Suntarattiwong P, Levy J, et al. Findings from a household randomized controlled trial of hand washing and face masks to reduce influenza transmission in Bangkok, Thailand. Influenza Other Respi Viruses. 2011;5(4):256–267. doi: 10.1111/j.1750-2659.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suess T, Buchholz U, Dupke S, et al. Shedding and transmission of novel influenza virus A/H1N1 infection in households--Germany, 2009. Am J Epidemiol. 2010;171(11):1157–1164. doi: 10.1093/aje/kwq071. [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto JD, Borse NN, Ta ML, et al. The effect of age on transmission of 2009 pandemic influenza A (H1N1) in a camp and associated households. Epidemiol. 2011;22(2):180–187. doi: 10.1097/EDE.0b013e3182060ca5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Boven M, Donker T, van der Lubben M, et al. Transmission of novel influenza A(H1N1) in households with post-exposure antiviral prophylaxis. PLoS One. 2010;5(7):e11442. doi: 10.1371/journal.pone.0011442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Gemert C, Hellard M, McBryde ES, et al. Intrahousehold Transmission of Pandemic (H1N1) 2009 Virus, Victoria, Australia. Emerg Infect Dis. 2011;17(9) doi: 10.3201/eid1709.101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carcione D, Giele C, Goggin LS, et al. Association between 2009 seasonal influenza vaccine and influenza-like illness during the 2009 pandemic: preliminary results of a large household transmission study in Western Australia. Euro Surveill. 2010;15(28) [PubMed] [Google Scholar]
- 46.Ghani AC, Baguelin M, Griffin J, et al. The Early Transmission Dynamics of H1N1pdm Influenza in the United Kingdom. PLoS Curr Influenza. 2009;1:RRN1130. doi: 10.1371/currents.RRN1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Sugimoto JD, Halloran ME, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326(5953):729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee DH, Kim CW, Kim JH, et al. Risk factors for laboratory-confirmed household transmission of pandemic H1N1 2009 infection. Am J Infect Control. 2010;38(10):e43–45. doi: 10.1016/j.ajic.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 49.Kelly H, Peck HA, Laurie KL, Wu P, Nishiura H, Cowling BJ. The age-specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLoS One. 2011;6(8):e21828. doi: 10.1371/journal.pone.0021828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klick B, Leung GM, Cowling BJ. Optimal design of studies of influenza transmission in households. I: Case-ascertained studies. Epidemiol Infect. 2011:1–9. doi: 10.1017/S0950268811000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cowling BJ, Chan KH, Fang VJ, et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med. 2009;151(7):437–446. doi: 10.7326/0003-4819-151-7-200910060-00142. [DOI] [PubMed] [Google Scholar]
- 52.Hayden FG, Belshe R, Villanueva C, et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis. 2004;189(3):440–449. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- 53.Hayden FG, Gubareva LV, Monto AS, et al. Inhaled zanamivir for the prevention of influenza in families. Zanamivir Family Study Group. N Engl J Med. 2000;343(18):1282–1289. doi: 10.1056/NEJM200011023431801. [DOI] [PubMed] [Google Scholar]
- 54.Monto AS, Pichichero ME, Blanckenberg SJ, et al. Zanamivir prophylaxis: an effective strategy for the prevention of influenza types A and B within households. J Infect Dis. 2002;186(11):1582–1588. doi: 10.1086/345722. [DOI] [PubMed] [Google Scholar]
- 55.Welliver R, Monto AS, Carewicz O, et al. Effectiveness of oseltamivir in preventing influenza in household contacts: a randomized controlled trial. JAMA. 2001;285(6):748–754. doi: 10.1001/jama.285.6.748. [DOI] [PubMed] [Google Scholar]
- 56.Klick B, Nishiura H, Ng S, et al. Transmissibility of seasonal and pandemic influenza in a cohort of households in Hong Kong in 2009. Epidemiology. 2011;22(6):793–796. doi: 10.1097/EDE.0b013e3182302e8e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cauchemez S, Bhattarai A, Marchbanks TL, et al. Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc Natl Acad Sci U S A. 2011;108(7):2825–2830. doi: 10.1073/pnas.1008895108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowling BJ, Fang VJ, Riley S, Malik Peiris JS, Leung GM. Estimation of the serial interval of influenza. Epidemiology. 2009;20(3):344–347. doi: 10.1097/EDE.0b013e31819d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klinkenberg D, Nishiura H. The correlation between infectivity and incubation period of measles, estimated from households with two cases. J Theor Biol. 2011;284(1):52–60. doi: 10.1016/j.jtbi.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 60.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274(1609):599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dingle JH, Badge GFWSJ. Illness in the Home. A Study of 25,000 Illnesses in a Group of Cleveland Families. Cleveland: The Press of Western Reserve University; 1964. [Google Scholar]
- 62.Poon LL, Chan KH, Chu DKW, et al. Viral genetic sequence variations in pandemic H1N1/2009 and seasonal H3N2 influenza viruses within an individual, a household and a community. J Clin Virol. 2011 doi: 10.1016/j.jcv.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Longini IM, Jr, Koopman JS. Household and community transmission parameters from final distributions of infections in households. Biometrics. 1982;38(1):115–126. [PubMed] [Google Scholar]
- 64.Ball F, Mollison D, Scalia-Tomba G. Epidemics with two levels of mixing. Ann App Prob. 1997;7(1):46–89. [Google Scholar]
- 65.Schnepf N, Resche-Rigon M, Chaillon A, et al. High burden of non-influenza viruses in influenza-like illness in the early weeks of H1N1v epidemic in France. PLoS One. 2011;6(8):e23514. doi: 10.1371/journal.pone.0023514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halloran ME, Preziosi MP, Chu H. Estimating Vaccine Efficacy From Secondary Attack Rates. J Am Stat Assoc. 2003;98:38–46. [Google Scholar]
- 67.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.