Abstract
In precancerous and cancerous lesions, excessive growth signals resulting from activation of oncogenes or loss of tumor suppressor genes lead to intensive replication stress, which is recognized by a high level of replication-associated DNA double-strand breaks (DSBs). However, the molecular mechanism by which cells alleviate excessive replication stress remains unclear. In this study, we report that the human nuclease/helicase DNA2 facilitates homologous recombination to repair replication-associated DNA double-strand breaks (DSBs), thereby providing cells with survival advantages under conditions of replication stress. The nuclease activity of DNA2 was required for DSB end resection, which allowed subsequent recruitment of RPA and RAD51 to repair DSBs and restart replication. More importantly, DNA2 expression was significantly increased in human cancers and its expression correlated with patient outcome. Our findings therefore indicate that enhanced activity of DSB resection likely constitutes one mechanism whereby precancerous and cancerous cells might alleviate replication stress.
Keywords: DNA2, DNA end resection, homologous recombination, DNA repair, replication stress
INTRODUCTION
Tumorigenesis is a multistep process by which normal cells successively acquire genetic alterations (1). Analyses of human tumors have shown that the presence of DNA damage, particularly DNA double-strand breaks (DSBs), distinguishes precancerous lesions and cancer from normal tissues (2, 3). Recent studies have indicated that oncogene-induced replication stress underlies DSB formation (4, 5). Specifically, activation of oncogenes provides cells with sustained proliferative signaling and leads to inappropriate DNA replication, which results in fork collapse and DSBs (6, 7).
There are two pathways for repairing DSBs: homologous recombination (HR) and nonhomologous end-joining (NHEJ) (8). NHEJ involves direct ligation of broken ends and primarily occurs in the G1 phase of the cell cycle. HR is considered to be more error-free repair mechanism that copies sequences from the homologous template to repair damaged DNA. It predominantly occurs in the S and G2 phases, when preferable template, sister chromatids, are available. Thereby, HR is a key pathway for repairing stalled and collapsed replication forks that occur spontaneously or are induced by topoisemarase I inhibitors such as camptotecin or polymerases inhibitors like aphidicolin (9). Repair of chromosomal breaks by HR is initiated by resection of the 5' strands that generates 3' single-strand DNA (ssDNA) tails at DSB ends (10, 11). Resection allows loading of single strand binding protein, RPA that is further replaced by key enzyme in HR, strand exchange protein RAD51 with the help of BRCA2 (12–14). Rad51 mediates homology search, and strand invasion at template sister chromatid which is followed by DNA synthesis and resolution of recombination intermediate that restores replication fork. Depletion of Rad51 leads to accumulation of unrepaired DSBs and cells death demonstrating the importance of HR for repair of spontaneous DNA breaks occurring during replication (15).
Continuous formation of DSBs induced by replication stress activates DNA damage response (DDR), which induces senescence or apoptosis and thus prevents precancerous lesions from progressing to malignant lesions (6, 7). Impairment of DDR (e.g., through loss of expression of signaling kinases ATM or CHK2) can lead to a breach of this anticancer barrier and tumor progression (4, 5). However, it remains unclear how precancerous and cancerous cells could cope with increased replication-associated DSBs and maintain their hyperactive DNA replication status.
In this study, we utilized a proteomic approach to investigate protein components preferentially associated with replication forks in the presence of oncogene activation. Here we report that human nuclease/helicase DNA2 is over-expressed in a variety of cancers and it plays an important role in alleviating replication stress likely by promoting DNA end resection and HR repair of replication-associated DSBs.
Materials and Methods
Cell Culture
U2OS and MCF10A cells were purchased from ATCC. U2OS cells were maintained in McCoy's 5A medium supplemented with 10% FBS. MCF10A cells were cultured in mammary epithelial growth medium containing insulin, hydrocortisone, epidermal growth factor, and bovine pituitary extract purchased from Clonetics. MCF7 cells were maintained in DMEM medium (Cellgro) with 10% FBS. Cell lines were validated by STR DNA fingerprinting using the AmpF_STR Identifiler kit according to manufacturer instructions (Applied Biosystems cat 4322288). The STR profiles were compared to known ATCC fingerprints, to the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (Nucleic Acids Research 37:D925–D932 PMCID: PMC2686526 and to the MD Anderson fingerprint database. The STR profiles matched known DNA fingerprints or were unique.MCF10A cells with stable cyclin E expression were generated by transfection with pcDNA3.1-cyclin E construct and maintained in the presence of 100 μg/mL G418. A series of pancreatic cell lines (HPDE, PD90, PD78, PD77 and PD74) representing RAS-mediated pancreatic cancer development were kindly provided by Dr. Michel J. Quellette (University of Nebraska Medical Center). Cells were maintained as previously described (16).
Antibodies, Immunoprecipitation, Chromatin Fractionation, and Western Blot Analysis
Antibodies used in the experiments include DNA2, H2AX, and rat anti-BrdU antibodies (Abcam), λ-H2AX (Upstate Biotechnology), RPA34 (Neomarkers), p-RPA34 (Bethyl Laboratories), Chk1, p-Chk1 (Ser345), and p-p53 (Cell Signaling), ORC2, PCNA, DNA polymerase δ, RFC2, RFC1, MCM2, and p21 (Santa Cruz Biotechnology), Rad51 (Ab-1), mouse anti-BrdU, and p53 antibodies (Calbiochem). The immunoprecipitation with anti-Flag affinity beads was done in U2OS cells transiently transfected with FLAG-tagged plasmids. Forty-eight hours later, whole cellular extracts were prepared with RIPA buffer and immunoprecipitated with anti-Flag M2 affinity gel (Sigma) overnight. Bead-bound immunocomplexes were eluted with 3xFlag peptide (Sigma) and subjected to SDS-PAGE. For reciprocal immunoprecipitation, whole cellular extracts were prepared in RIPA buffer as indicated above and precleaned with protein A/G plus-agarose beads (Santa Cruz). Then cellular extracts were subjected to incubation with antibodies against PCNA and DNA polymerase δ (2 μg/mg of cell lysis) overnight and then incubated with protein A/G agarose beads for 4 hr at 4°C. The immunocomplex was eluted in loading buffer by boiling at 95°C for 5 min. The preparation of chromatin fractions and Western blot analysis, including the conditions for RPA analysis, were as previously described (17).
Statistical Analysis
All statistical analysis was done using one-tailed Student's t-test.
Additional methods are included in Supplementary Materials.
RESULTS
DNA2 forms a complex with replication factors and prevents accumulation of replication-associated DSBs
Based on previous findings that the activation of oncogene RAS causes replication stress and replication-associated DSBs, we selected the MCF10A and MCF10AT cell lines for proteomic analysis (4, 5, 18). MCF10A cells are immortalized normal breast epithelial cells. MCF10AT cells are derived from MCF10A cells by forced expression of oncogenic H-RAS. MCF10A cells do not grow in immunocompromised mice. In contrast, MCF10AT cells form the lesions that resemble the progression of breast cancer. Thus, these cell lines provided us with a model system to study the genetic alterations promoting carcinogenesis initiated by oncogene activation.
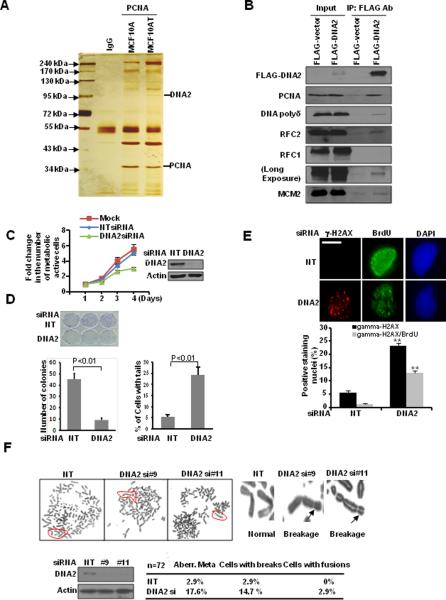
Replication forks are composed of many proteins. PCNA, DNA polymerase processivity factor, encircles DNA and orchestrates replication-linked processes by recruiting crucial players to the replication fork (19).Thereby, we used PCNA as our bait to isolate replication factor-associated protein complexes. Among many known proteins involved in DNA replication including RFC factors and MCM proteins, we found DNA2 had higher abundance in MCF10AT cells than in MCF10A cells (Fig. 1A). We then performed immunoprecipitation (IP) analysis to confirm DNA2 as a component of replication factor-associated protein complex (Fig. 1B and Supplemental Fig. S1A). This result is consistent with previous studies in yeast and Xenopus showing the involvement of DNA2 helicase/nuclease in DNA replication and repair (20, 21).
Figure 1. DNA2 associates with replication forks and regulates replication-associated DSBs.
A, Cell extracts were prepared from MCF10A and MCF10AT cells for IP. B, Co-IP of DNA2 in U2OS cells transfected with FLAG-vector or FLAG-DNA2. C and D, U2OS cells were transfected with control siRNA (NT) or DNA2 siRNA (SMARTpool). Each value represents the mean ± SEM from three independent experiments. C, MTT assay each day relative to day 1. D, Left panel: clonogenic assay. Right panel: Quantitative analysis of comet assay. At least 100 cells were scored in each sample. U2OS cells were transfected with DNA2 siRNA. E, Seventy-two hours later, cells were pulsed with BrdU (10 μM) for 30 min and then stained. (Top) Representative images (scale bar: 5 μm). (Bottom) At least 50 cells were scored in each sample (** p<0.01). F, (Top) Cells were analyzed by metaphase spreads. (Bottom) Quantitative summary. Western blot analyses demonstrating effective DNA2 knockdown are shown next to the graphs.
In humans, DNA2 is localized in both mitochondria and nuclei (22, 23). While the mitochondrial function of DNA2 was well documented, its nuclear functions remain unknown. We found that DNA2 depletion impaired both the number of cells with active metabolic activity and cell survival (Fig 1C, D). By neutral comet assay, we observed that DNA2-knockdown cells had a significantly higher proportion of cells with comet tails (Fig. 1D and Supplementary Fig. S1B), suggesting that DNA2 is required to prevent accumulation of endogenous DSBs.
We next examined whether DSBs present in DNA2-deficient cells were formed in cells with ongoing replication. γ–H2AX is a marker of DSBs (24). BrdU incorporation represents actively replicating cells. We found that DNA2-deficient cells had a significantly higher proportion of cells with γ-H2AX staining, and 60% of these cells also showed BrdU incorporation (Fig. 1E), which was significantly higher than that expected from a normal replication process (25). To further confirm that DNA2-knockdown-induced replication-dependent DSBs, we showed that inhibition of replication by aphidicolin significantly decreased the number of cells with positive γ-H2AX staining (Supplemental Fig. S1C). Additionally analysis of metaphase spreads showed that DNA2-deficient cells were significantly more likely to exhibit chromosomal breakage (Fig. 1F). These findings supported an important role of DNA2 in regulating accumulation of replication-associated DSBs and chromosomal stability.
DNA2 accumulates at restarted replication forks and promotes HR repair
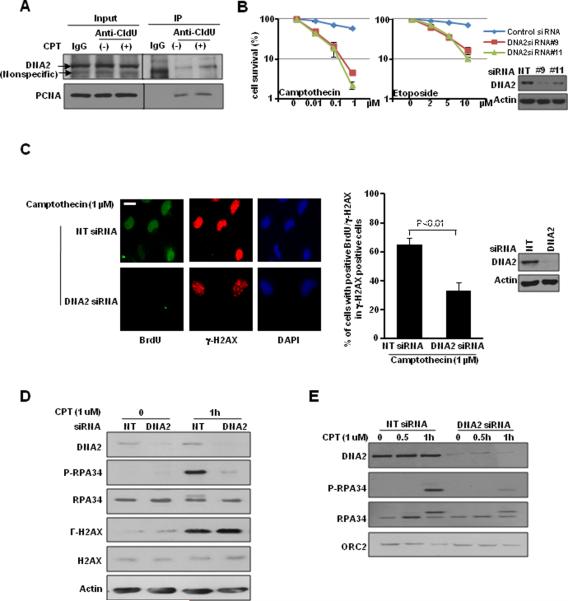
To confirm the role of DNA2 in response to replication-associated DSBs, we pulse-labeled newly synthesized DNA with a thymidine analog, CIdU. We performed IP with antibody against CIdU to detect the protein complexes associated with newly synthesized DNA. Interestingly, we observed that the association of DNA2 with replication forks was indeed enhanced in the presence of CPT, which causes replication-associated DSBs (Fig. 2A). In addition, we found that DNA2-depleted cells were more sensitive to topoisomerases inhibitors generating replication-associated DSBs, CPT and etoposide (Fig. 2B). Together, these data suggested that DNA2 accumulates at replication forks and plays a functional role in response to replication-associated DSBs.
Figure 2. DNA2 accumulates at restarted replication forks and promotes DSB end resection.
A, U2OS cells were treated with CPT (1 μM) for 1 hr and pulsed with CIdU for 30 min prior to IP analysis. PCNA was used as a control. B, U2OS cells were transfected with control siRNA or DNA2 siRNAs (#9 and #11) and then treated with CPT (Left) or etoposide (Right). U2OS cells were transfected with control siRNA or DNA2 siRNAs. C, Forty-eight hours later, cells were treated with or without CPT (1 μM) for 1 hr. Cells were labeled with BrdU (10 μM) for 30 min. γ-H2AX and BrdU were co-stained in native conditions. (Left) Representative images (scale bar: 5 μm). (Right) At least 50 cells were scored in each sample. Each value represents the mean ± SEM from two independent experiments. D and E, cells were treated with CPT (1 μM) for 1 hr. (D) Total cell lysates and (E) chromatin-enriched fractions were analyzed. Western blot analyses demonstrating effective DNA2 knockdown are shown next to the graphs.
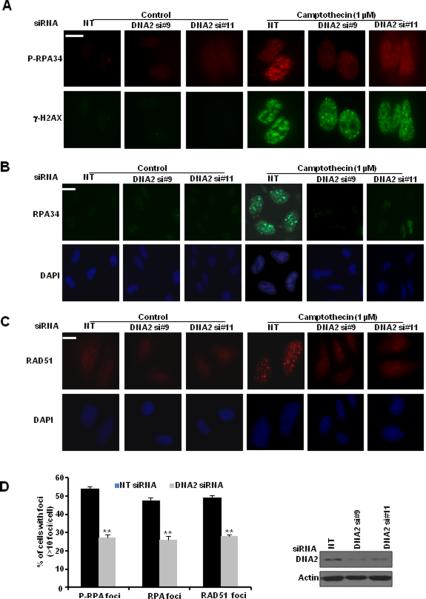
Several studies implicated yeast and Xenopus DNA2 in the initial step of HR, resection of DSBs (26–30). Also purified human DNA2 nuclease promotes DSB end resection (31). These reports, together with our observation led us to hypothesize that DNA2 may prevent the accumulation of replication-associated DSBs by promoting HR repair. We first examined ssDNA formation at DSBs in DNA2-depleted cells. After cells were labeled with BrdU, we stained nondenatured BrdU, which is located only at ssDNA (32, 33). We found reduced CPT-triggered ssDNA formation in DNA2-depleted cells (Fig. 2C) suggesting DNA2's role in DSB end resection. Next, we found that depletion of DNA2 impaired phosphorylation of RPA34 as measured by phospho-RPA34 antibody and by a slower migration of RPA34, but it did not affect γ-H2AX formation induced by CPT (Fig. 2D). In the presence of DSBs, both H2AX and RPA34 phosphorylation are regulated by kinases ATM, ATR, and DNA-PK (24, 34, 35). Because RPA is a ssDNA-binding protein and we did not observe impairment of H2AX phosphorylation, we reasoned that reduced RPA34 phosphorylation might be due to inefficient generation of ssDNA and consequently impaired recruitment of RPA34 to DSBs, where it is highly accessible to the kinases, rather than to impaired function of its upstream kinases. Indeed, chromatin fractionation assay showed that in DNA2-deficient cells, binding of both phosphorylated RPA34 and total RPA34 to chromatin was significantly reduced (Fig. 2E). We also found that DNA2 depletion resulted in reduced foci formation of both phosphorylated RPA34 and total RPA34 (Fig. 3A, B, D), indicating impaired RPA34 recruitment to DNA damage sites. Consistent with this result, we observed significantly reduced activation of CHK1 (Supplemental Fig. S2A) and reduced foci formation of RAD51 (Fig. 3C, D), which are recruited to DSB after RPA. These data suggested that DNA2 facilitates the recruitment of HR repair factors to DSBs.
Figure 3. DNA2 depletion impairs the recruitment of RPA and RAD51 to replication-associated DSBs.
U2OS cells were transfected with control siRNA or DNA2 siRNAs (#9 and #11). Forty-eight hours later, cells were treated with CPT (1 μM) for 1 hr and co-stained with the indicated antibodies: A, p-RPA34; B, RPA34; C, RAD51; and D, quantitative results. At least 50 cells (>10 foci/cell) in each sample were scored (** p<0.01) (scale bar: 5 μm). Western blot analyses demonstrating effective DNA2 knockdown are shown next to the graph.
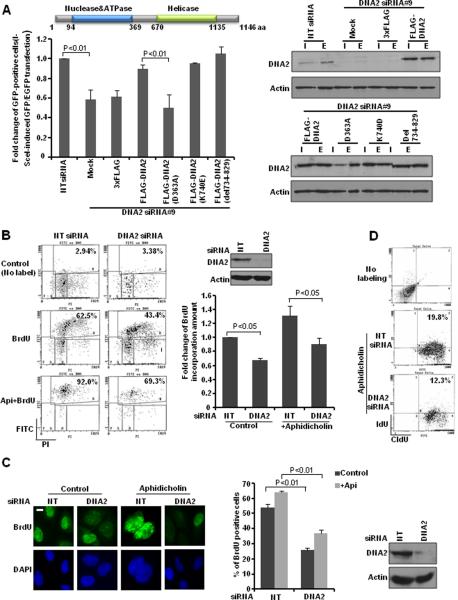
Next, we used a I-SceI inducible recombination assay to assess whether DNA2 depletion affects HR repair (36) (Supplemental Fig. S2B). We found that DNA2 knockdown significantly reduced HR repair efficiency (Fig. 4A). DNA2 contains both a nuclease/ATPase domain and a helicase domain (37). We conducted rescue experiments with RNAi-resistant wild-type, nuclease-dead (D363A) or helicase-dead DNA2 (K740E) mutants (37). As the mitochondrial localization signal was mapped to amino acids from 734 to 829 (22), we made a DNA2 construct with a specific deletion of this region (DNA2 del734-829), which also disrupts the helicase domain. The abrogation of mitochondrial localization of this construct was confirmed by a mitochondria marker, mtHSP70 (Supplementary Fig. S2C). In the rescue experiments, we found that nuclease activity of DNA2 is required for its function in HR repair, which is independent of its mitochondria localization (Fig. 4A). The effect of DNA2 on HR repair was not due to the changes in cell cycle distribution (Supplementary Fig. S2D). We then tested whether the DNA2 mutants would cause dominant-negative effects when they were overexpressed. As we expected, only the nuclease-dead DNA2 mutant impaired cell survival and HR repair (Supplemental Fig. S3A and B).
Figure 4. DNA2 facilitates HR repair and replication fork restart.
A, DRGFP cells were transfected with the indicated constructs. (Left) Quantitative summary of at least three independent experiments. Each value is relative to the percentage of GFP-positive cells in I-SceI-transfected control cells, which was set to 1 and represents the mean ± SD. A schematic diagram of DNA2 structure is shown at the top. (Right) Western blot analyses demonstrating effective transfection (I: I-SceI-transfected samples; E: EGFP-transfected samples as controls to normalize transfection efficiency). B, Forty-eight hours after transfection with control or DNA2 siRNA, U2OS cells were untreated or treated with aphidicolin (Api) (5 μM) for 17 hr. Then cells were released and labeled with BrdU (10 μM) for 30 min. (Left) Representative flow cytometry profile. (Right) Quantitative summary. Each value is relative to the percentage of BrdU-positive cells in cells with control siRNA transfection, which was set to 1 and represents the mean ± SD. C, U2OS cells were transfected with control siRNA or DNA2 siRNA. Forty-eight hours later, cells were treated or untreated with aphidicholin (5 μM) overnight. (Left) Representative images of BrdU incorporation detected in denatured conditions 30 min after BrdU (10 μM) labeling (scale bar: 5 μm). (Right) At least 50 cells were scored in each sample. Each value represents the mean ± SEM from two independent experiments. D, U2OS cells were transfected with control or DNA2 siRNA. Forty-eight hours later, cells were labeled with CIdU (25 μM) for 30 min and then were treated with camptothecin (1 μM) for 1 hr and released in culture medium containing IdU (250 μM) for 1 hr. Cells were stained with antibodies recognizing IdU (red) and CIdU (green). The percentage of double-positive cells is indicated. Quantitative summary is shown in Supplementary Fig. S4A. Western blot analyses demonstrating effective DNA2 knockdown are shown next to the graphs.
DNA2 is required for restart of replication forks
Given that DNA2 has enhanced association with replication forks and can promote repair of DSBs by HR, we proposed that DNA2 might enhance cellular tolerance of replication-associated DSBs and promote the restart of stalled or collapsed replication forks. To test this hypothesis, we performed three sets of experiments. Firstly, we analyzed the BrdU incorporation by flow cytometry after the release from aphidicolin treatment. We found that DNA2 deficiency impaired normal replication and had a significant impact on the restart of DNA replication after aphidicolin treatment (Fig. 4B). Secondly, we used immunofluorescent staining to detect BrdU incorporation. We observed that DNA2 was required for promoting DNA replication and restart of DNA replication after the removal of replication-stress-induced factor aphidicolin (Fig. 4C). Thirdly, we tested the efficiency of restart of replication forks in the absence of DNA2. We first labeled cells with CIdU and then labeled cells with IdU after treatment with aphidicolin. CIdU incorporation indicates unperturbed DNA synthesis (38). In contrast, IdU incorporation correlates with DNA synthesis after the release from DNA replication inhibition. Compared with control cells, DNA2-depleted cells had a reduced proportion of cells with double staining, suggesting impaired restart of replication forks after treatment with replication-stress-inducing stimuli (Fig. 4D, Supplementary Fig. S4A).
DNA2 enhances cellular tolerance of replication-associated DSBs in the context of oncogene activation
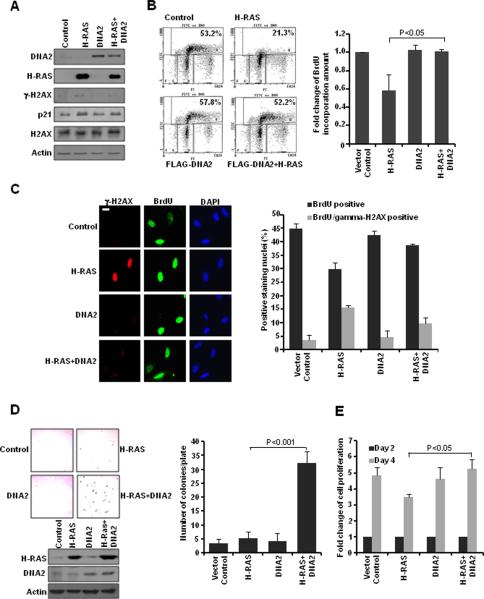
Excessive growth signaling induced by oncogene activation is one of the major sources of replication-associated DSBs in cancer cells (2, 4, 5). To elucidate the pathophysiological relevance of DNA2's function, we asked whether DNA2 could increase cellular tolerance of replication-associated DSBs induced by oncogene activation. We ectopically expressed H-RAS (V12) and DNA2 in cells and observed that H-RAS expression induced γ-H2AX and p21 formation, which was reduced in the presence of DNA2 (Fig. 5A). Next we found that H-RAS activation led to a more than 40% reduction in the number of BrdU-positive cells (Fig. 5B). This impaired DNA replication was rescued by co-expression of DNA2 (Fig. 5B). We then examined BrdU incorporation and γ-H2AX formation to test whether DNA2 might reduce accumulation of replication-associated DSBs induced by oncogene activation. The impaired DNA replication in H-RAS-expressing cells was accompanied by increased γ-H2AX foci formation in BrdU-positive cells, which indicated the presence of replication-associated DSBs (Fig. 5C). Interestingly, when DNA2 was co-expressed with H-RAS, cells showed significantly reduced γ-H2AX foci formation in BrdU-positive cells, suggesting reduced levels of replication-associated DSBs. These cells consequently had increased BrdU incorporation, similar to that in the control cells (Fig. 5C).
Figure 5. DNA2 alleviates replication-associated DSBs induced by oncogene activation.
U2OS cells were transfected with the indicated constructs. A, Western blot analyses. B, Forty-eight hours later, cells were pulsed with BrdU (10 μM) for 30 min. (Left) Flow cytometry profile. (Right) Quantitative summary. Each value is relative to the percentage of BrdU-positive cells in the control cells, which was set to 1 and represents the mean ± SD. C, Seventy-two hours later, cells were labeled with BrdU (10 μM) for 30 min before fixation. γ-H2AX and BrdU were co-stained in denatured conditions. (Left) Representative images (scale bar: 5 μm). (Right) At least 50 cells were scored in each sample. Each value represents the mean ± SEM from two independent experiments. D, Soft-agar assay was performed in MCF10A cells transfected with the indicated constructs. (Left) Representative images and Western blot analyses demonstrating efficient transfection. (Right) Quantitative summary represents the mean ± SD. E, Cell growth was analyzed by MTT assay. Each value is relative to the absorbance measured on day 2 (48 hr after transfection), which was set to 1 and represents the mean ± SD.
Based on these observations, we tested whether co-expression of DNA2 would potentiate oncogenic effects of H-RAS activation. By using soft-agar assay, we observed that co-expression of H-RAS and DNA2 significantly increased the number of colonies (Fig. 5D). Next, we found that H-RAS expression reduced cell proliferation and cells with DNA2 and H-RAS co-expression had increased cell proliferation (Fig. 5E). This result was further confirmed by using a second cell line expressing a different oncogene, cyclin E (Supplemental Fig. S3C). To summarize, our data revealed that an increase in DNA2 expression level may promote HR repair and reduce accumulation of replication-associated DSBs induced by oncogene activation.
Clinical relevance of DNA2 in cancer
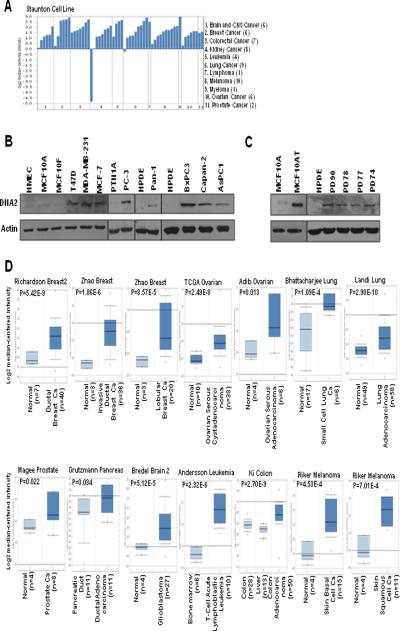
Given the function of DNA2 in alleviating replication stress-induced by oncogene activation, we sought to address the clinical relevance of DNA2 in human cancers. First, by both Oncomine database search (39) and Western blot analysis, we found that cancer cells exhibited increased DNA2 mRNA expression (Fig. 6A) and protein expression (Fig. 6B). DNA2 expression was also upregulated in MCF10AT cells, which represent atypical hyperplasia, a premalignant disease observed early in the natural course of breast cancer development (Fig. 6C). We further examined DNA2 expression in a cell model representing a series of transitions from normal pancreatic ductal cells to cancer cells due to activating K-RAS (16). Again, we found that overexpression of DNA2 occurred at an early stage of transformation (Fig. 6C). More importantly, we found that DNA2 mRNA levels were significantly increased in a wide range of cancer types reported from independent research groups in the Oncomine database (39) (Fig. 6D).
Figure 6. Enhanced DNA2 expression in human cancers.
A, DNA2 mRNA levels in cancer cell lines. B and C, Cell lysates from the indicated cancer cell lines were subjected to Western blot analyses. PD74, PD77, PD78, and PD90 cells represent a series of steps in oncogenic transformation of human pancreatic ductal epithelial (HPDE) cells. D, Box plots showing DNA2 mRNA expression analyses. The mean expression values of DNA2 are shown for each group. Ca, carcinoma.
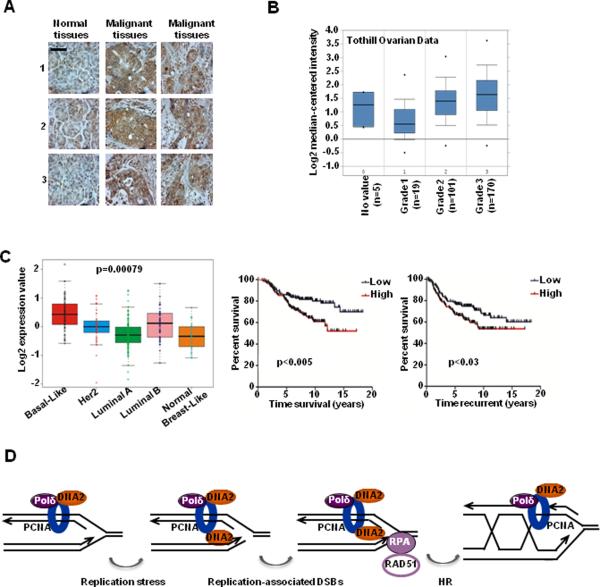
Pancreatic ductal adenocarcinoma has been found to almost always contain K-RAS mutations (>95% of tumors) (40). We specifically tested DNA2 expression in human tissues from pancreatic ductal adenocarcinoma. As we anticipated, DNA2 expression was increased in cancer tissues compared to adjacent normal tissues (Fig. 7A). We also found that DNA2 expression was positively correlated with the histologic grade of ovarian cancer (Fig. 7B). This finding suggested that elevated DNA2 expression might be functionally associated with increased intrinsic genomic instability during cancer development. To further test this possibility, we examined whether DNA2 expression exhibited a distinct pattern in different subtypes of breast cancer. Based on gene expression profiles, breast cancer can be divided into five subtypes: basal-like, Her2-positive, luminal A, luminal B, and normal breast-like (41). Morphologically, basal-like breast cancers have higher histologic grade than the other subtypes, and molecularly, basal-like breast cancers exhibit a higher degree of genomic instability, which is manifested by increased numbers of mutations, translocations, and single-nucleotide polymorphisms (42). As we expected, DNA2 expression in breast cancer cohort (NKI cohort, n = 295) was significantly higher in basal-like breast cancer than in the other subtypes (Fig. 7C). Indeed, in breast cancer patients, DNA2 expression was positively correlated with the likelihood of breast cancer metastasis and was inversely correlated with the duration of overall patient survival (Fig. 7C).
Figure 7. Clinical relevance of DNA2 in cancer.
A, DNA2 protein expression in pancreatic cancer tissue samples (scale bar: 200 μm). B, DNA2 expression correlates with tumor grade in ovarian cancer. C, (Left) Box plot showing gene expression levels among breast cancer subtypes (295 breast cancers). Kaplan-Meier survival curves are shown for overall survival time (Middle) and time to recurrence (Right) stratified by expression levels of DNA2. D, Proposed model for DNA2-mediated HR repair of replication-associated DSBs.
To further determine the biological effects of enhanced expression of DNA2 in cancer cells, we depleted DNA2 in breast cancer MCF-7 cells. DNA2 knockdown significantly increased DSB formation as and reduced cell proliferation (Supplementary Fig. S4B). Interestingly, in another cancer cell line U2OS cells which have competent DNA damage response, DNA2-deficient cells were remarkably larger than control cells (Supplementary Fig. S4C) and exhibited a flat morphological change. This observation led us to test whether DNA2 deficiency activates the senescence pathway, which permanently withdraws cells from the cell cycle. Compared to control cells, DNA2-depleted cells showed activation of γ-H2AX, phosphorylation and stabilization of p53, and activation of p21 (Supplementary Fig. S4D). DNA2 knockdown induced senescence as shown by β-galactosidase staining (Supplementary Fig. S4E). Notably, the number of senescent cells was remarkably reduced in the presence of aphidicolin (Supplementary Fig. S4E). Because DNA2 has been reported to regulate mitochondrial DNA replication and repair (22), we excluded the possibility that DNA2 depletion could cause mitochondria dysfunction and produce higher levels of reactive oxygen species (ROS), which could also contribute to cellular senescence (Supplementary Fig. S5).
DISCUSSION
In summary, we propose a model in which DNA2 is associated with factors involved in DNA replication and accumulates at stalled or collapsed replication forks. One likely function of DNA2 nuclease is in ssDNA formation at stalled replication forks, which allows RPA and RAD51 loading and subsequent HR repair to restart the stalled replication forks. Thereby, DNA2 can alleviate replication stress, primarily by facilitating HR repair of replication-associated DSBs (Fig. 7D). Our finding that DNA2 interacts with polymerase δ, which is thought to be the main polymerase involved in lagging strand synthesis, is consistent with previous observations of DNA2 function in processing Okazaki fragment in yeast (43, 44). It is notable that compared to leading strand synthesis, lagging strand synthesis is associated with a greater risk of aberrant replication and genome instability because a large number of Okazaki fragments need to be processed and ligated (20). It is very likely that DNA2, besides resection, has a role in preventing excessive DNA damage by processing 5' flaps formed during lagging strand synthesis or during HR. It is believed that DSBs are initially sensed by ATM, which is followed by end resection generating ssDNA for RPA loading and activation of ATR (35). As Xenopus DNA2 is found to be in a complex with ATM (21), whether the nuclease activity of DNA2 and the accumulation of DNA2 at DSB ends require ATM signaling requires further investigation.
Our data link the evolutionarily conserved function of DNA2 in HR repair to its role in alleviating both chemical- and oncogene-induced replication stress (2–5). During tumorigenesis, excessive growth signals often lead to hyperproliferation of cancer cells, which results in replication stress and increased formation of replication-associated DSBs. In our study, we have identified that enhanced DSB resection activity such as mediated by increased DNA2 expression may facilitate HR repair, which constitutes a key step to enhance cellular tolerance of replication-associated DSBs and provides cancer cells with a survival advantage. Currently by using a mouse genetic interaction approach, we are establishing a DNA2 conditional knockout mouse model to test this hypothesis in vivo.
We speculate that our findings may have significant clinical implications for cancer management. First, DSBs associated with oncogene-induced replication stress are observed in large fractions of early lesions from various human cancer tissues. Activation of the DSB end resection pathway such as DNA2 overexpression might serve as a marker for genetic alterations in premalignant lesions that promote tumor progression. Second, our findings raise the intriguing possibility that nucleases/helicases involved in DSB end resection, which are highly expressed in a wide range of human cancers, could serve as a new class of therapeutic targets. Indeed DNA nucleases/helicases are intensively studied as potential anti-cancer targets. Lately a small molecule inhibitor of the WRN helicase was identified, therefore similar strategy should be possible for DNA2 enzyme (45–48). Third, recent studies showed excellent response of BRCA1/BRCA2-mutated tumors with HR repair deficiency to Poly ADP ribose polymerase (PARP) inhibitors (49, 50). Currently, multiple clinical trials are testing the efficacy of PARP inhibitors in treating triple-negative breast cancer, which clinically and molecularly resembles BRCA1/BRCA2-mutated tumors (51, 52). Our data showed that basal-like breast cancer, which largely overlaps with triple-negative breast cancer, had significantly higher expression levels of DSB end resection factor DNA2. These factors and the activity of DSB end resection might provide a candidate biomarker to predict the response of triple-negative breast cancer to PARP inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Deming for proofreading the manuscript; M. Jasin for reagents; and M. D. Anderson Cancer Center core facilities for mass-spec, FACS and Molecular Cytogenetics.
GRANT SUPPORT This work was in part supported by NCI grant R01 CA112291 and DoD Era of Hope Scholar Award (W81XWH-10-1-0558) to S.-Y.L., NIH grant GM080600 to G.I., postdoctoral fellowship from Susan Komen Foundation for the Cure to G. P. and NCI grant K99 CA149186 to G. P.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST No potential conflicts of interest were disclosed.
REFERENCES
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004 Aug;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005 Apr 14;434(7035):864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 3.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005 Apr 14;434(7035):907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 4.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006 Nov 30;444(7119):633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 5.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006 Nov 30;444(7119):638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 6.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007 Dec 10;26(56):7773–9. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 7.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008 Mar 7;319(5868):1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 8.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 9.Helleday T. Amplifying tumour-specific replication lesions by DNA repair inhibitors - a new era in targeted cancer therapy. Eur J Cancer. 2008 May;44(7):921–7. doi: 10.1016/j.ejca.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 10.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009 Sep 2;8(9):983–95. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu H, Raynard S, Sung P. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 2009 Jul 1;23(13):1481–6. doi: 10.1101/gad.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010 Oct 7;467(7316):678–83. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010 Oct;17(10):1260–2. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010 Oct;17(10):1263–5. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998 Jan 15;17(2):598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007 Mar 1;67(5):2098–106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 17.Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009 Jul;11(7):865–72. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996 Jan;148(1):313–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007 May 18;129(4):665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit Rev Biochem Mol Biol. 2010 Apr;45(2):71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 21.Wawrousek KE, Fortini BK, Polaczek P, Chen L, Liu Q, Dunphy WG, Campbell JL. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle. 2010 Mar 23;9(6) doi: 10.4161/cc.9.6.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008 Nov 7;32(3):325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009 Aug;29(15):4274–82. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004 Aug-Sep;3(8–9):959–67. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009 Oct 15;23(20):2405–14. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010 Sep 2;467(7311):112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010 Sep 2;467(7311):108–11. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008 Sep 19;134(6):981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010 Oct 6;29(19):3370–80. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J. 2010 Sep 1;29(17):2864–74. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011 Feb 15;25(4):350–62. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007 Nov 22;450(7169):509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009 May 21;459(7245):460–3. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999 Mar 1;18(5):1397–406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakasai R, Shinohe K, Ichijima Y, Okita N, Shibata A, Asahina K, Teraoka H. Differential involvement of phosphatidylinositol 3-kinase-related protein kinases in hyperphosphorylation of replication protein A2 in response to replication-mediated DNA double-strand breaks. Genes Cells. 2006 Mar;11(3):237–46. doi: 10.1111/j.1365-2443.2006.00942.x. [DOI] [PubMed] [Google Scholar]
- 36.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999 Oct 15;13(20):2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34(6):1865–75. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009 Sep 2;28(17):2601–15. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004 Jan;Feb;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010 Oct;10(10):683–95. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 42.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010 Nov 11;363(20):1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 43.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009 Feb 13;284(7):4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart JA, Miller AS, Campbell JL, Bambara RA. Dynamic removal of replication protein A by Dna2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J Biol Chem. 2008 Nov 14;283(46):31356–65. doi: 10.1074/jbc.M805965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aggarwal M, Brosh RM., Jr. Hitting the bull's eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J Cell Biochem. 2009 Apr 1;106(5):758–63. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1525–30. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arai A, Chano T, Futami K, Furuichi Y, Ikebuchi K, Inui T, Tameno H, Ochi Y, Shimada T, Hisa Y, Okabe H. RECQL1 and WRN proteins are potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Res. 2011 Jul 1;71(13):4598–607. doi: 10.1158/0008-5472.CAN-11-0320. [DOI] [PubMed] [Google Scholar]
- 48.Gupta R, Brosh RM., Jr. Helicases as prospective targets for anti-cancer therapy. Anticancer Agents Med Chem. 2008 May;8(4):390–401. doi: 10.2174/187152008784220339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005 Apr 14;434(7035):917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 50.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005 Apr 14;434(7035):913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 51.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011 Jan 20;364(3):205–14. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 52.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009 Jul 9;361(2):123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.