Abstract
BACKGROUND & AIMS
Zinc-fingers and homeoboxes 2 (ZHX2) represses transcription of several genes associated with liver cancer. However, little is known about the role of ZHX2 in development of hepatocellular carcinoma (HCC). We investigated the mechanisms by which ZHX2 might affect proliferation of HCC cells.
METHODS
We overexpressed and knocked down ZHX2 in HCC cells and analyzed the effects on proliferation, colony formation, and the cell cycle. We also analyzed the effects of ZHX2 overexpression in growth of HepG2.2.15 tumor xenografts in nude mice. Chromatin immunoprecipitation and luciferase reporter assays were used to measure binding of ZHX2 target promoters. Levels of ZHX2 in HCC samples were evaluated by immunohistochemistry.
RESULTS
ZHX2 overexpression significantly reduced proliferation of HCC cells and growth of tumor xenografts in mice; it led to G1 arrest and reduced levels of cyclins A and E in HCC cell lines. ZHX2 bound to promoter regions of CCNA2 (which encodes Cyclin A) and CCNE1 (which encodes cyclin E) and inhibited their transcription. Knockdown of cyclin A or cyclin E reduced the increased proliferation mediated by ZHX2 knockdown. Nuclear localization of ZHX2 was required for it to inhibit proliferation of HCC cells in culture and in mice. Nuclear localization of ZHX2 was reduced in human HCC samples, even in small tumors (diameter<5 cm), compared to adjacent non-tumor tissues. Moreover, reduced nuclear levels of ZHX2 correlated with reduced survival times of patients, high levels of tumor microvascularization, and hepatocyte proliferation.
CONCLUSIONS
ZHX2 inhibits HCC cell proliferation, by preventing expression of cyclins A and E, and reduces growth of xenograft tumors in mice. Loss of nuclear ZHX2 might be an early step in the development of HCC.
Keywords: mouse model, carcinogenesis, shRNA, CCK-8
Hepatocellular carcinoma (HCC) is the fifth most common cancer and third most common cause of cancer deaths worldwide1. Environmental factors, most notably HBV and HCV infection and chronic alcohol exposure, are known risk factors of HCC development1,2. A number of genetic loci that contribute to HCC progression have also been identified2,3. Numerous studies have investigated transcriptional changes in HCC, including microarray analyses of global gene expression changes. While differences in expression of a number of liver-enriched transcription factors have been reported, consistent differences have not always been observed and it is not clear which of these are relevant to HCC progression4. However, several genes that are expressed abundantly in the fetal liver, silenced at birth, and reactivated in HCC have been identified4, including alpha-fetoprotein (AFP), H19 and glypican 3 (GPC3)5-8. A better understanding of how these genes are reactivated in HCC may elucidate transcription changes that occur during liver cancer progression.
While the AFP, H19 and GPC3 genes are silenced at birth in most mouse strains, these three genes continue to be expressed in the adult liver of BALB/cJ mice9. Further studies indicated that this incomplete repression of these three genes in BALB/cJ mice is due to a natural mutation in the Zinc Fingers and Homeoboxes 2 (ZHX2) gene9,10. More recent studies indicate that ZHX2 also regulates hepatic enzymes involved in plasma lipid homeostasis, including lipoprotein lipase11. ZHX2 is a member of a small family which also includes ZHX1 and ZHX312-14. These proteins are predicted to contain two zinc-finger (Znf) and four or five homeodomains (HDs), motifs which could confer protein interaction and nucleic acid binding activities. Two-hybrid studies indicate that ZHX proteins can form homodimers as well as heterodimers with each other and with the A subunit of nuclear factor Y (NF-YA)13-15. Current studies suggest that ZHX proteins are ubiquitously expressed and found primarily in the nucleus, where they function as transcriptional repressors14,16. We previously showed that ZHX2 reduces AFP secretion17 and GPC3 expression (unpublished data) in human HCC cell lines. Cotransfection assays by us and others have also shown that ZHX2 can repress the promoters of AFP and the NF-YA-regulated genes cdc25C and Hexokinase II (HKII)14, 17, 18.
Several studies investigating ZHX2 and HCC have provided conflicting data. Using methylation-sensitive restriction fingerprinting, Lv et al., demonstrated hypermethylation of the ZHX2 promoter in some HCC samples and HepG2 cells which correlated with the lack of ZHX2 expression. This silencing of expression suggests that ZHX2 might function as a tumor suppressor19. In contrast, using immunohistochemical analysis, Hu, et al., reported increased ZHX2 staining in HCC samples compared to normal liver; this study also noted higher ZHX2 expression in poorly differentiated and metastasis samples. This data is consistent with ZHX2 having tumor promoting properties20.
In the present study, we investigated the role of ZHX2 in the growth of liver cell lines both in vitro and in vivo. Our data showed that ZHX2 inhibits HCC cells growth. We also showed that ZHX2 represses Cyclin A and Cyclin E expression, which might account for the growth inhibitory properties of ZHX2. Nuclear localization of ZHX2 is critical for its inhibitory effects. This data is supported by analysis of clinical samples, which show decreased nuclear expression of ZHX2 in HCC samples compared to adjacent non-tumor tissue.
Materials and Methods
Cell lines, plasmids and siRNAs
The human HCC cell lines HepG2, SMMC7721 and QSG7701, Chinese hamster ovary (CHO) cells and human embryonic kidney 293 (HEK293) cells were purchased from the Shanghai Cell Collection (Chinese Academy of Sciences). The HepG2.2.15 cell line was obtained from the Shandong Academy of Medical Sciences. These cells were maintained as described previously17.
ZHX2 expression vectors pcZHX2 (full-length ZHX2 with a carboxy-terminal HA tag) and pZHX2 (ZHX2-EGFP fusion protein) and shRNA vectors against human ZHX2 (pS1674, pS2360) were described previously17. Truncated forms of human ZHX2 containing HD1 and HD2 in which the nuclear localization signal (NLS) was present or absent [ZHX2(242-446) and ZHX2(242-439), respectively] were generated by PCR amplification of pZHX2 using primers shown in Supplementary Figure S2 and cloned into pEGFP-N1 (Invitrogen). The luciferase reporter plasmids pGL3-Ap and pGL3-Ep were constructed by cloning the promoter regions of human Cyclin A (-505 to +361, the transcription initiation site designated as +1) and Cyclin E (-402 to +72), respectively, into the promoterless pGL3-basic vector (Promega)21, 22. The siRNAs against Cyclin A, Cyclin E and Cyclin D1 (Table S2) were synthesized by the Shanghai Genepharma Co.
Analysis of cell proliferation, cell cycle and in vivo tumor growth
Cell viability was measured using the Cell Counting Kit-8 (CCK-8, Beyotime, China) and standard colony formation assays were used to measure cell proliferation. Each experiment was repeated 3-4 times. For cell cycle analysis, cells were collected 48 hrs after transfection with indicated plasmids, stained with propidium iodide (PI, Sigma) and assayed using a Beckman Coulter Flow Cytometer (Fullerton).
Male BALB/c nude mice (4~6 weeks of age) were purchased from the Animal Research Committee of the Institute of Biology and Cell Biology (Shanghai, China) and housed in the Shandong University School of Medicine animal facility according to protocols approved by the Shandong University Animal Care Committee. HepG2.2.15 cells (1×107) were transplanted subcutaneously into nude mice. After reaching a diameter of 0.5 cm, tumors were injected with plasmid (20μg/100μl) every fourth day for a total of 3-4 injections. Tumor size was monitored every other day. Mice were sacrificed four days after the final injection and the tumors were isolated and weighed. Animal experiments were repeated at least twice and six mice were included in each cohort. Cell proliferation in each tumor was assayed by immunoperoxidase staining with an anti-Ki-67 antibody (ab15580; Abcam). Eight fields of roughly 1000 tumor cells for each section were scored independently by three pathologists.
Western blotting
Cytoplasmic, nuclear or whole cell extracts were prepared and analyzed by western blotting as previously described using anti-ZHX2 (Abcam), anti-cyclin A (4656, Cell Signal Technology), anti-cyclin E (sc-25303, Santa Cruz), anti-cyclin D1 (ab6152, Abcam), anti-p21 (sc-6246, Santa Cruz), anti-p27 (ab32034, Abcam), anti-GFP (AG281, Beyotime), anti-Histone H2A.X (BS5524, Bioworld Technology, Inc), anti-Lamin A/C (BS1446, Bioworld Technology, Inc) and anti-β-actin (Sigma)17.
Transfections, fluorescent staining and luciferase assays
CHO and HEK293 cells transfected with indicated plasmids were stained with DAPI (Sigma, USA) and observed for GFP and DAPI using fluorescence microscopy (Olympus, Japan). HepG2 cells were co-transfected with reporter plasmids (0.25 μg) and expression plasmids (0.75 μg) using Lipofectamine™ 2000. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega)17.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed with ZHX2-HA (pcZHX2)-transfected HepG2 cells. Briefly, transfected cells were fixed in 1% formaldehyde after 48 hrs and sonicated to shear DNA to 200~1000 bp. Supernatants obtained after centrifugation at 13,000×g for 10 min were used for immunoprecipitations using an anti-HA antibody (ab9110; Abcam) or control IgG. Immunoprecipitated DNA was used for PCR amplification. Total cellular DNA was used as input control.
Patient samples and immunohistochemical staining
Eighty-two tumor tissues and seventy-eight adjacent non-tumor tissues were collected from patients with primary HCC who underwent surgery between Oct. 30, 2010 and Aug. 31, 2011 at Qilu Hospital and Shandong Provincial Hospital, Shandong University (Supplementary Table S1). Cell differentiation-based HCC tumor grading was determined as described by Edmondson and Steiner23. Liver fibrosis was staged using the Ishak score and samples with a score ≥5 were classified as cirrhotic24. The presence of HBV antigens was determined by ELISA. None of the patients were positive for HCV or HIV, consumed excessive alcohol or received chemotherapy prior to surgery. Informed consent was obtained from all patients before the study was initiated with approval of the Shandong University Medical Ethics Committee in accordance with the Declaration of Helsinki. Tissue microarrays were purchased from the Shanghai Outdo Biotech Company.
Immunohistochemical staining was performed according to standard protocols using the following antibodies: anti-ZHX2 (ab56886; Abcam), anti-Ki-67 (ab15580; Abcam), anti-Cyclin A (#4656, Cell Signaling Technology), anti-Cyclin E (sc-25303, Santa Cruz Biotechnology), and anti-CD31 (#3528, Cell Signaling Technology). Eight fields of ~1000 cells from each tumor and non-tumor sections were counted independently by three pathologists. Cytoplasmic and nuclear ZHX2 staining were reported separately according to the German semi-quantitative scoring system25, 26. Briefly, each sample was scored according to staining intensity (no staining = 0; weak staining = 1; moderate staining = 2; strong staining = 3) and the number of stained cells (0% = 0; 1-25% = 1; 26-50% = 2; 51-75% = 3; 76-100% = 4). Final immunoreactive scores were determined by multiplying the staining intensity by the number of stained cells, with minimum and maximum scores of 0 and 12, respectively27. Levels of hepatocyte proliferation and intratumoral microvascular density (MVD) were identified by Ki-67 and CD31 staining respectively. For quantization of MVD, the average numbers of CD31 positive vessels from three areas of maximal vascular density (vascular hotspots) were counted for each section28.
Statistical Analysis
GraphPad Prism (GraphPad Software, San Diego, CA) was used for data analysis. The Student t-test, Mann-Whitney U test or one-way ANOVA was applied to determine significant differences between groups. Two-way ANOVA was applied to determine significant differences between different treatments, in different cell cohorts or at different time points. The statistical correlation between the clinical parameters of HCC and the ZHX2 staining levels in tissue sections was analyzed by the Chi-square test. Survival differences were analyzed using the log-rank test. In these analyses, p values <0.05 were considered to be significant.
Results
ZHX2 inhibited the proliferation of HCC cell lines both in vitro and in vivo
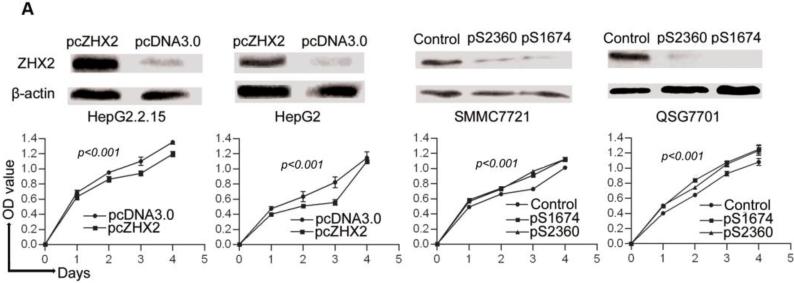
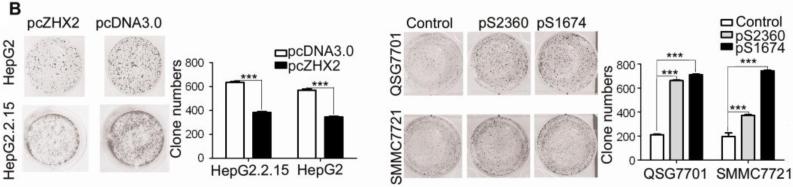
Previous studies provided conflicting data regarding a potential role for ZHX2 in HCC progression. To investigate further the role of ZHX2 in hepatocarcinogenesis, we measured the growth of hepatoma cell lines in which ZHX2 levels were modulated. The HCC cell lines HepG2 and HepG2.2.15 have low endogenous ZHX2 levels. In both cell lines, ZHX2 overexpression reduced proliferation over a four-day period (Fig. 1A). Consistent with this data, reducing ZHX2 levels by transfecting shRNAs pS1674 and pS2360 in HCC cell lines SMMC7721 and QSG7701, which have high endogenous ZHX2 levels, significantly enhanced cell proliferation (Fig. 1A). The inhibitory properties of ZHX2 were also analyzed using colony formation assay. ZHX2 overexpression led to a significant decrease in the number of colonies formed when assayed ten days after transfection in both HepG2 and HepG2.2.15 cell lines (Fig. 1B). The shRNA-mediated knockdown of ZHX2 significantly increased the number of colonies formed in both the QSG7701 and SMMC7721 cell lines (Figure 1B). Taken together, these results indicate that ZHX2 inhibits the proliferation of HCC cell lines.
Figure 1. Elevated ZHX2 levels inhibit growth of HCC both in vitro (A,B) and in vivo (C,D,E,F).
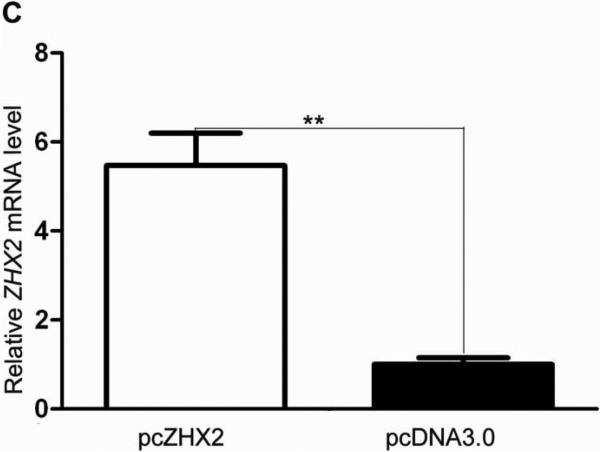
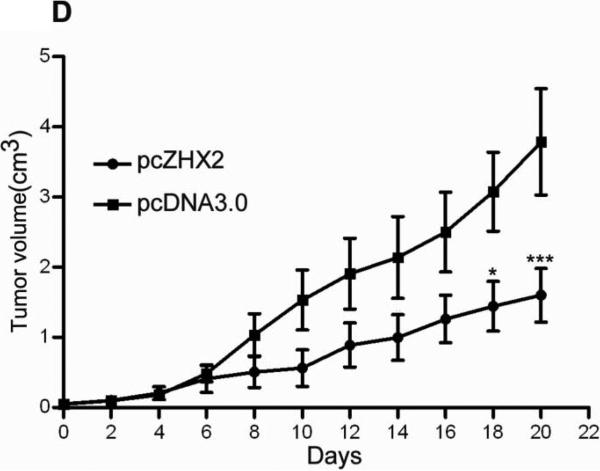
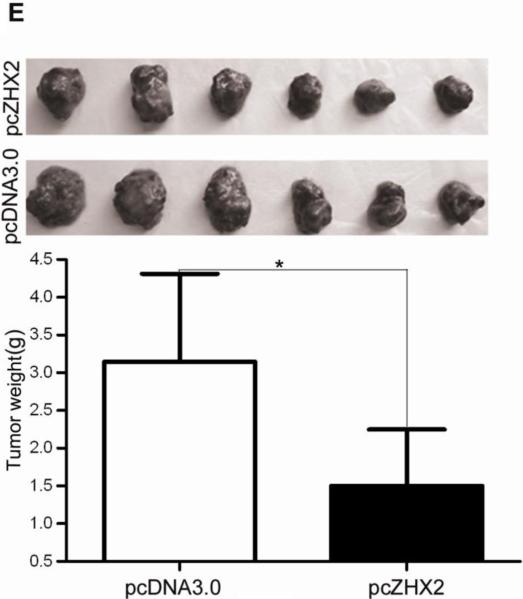
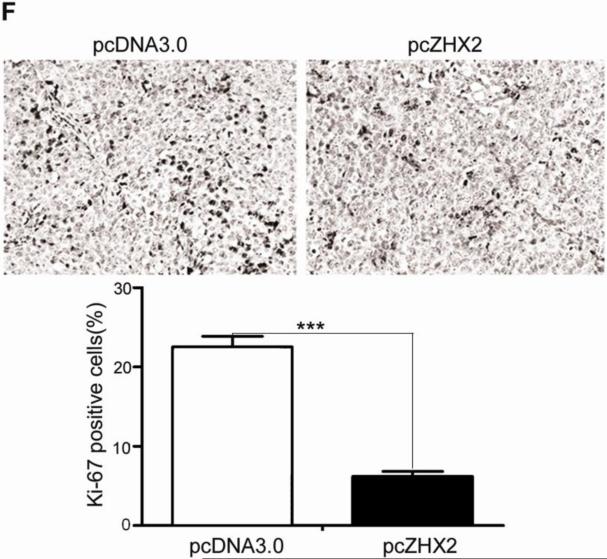
(A) Proliferation of HepG2.2.15 and HepG2 cells transfected with pcZHX2 or control pcDNA3.0 and SMMC7721 and QS7701 cells transfected with shRNAs pS2360 or pS1674. Cells were assayed over a 4-day period and data shown is mean±SD from four experiments. ZHX2 and β-actin levels in cell lines were determined by western blot. (B) Colony formation HCC cell lines transfected with pcZHX2 or ZHX2 shRNAs as described in 1A. Plates are shown on the left; the statistical results are shown in the right (Mean±SD from three experiments) * * *, p<0.001. (C) Real-time RT-PCR analysis of ZHX2 mRNA in pcZHX2-injected tumors and control pcDNA3.0-injected tumors. (Mean ±SD, n=6). * *, p<0.01. (D) Xenograft tumor growth determined over a 20 day period. (Mean±SD, n=6); *, p<0.05, * * *, p<0.001. (E) Weight of pcZHX2- and pcDNA3.0-injected tumors at time of sacrifice. (Mean±SD, n=6); *, p<0.05. Images of tumors from each group is shown on top. (F) Immunohistochemical staining of Ki-67 of pcDNA3.0-and pcZHX2-injected tumors; statistical results were shown in the bottom. (Mean±SD, n=6); * * *, p<0.001.
The influence of ZHX2 on tumor growth was evaluated further by measuring growth of subcutaneous HepG2.2.15 xenografts in nude mice. After reaching a diameter of 0.5 cm, tumors were injection with pcDNA3.0 or pcZHX2. Real-time RT-PCR analysis demonstrated that pcZHX2-injected tumors had increased ZHX2 mRNA levels compared to pcDNA3.0-injected tumors (Fig. 1C). Moreover, injection of pcZHX2 significantly inhibited the tumor growth over the course of the experiment (Fig.1D). Consistent with this finding, the weight of ZHX2-injected tumors at the time of sacrifice was less than half of pcDNA3.0-injected control tumors (Fig. 1E). Immunohistochemical analysis demonstrated less Ki-67 staining in pcZHX2-treated tumors (Fig. 1F), indicative of reduced proliferation. Taken together, these in vitro and in vivo studies indicate that ZHX2 inhibits cell growth and consistent with ZHX2 functioning as a tumor suppressor.
ZHX2 induces G1 arrest and represses Cyclin A and Cyclin E expression
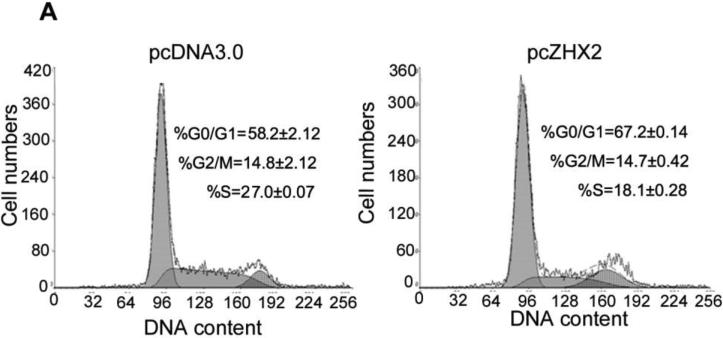
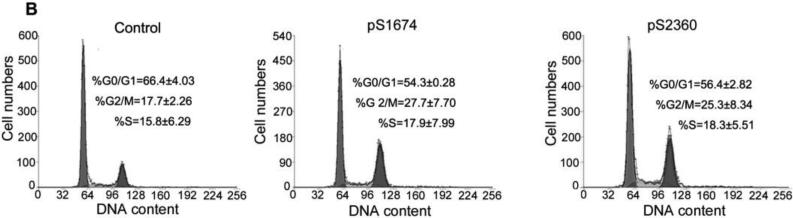
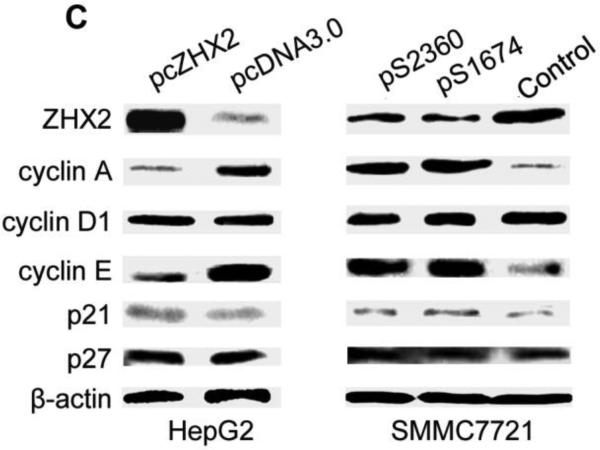
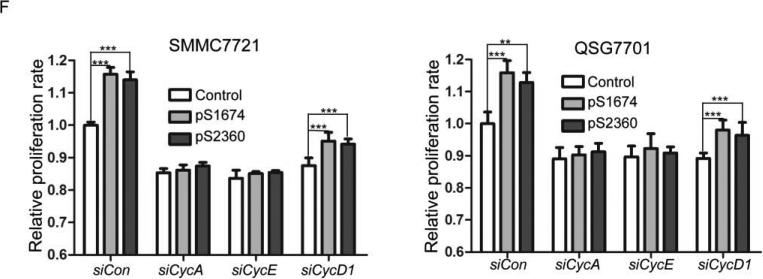
To explore the mechanism by which ZHX2 inhibits cell growth, cell cycle analysis was performed by PI staining and flow cytometry of HCC cell lines in which ZHX2 levels were increased or decreased. Transfection of pcZHX2 in HepG2 cells (Fig. 2A and HepG2.2.15 (data not shown) increased the percentage of G0/G1 cells and decreased the percentage of S phase cells. In a reciprocal experiment, shRNA-mediated reduction of ZHX2 in SMMC7721(Fig. 2B) and QSG7701 cells (data not shown) by pS1674 or pS2360 decreased and increased the percentage of cells in G0/G1 and G2/M, respectively. Analysis of potential cell cycle regulators indicated that ZHX2 overexpression in HepG2 cells decreased Cyclin A and Cyclin E but had no effect on Cyclin D1, p21, or p27 protein levels (Fig. 2C). The shRNA-mediated reduction of ZHX2 in SMMC7721 cells led to increased Cyclin A and Cyclin E levels whereas Cyclin D1, p21, and p27 levels remained unchanged (Fig. 2C). These data suggest that ZHX2 influences cell cycle progression by reducing Cyclin A and/or Cyclin E levels.
Figure 2. ZHX2 induces G1 arrest and represses Cyclin A and Cyclin E expression.
(A) HepG2 cells transfected with pcZHX2 or pcDNA3.0 or (B) SMMC7721 cells transfected with scrambled control vector or pS1764 or pS2360 shRNA vectors were analyzed by PI staining and flow cytometry. A representative plot from one experiment and Mean±SD from three experiments are shown. (C) Western blot to monitor levels of ZHX2, Cyclin A, Cyclin D1, Cyclin E, p21, p27 and β-actin in transfected HCC cell lines described in (A) and (B). The experiments were repeated for four times and one representative result is shown.
ZHX2 inhibits cell proliferation by reducing Cyclin A and Cyclin E transcription
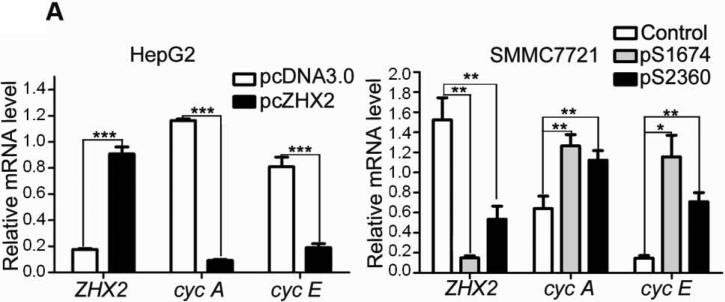
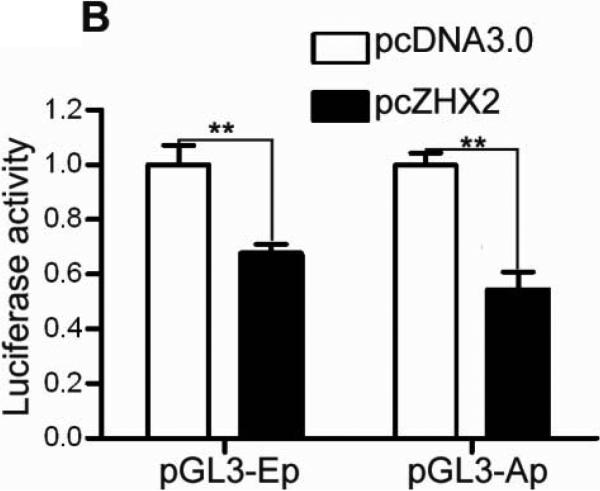
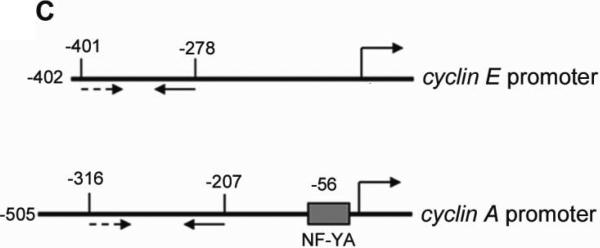
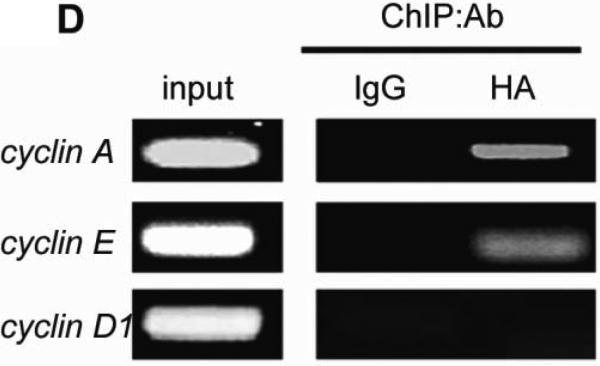
Previous studies suggest that ZHX2 functions as a transcriptional repressor14. Since our western data indicated that Cyclin A and Cyclin E levels inversely correlated with ZHX2 levels, we considered whether ZHX2 represses the transcription of these genes. Real-time RT-PCR analysis showed that Cyclin A and Cyclin E steady state mRNA levels were dramatically decreased when ZHX2 levels were elevated in HepG2 cells and increased when ZHX2 levels were reduced in SMMC7721 cells (Fig. 3A). To explore further this regulation, the Cyclin A and Cyclin E promoters were fused to a luciferase reporter gene (Figure 3C). Transient co-transfections demonstrated that ZHX2 repressed the activities of both promoters (Fig. 3B). To determine if this regulation involved ZHX2 binding to these promoters, ChIP assays were performed in HepG2 cells transfected with HA-tagged ZHX2. This data indicated that ZHX2 binds the promoter regions of both Cyclin A and Cyclin E but not Cyclin D1 (Fig. 3D), suggesting that ZHX2 represses Cyclin A and Cyclin E expression by binding, directly or indirectly, to the promoters of these genes.
Figure 3. ZHX2 binds to and represses activity of the Cyclin A and Cyclin E promoters.
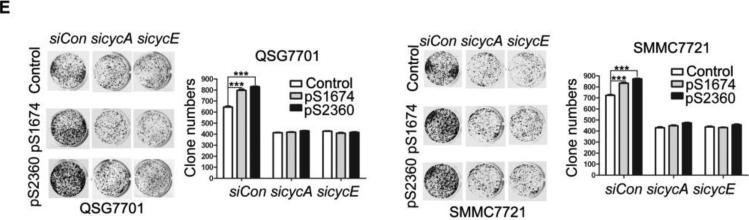
(A) Real-time RT-PCR analysis of ZHX2, Cyclin A and Cyclin E mRNA levels in HepG2 cells transfected with pcZHX2 or pcDNA3.0 or SMMC7721 cells transfected with shRNAs. Data (mean±SD) of three experiments are shown. *, p<0.05, * *, p<0.01, * * *, p<0.001. (B) Inhibition of the pGL3-Ep and pGL3-Ap luciferase reporter genes by ZHX2 in HepG2 cells. Data (mean±SEM) of four experiments are shown. * *, p<0.01. (C) Diagram of the Cyclin E and Cyclin A promoters, extending to -402 and -505, respectively, including the NF-Y site in the cyclin A promoter34 and location of the primers (solid or dotted lines under promoters) used for ChIP analysis. (D) ChIP analysis of DNA from HepG2 cells transfected with ZHX2-HA. PCR amplification of HA-immunoprecipitated DNA using the primers shown in (C) shows ZHX2 binding to Cyclin A and Cyclin E promoters but not Cyclin D1. One of three independent experiments is shown. (E) Colony formation of QSG7701 and SMMC7721 cells transfected with shRNAs against ZHX2 (pS1674 or pS2360) and siRNAs agaist Cyclin A (sicycA) or Cyclin E (sicycE). One representative plate of each group is shown on the left; results from three independent experiments are shown in the right. (mean±SD) * * *, p<0.001. (F) Proliferation of QSG7701 and SMMC7721 cells that were co-transfected with ZHX2 shRNAs along with siRNAs for Cyclin A, Cyclin E or Cyclin D1. Cells were measured using CCK-8 48h after cotransfection. Mean±SD of three experiments is shown. ; * * *, p<0.001.
The growth inhibitory properties of ZHX2 may be due to its ability to repress Cyclin A and Cyclin E. If so, reducing these cyclins should overcome the effects of reducing ZHX2 levels. To test this possibility, Cyclin A and Cyclin E protein levels were reduced by siRNAs in the same cells where ZHX2 levels were knocked down. Indeed, we found that the siRNAs against these cyclins abrogated the accelerated growth mediated by the ZHX2 shRNAs in SMMC7721 or QSG7701 cells in both colony growth and cell proliferation assays (Figs. 3E and 3F). In contrast, the increased proliferation of cells treated with shRNAs against ZHX2 was not overcome by reducing Cyclin D1 levels (Fig. 3F). These data support the possibility that ZHX2 inhibits HCC cell growth, at least in part, by inhibiting Cyclin A and Cyclin E expression.
Nuclear but not cytoplasmic ZHX2 expression is decreased in HCC tissues
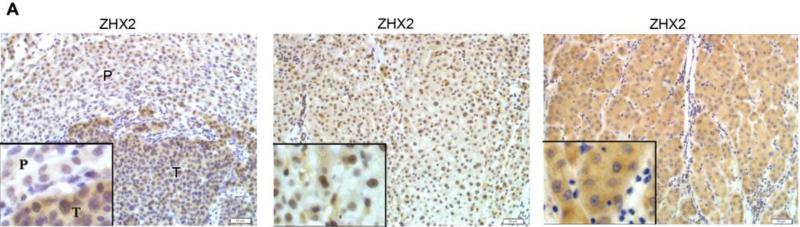
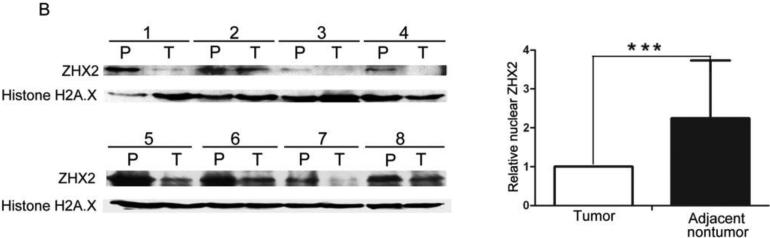
Previous studies provided conflicting data regarding ZHX2 expression in HCC20. To explore further ZHX2 expression in liver cancer, we stained for ZHX2 in both HCC and adjacent non-tumor regions (Figure 4A). ZHX2 staining was observed in both regions. However, the percentage of positive nuclear staining (score of 4-12) was significantly higher in adjacent non-tumor regions compared to that in tumors (p = 0.0282; Table 1). In contrast, the percentage of positive cytoplasmic ZHX2 staining in tumors was significantly higher than that in adjacent non-tumor sections (p=0.0190; Table 1). Overall, there was no difference in total ZHX2 expression (nuclear and cytoplasmic staining) between tumor and adjacent non-tumor groups (Table 1). This difference in nuclear/cytoplasmic ZHX2 staining is supported by western blot analysis of nuclear extracts from tumors and adjacent non-tumor tissue, with nuclear ZHX2 being lower in tumor nuclei (Fig. 4B). Interestingly, decreased nuclear ZHX2 staining was also observed in tumors with diameter less than 5 cm (Table 1).
Figure 4. ZHX2 expression in HCC and correlation with clinical parameters.
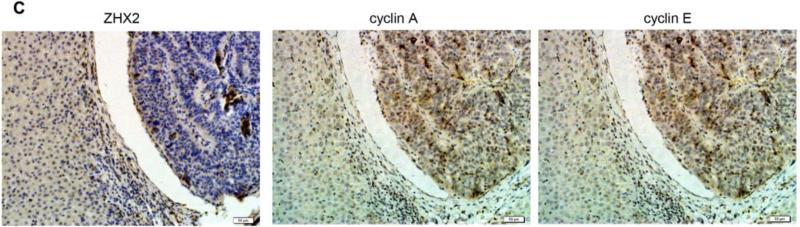
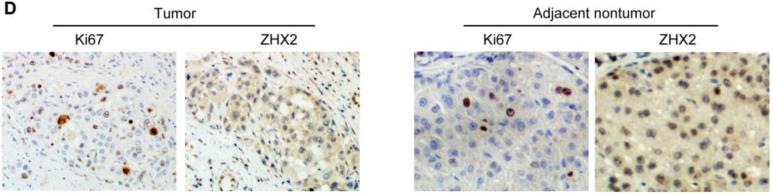
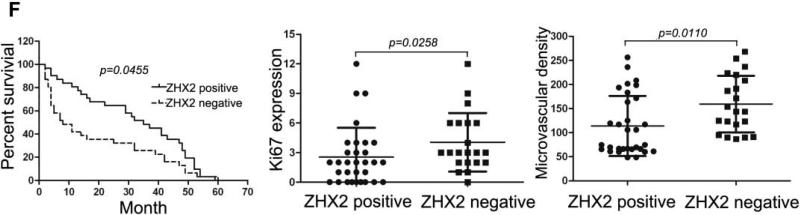
(A) Immunohistochemical staining of ZHX2 in HCC sections (left and right panels) and non-tumor liver sections (middle panel). T=tumor and P=adjacent non-tumor. Original magnification 200×. (B) Western blot analysis of ZHX2 levels in nuclear extracts of adjacent non-tumor (P) and tumor (T) samples from patients with HCC. Histone H2A.X was used as a control. Statistical data is shown at right. * * *, p<0.001. (C) Immunohistochemical staining of ZHX2 (left), Cyclin A (middle) and Cyclin E (right) in adjacent sections of a cancer biopsy from one patients. Original magnification 200×. (D) Immunohistochemical staining of ZHX2 and Ki-67 (marker of cell proliferation) in continuous biopsies (tumor sections and adjacent nontumor sections). (E) Statistical analysis of ZHX2 nuclear expression in poor, moderate or highly differentiated HCC samples. The immunoreactive score is shown as median±SD. * *, p<0.01. (F) Nuclear ZHX2 expression correlates with overall survival (left), Ki-67 (middle), and intratumoral microvascular density (right).
Table 1.
Detection of ZHX2 expression in clinical specimens.
| No. of cases | ZHX2 expression |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nuclear staining |
Cytoplasmic staining |
Nuclear plus cytoplasmic staining |
|||||||||
| Positive (4-12) | Negative (0-3) | Median±SD (range) | Positive (4-12) | Negative (0-3) | Median±SD (range) | Positive (4-12) | Negative (0-3) | Median±SD (range) | |||
| All specimens | Cancer | 82 | 44 (53.7%) | 38 (46.3%) | 4±1.2 (0-12) | 39 (47.6%) | 43 (52.4%) | 3±3.5 (0-12) | 37 (45.1%) | 45 (54.9%) | 3±3.4 (0-12) |
| Noncancer | 78 | 55 (70.5%) | 23 (29.5%) | 4±4.1 (0-12) | 23 (29.5%) | 55 (70.5%) | 0±2.1 (0-8) | 35 (44.9%) | 43 (55.1%) | 3±2.7 (0-10) | |
| P value | 0.0282a | 0.0523b | 0.0190a | 0.0011b | 0.97641a | 0.8351b | |||||
| Specimens with small tumor (≤5cm) | Cancer | 51 | 25 (49%) | 26 (51%) | 3.5±4.1 (0-12) | 21 (41.2%) | 30 (58.8%) | 3±3.5 (0-12) | 19 (37.3%) | 32 (62.7%) | 3.2±3.1 (0-12) |
| Noncancer | 48 | 33 (68.8%) | 15 31.2%) | 5.1±3.7 (0-12) | 10 (20.8%) | 38 (79.2%) | 1.2±1.7 (0-4) | 18 (37.5%) | 30 (62.5%) | 3.1±2.4 (0-8) | |
| P value | 0.0464a | 0.0428b | 0.0292a | 0.0017b | 0.9799a | 0.8876b | |||||
p values were obtained from the Chi-square test.
p values were obtained from the non-parametric test.
Consistent with our in vitro data with HCC cell lines, we found that decreased nuclear ZHX2 expression was accompanied by increased expression of Cyclin A and Cyclin E in continuous tissue sections from same patients (Figure 4C), suggesting the inhibitory effect of ZHX2 on cell proliferation in vivo. In addition, less nuclear ZHX2 expression correlated with greater Ki-67+ nuclei in HCC sections (Fig. 4D). Moreover, ZHX2 nuclear expression was significantly higher in well-differentiated tumor tissues (84.6%) than that in moderate (53.5%) or poorly differentiated (34.6%) ones (Fig. 4E, p=0.0006), suggesting a correlation of nuclear ZHX2 with HCC progression. To further test this, tissue arrays containing 106 cores were analyzed for ZHX2 expression (supplementary Table S3). Reduced nuclear ZHX2 expression was observed in liver cancer samples, including specimens from small tumors (diameter ≤5cm). In addition, decreased nuclear staining of ZHX2 significantly correlated with reduced overall survival times of patients. Consistently, levels of hepatocyte proliferation (Ki-67 as the marker) and intratumoral MVD(CD31 as the marker) were significantly higher in tissues without nuclear ZHX2 expression compared to that with higher ZHX2 expression (Fig. 4F). Taken together, our data demonstrate that reduced nuclear ZHX2 levels correlates with HCC progression and that this reduction likely occurs at an early stage of liver cancer.
Nuclear ZHX2 localization is essential for inhibition of cell proliferation
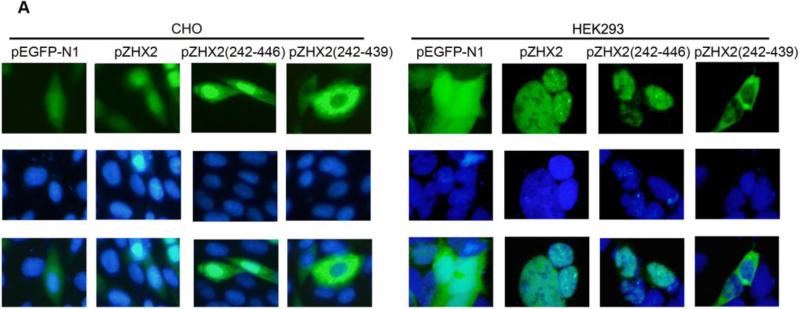
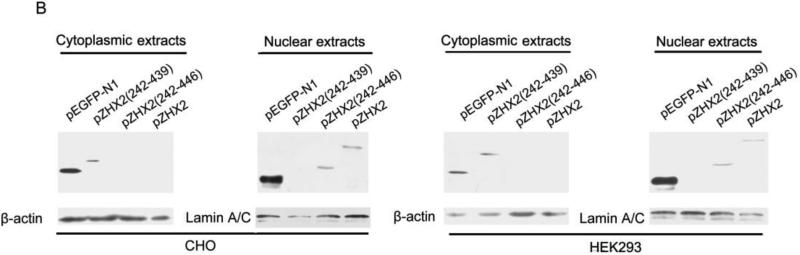
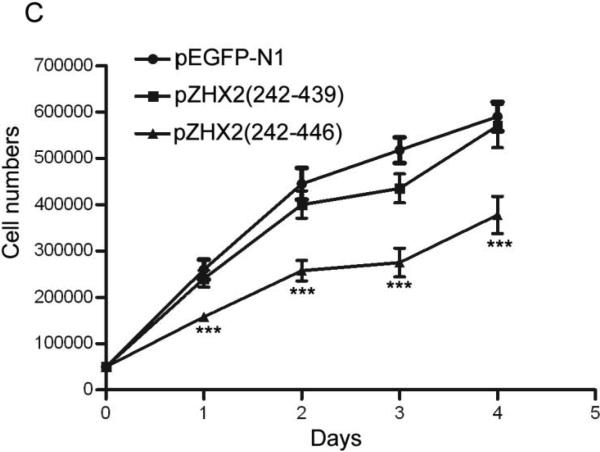
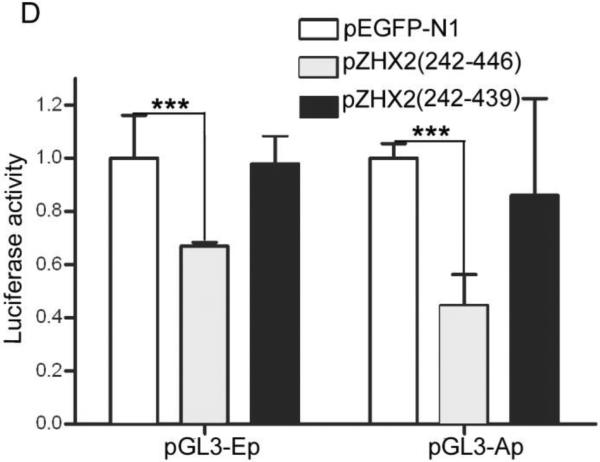
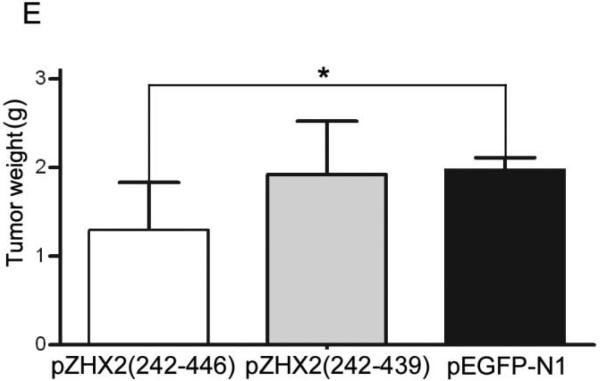
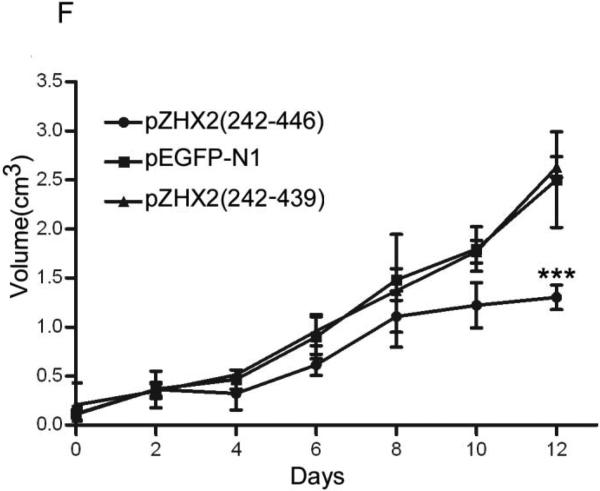
Our data showing decreased nuclear ZHX2 levels in HCC tissues (Figure 4 and Table 1) led us to hypothesize that ZHX2 must localize to the nucleus to inhibit cell growth. To directly test this possibility, we generated ZHX2-EGFP fusion proteins, including full length ZHX2 (pZHX2), pZHX2(242-446), which contains the ZHX2 dimerization domain and NLS, and pZHX2(242-439), which contains the dimerization domain without the NLS. Constructs were transiently transfected into 293 cells and CHO cells. By both fluorescence staining and western blot with cytoplasmic and nuclear extracts, we found that EGFP fusion proteins with full-length ZHX2 and ZHX2(242-446) were localized mainly to the nucleus, whereas ZHX2(242-439) and the control pEGFP-N1 were found predominantly in the cytoplasm. (Fig. 5A and 5B). In addition, pZHX2(242-446), but not ZHX2(242-439), inhibited the proliferation of HepG2 cells (Fig. 5C) and activity of pGL3-Ep and pGL3-Ap reporter constructs (Fig. 5D). Moreover, injection of ZHX2(242-446) but not ZHX2(242-439) inhibited the weight (Fig. 5E) and growth (Fig. 5F) of HepG2.215 tumors in our xenograft model. Collectively, these data indicate that nuclear ZHX2 localization is essential for its ability to inhibit cell growth in vitro and in vivo.
Figure 5. Nuclear ZHX2 localization is essential for growth inhibition in vitro and in vivo.
CHO and 293 cells were transiently transfected with pZHX2(242-446), pZHX2(242-439), full-length pZHX2 and pEGFP-N1. (A) ZHX2 localization was determined by fluorescence microscopy of EGFP. DAP (blue) was used to stain nuclei. Original magnification 200×. (B) Western blot analysis of EGFP and EGFP fusion proteins in cytoplasm and nuclei. (C) Proliferation of HepG2 for four days after transfection with pZHX2(242-439), pZHX2(242-446) and pEGFP-N1. * * *, p<0.001. (D) Activity of the pGL3-Ep and pGL3Ap in HepG2 cells co-transfected with pZHX2(242-439), pZHX2(242-446) and pEGFP-N1. Data shown is mean±SD of three independent experiments. * *, p<0.01 * * *, p<0.001. (E and F) Tumors of HepG2.2.15 cells grown in nude mice were injected with pZHX2(242-439), pZHX2(242-446) or pEGFP-N1. (E) Weights of tumors were determined after sacrifice. (mean±SD ; n=6) *, p<0.05. (F) Tumor volume calculated every other day over 12 days (mean±SD ; n=6). * * *, p<0.001.
Discussion
ZHX2 has been described as a transcriptional repressor, possibly through its interactions with NF-YA14. Based on changes in its expression, ZHX2 has been implicated in several human diseases, including podocyte disease and multiple myeloma29, 30. Conflicting data exists regarding ZHX2 in HCC19, 20. Here, we demonstrate that nuclear ZHX2 levels are reduced in HCC, including small tumors (≤5 cm in diameter), suggesting that the loss of nuclear ZHX2 is an early event in HCC progression. Moreover, we show that ZHX2 significantly inhibits the growth of HCC cell lines in vitro and in vivo. Analysis of clinical HCC samples showed a significant correlation between reduced nuclear ZHX2 and poor overall survival as well as increased levels of microvascularization and hepatocyte proliferation. Finally, studies in BALB/cJ mice indicate that ZHX2 is a negative regulator of the tumor markers AFP, GPC3 and H19 in the adult liver9, 10. Taken together, these data suggests that ZHX2 functions to suppress HCC growth and is consistent with the previous report which identified hypermethylation in the ZHX2 promoter, along with silencing of ZHX2 expression, in HCC tissues19.
One important question is how ZHX2 inhibits the growth of HCC cell lines. Our data suggests that this may occur through its transcriptional repression of Cyclin A and Cyclin E, both of which are key cell cycle regulators. Cyclin A is associated with CDK2 and responsible for the control of S phase progression and the G2-M transition31. Ectopic Cyclin E overexpression can accelerate cell cycle progression from the G1 to S phase and reinforce the loss of growth control32. A previous report showed ZHX2-mediated repression of the proliferation-related gene cdc25C14. Since Cyclin A, Cyclin E and cdc25C function in many cells and ZHX2 is ubiquitously expressed, we hypothesize that the growth inhibitory effect of ZHX2 would not be restricted to HCC but could act in multiple cell types. This possibility is supported by a recent report suggesting that the loss of ZHX2 expression confer myeloma cells a stem cell-like phenotype resulting in a resistance to chemotherapy33. Therefore, identification of novel genes regulated by ZHX2 will not only provide insight into tumorigenesis but also provide new targets for tumor therapy and diagnosis.
Using luciferase reporter genes, we show that ZHX2 controls the Cyclin A and Cyclin E promoters. This is consistent with a recent report by Gargalovic, et al., which reported increased hepatic Cyclin E expression in BALB/cJ mice, which have a mutated ZHX2 gene11. Our ChIP data indicates that ZHX2 can bind the Cyclin A and Cyclin E promoters and this is the first direct demonstration of ZHX2 binding to a target promoter. ZHX2, like other ZHX proteins, is thought to regulate at least some target genes in a NF-Y dependent manner14, 18. NF-YA binds to the CCAAT element at -52 of the Cyclin A promoter34, 35. Whether NF-Y or the CCAAT element in the Cyclin A promoter is required for ZHX2-mediated repression is not known. Also, there is no evidence of NF-YA binding to the promoters of the Cyclin E or AFP genes. While ZHX2 is considered to function at the transcriptional level14, 17, there is evidence that it also acts post-transcriptionally9,36,37. Understanding this aspect of ZHX2-mediated regulation will require further study.
Previous studies localized the NLS of ZHX2 to amino acids 317-44614. Using EGFP fusion proteins, our data indicate that ZHX2(242-446) is localized to the nucleus whereas ZHX2(242-439) remains cytoplasmic. The cytoplasmic form of ZHX2 no longer inhibited Cyclin A and Cyclin E promoters or reduced cell proliferation in HCC cell lines or tumor growth in nude mice, whereas the nuclear form ZHX2 (242-446) retained these functions. These data indicate that nuclear location of ZHX2 is required for its growth inhibitory properties. This is consistent with our analysis of clinical samples, in which we detected decreased nuclear ZHX2 expression at the level of protein but not total ZHX2 protein (Table 1) and mRNA (supplementary Fig. S1) in HCC compared to non-tumor tissues. This might account for the failure to identify ZHX2 as an HCC relevant gene by transcriptome analysis38. The loss of nuclear localization might occur during the early stages of tumorigenesis since this was observed in small liver tumors (diameter of ≤5cm). This change of ZHX2 expression pattern might be, at least partially, responsible for increased proliferation and tumor development. This is supported by the correlation of nuclear ZHX2 level with HCC progression markers, including disease grade of liver cancer, patients’ overall survival, levels of tumor microvascularization and hepatocytes proliferation, detected in HCC tissues.
In conclusion, our study indicates that ZHX2 controls cell proliferation in a manner that may involve regulation of cyclin A and cyclin E expression. These data provide new insight into the mechanisms by which ZHX2 might function as a tumor suppressor in liver cancer. Also, these studies with truncated forms of ZHX2 indicate that homeodomains 1 and 2 are sufficient for the growth inhibitory properties of ZHX2. Further studies will be required to investigate the function of other domains of ZHX2, including two zinc fingers and two additional homeodomains.
Supplementary Material
Acknowledgments
Grant Support: This work was supported in part by grants from the NSFC of China (No. 30972753), Programme for NCET-10-0524, Independent Innovation Foundation of Shandong University (IIFSDU) and Shandong Provincial Nature Science Foundation for Distinguished Young Scholars (No. JQ200907). BTS acknowledges support from the National Institutes of Health grant DK95866.
Abbreviations
- AFP
α-fetoprotein
- ChIP
chromatin immunoprecipitation
- HDs
homeodomains
- MM
multiple myeloma
- RT-PCR
reverse transcription-polymerase chain reaction
- MVD
microvascular density
- Znf
zinc-finger
- ZHX2
zinc-fingers and homeoboxes 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest
The authors declare no conflict interest.
Author Contribution:
XUETIAN YUE : Performed experiments, including Western blots, cell proliferation and colony formation assays, fluorescence staining, luciferase assays and ChIP; drafting of the manuscript. ZHENYU ZHANG: Performed in vivo studies.
XIAOHONG LIANG: Performed clinical studies, participated in data interpretation and drafting of the manuscript.
LIFEN GAO: Performed statistical analysis and supervised aspects of the study.
XIAONING ZHANG: Performed recombinant DNA work involved in vector preparation.
DI ZHAO: Performed cell cycle assays.
XIAO LIU: Performed cell culture and fluorescent staining of cells.
HONGXIN MA and MIN GUO: Performed cell proliferation assays and managed animals.
BRETT T. SPEAR and YAOQIN GONG: Provided essential experimental reagents and participated in writing of manuscript, including revisions.
CHUNHONG MA: Primarily responsible for studies described here, including study concept, experimental design and critical revision of the manuscript for intellectual content; performed statistical analysis; obtained funding for studies described here.
References
- 1.Bosch FX, Ribes J, Diaz M, et al. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Mason AC. Risk factors for the rising rates of primary liver cancer in the United States. Arch Intern Med. 2000;160:3227–30. doi: 10.1001/archinte.160.21.3227. [DOI] [PubMed] [Google Scholar]
- 3.Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–7. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Peterson ML, Ma C, Spear BT. Zhx2 and Zbtb20: novel regulators of postnatal alpha-fetoprotein repression and their potential role in gene reactivation during liver cancer. Semin Cancer Biol. 2011;21:21–7. doi: 10.1016/j.semcancer.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abelev GI. Alpha-fetoprotein in ontogenesis and its association with malignant tumors. Adv Cancer Res. 1971;14:295–358. doi: 10.1016/s0065-230x(08)60523-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang XY, Degos F, Dubois S, et al. Glypican-3 expression in hepatocellular tumors: diagnostic value for preneoplastic lesions and hepatocellular carcinomas. Hum Pathol. 2006;37:1435–41. doi: 10.1016/j.humpath.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Ariel I, Miao HQ, Ji XR, et al. Imprinted H19 oncofetal RNA is a candidate tumour marker for hepatocellular carcinoma. Mol Pathol. 1998;51:21–5. doi: 10.1136/mp.51.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abelev GI, Eraiser TL. Cellular aspects of alpha-fetoprotein reexpression in tumors. Semin Cancer Biol. 1999;9:95–107. doi: 10.1006/scbi.1998.0084. [DOI] [PubMed] [Google Scholar]
- 9.Spear BT, Jin L, Ramasamy S, et al. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci. 2006;63:2922–38. doi: 10.1007/s00018-006-6258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perincheri S, Dingle RW, Peterson ML, et al. Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci U S A. 2005;102:396–401. doi: 10.1073/pnas.0408555102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gargalovic PS, Erbilgin A, Kohannim O, et al. Quantitative trait locus mapping and identification of Zhx2 as a novel regulator of plasma lipid metabolism. Circ Cardiovasc Genet. 2010;3:60–7. doi: 10.1161/CIRCGENETICS.109.902320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthelemy I, Carramolino L, Gutierrez J, et al. zhx-1: a novel mouse homeodomain protein containing two zinc-fingers and five homeodomains. Biochem Biophys Res Commun. 1996;224:870–6. doi: 10.1006/bbrc.1996.1114. [DOI] [PubMed] [Google Scholar]
- 13.Kawata H, Yamada K, Shou Z, et al. The mouse zinc-fingers and homeoboxes (ZHX) family; ZHX2 forms a heterodimer with ZHX3. Gene. 2003;323:133–40. doi: 10.1016/j.gene.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Kawata H, Yamada K, Shou Z, et al. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem J. 2003;373:747–57. doi: 10.1042/BJ20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird LE, Ren J, Nettleship JE, et al. Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains. BMC Struct Biol. 2010;10:13. doi: 10.1186/1472-6807-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada K, Kawata H, Shou Z, et al. Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3. Biochem J. 2003;373:167–78. doi: 10.1042/BJ20021866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Luan F, Liu H, et al. ZHX2 is a repressor of alpha-fetoprotein expression in human hepatoma cell lines. J Cell Mol Med. 2008;12:2772–80. doi: 10.1111/j.1582-4934.2008.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada K, Ogata-Kawata H, Matsuura K, et al. ZHX2 and ZHX3 repress cancer markers in normal hepatocytes. Front Biosci. 2009;14:3724–32. doi: 10.2741/3483. [DOI] [PubMed] [Google Scholar]
- 19.Lv Z, Zhang M, Bi J, et al. Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular carcinoma. Am J Clin Pathol. 2006;125:740–6. doi: 10.1309/09B4-52V7-R76K-7D6K. [DOI] [PubMed] [Google Scholar]
- 20.Hu S, Zhang M, Lv Z, et al. Expression of zinc-fingers and homeoboxes 2 in hepatocellular carcinogenesis: a tissue microarray and clinicopathological analysis. Neoplasma. 2007;54:207–11. [PubMed] [Google Scholar]
- 21.Henglein B, Chenivesse X, Wang J, et al. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci U S A. 1994;91:5490–4. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A. 1995;92:12146–50. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Ishak KG. Pathologic features of chronic hepatitis. A review and update. Am J Clin Pathol. 2000;113:40–55. doi: 10.1309/42D6-W7PL-FX0A-LBXF. [DOI] [PubMed] [Google Scholar]
- 25.Han CP, Kok LF, Wang PH, et al. Scoring of p16(INK4a) immunohistochemistry based on independent nuclear staining alone can sufficiently distinguish between endocervical and endometrial adenocarcinomas in a tissue microarray study. Mod Pathol. 2009;22:797–806. doi: 10.1038/modpathol.2009.31. [DOI] [PubMed] [Google Scholar]
- 26.Kamoi S, AlJuboury MI, et al. Immunohistochemical staining in the distinction between primary endometrial and endocervical adenocarcinomas: another viewpoint. Int J Gynecol Pathol. 2002;21:217–23. doi: 10.1097/00004347-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Han CP, Lee MY, Tzeng SL, et al. Nuclear Receptor Interaction Protein (NRIP) expression assay using human tissue microarray and immunohistochemistry technology confirming nuclear localization. J Exp Clin Cancer Res. 2008;27:25. doi: 10.1186/1756-9966-27-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidner N. Current pathologic methods for measuring intratumoral microvessel density within breast carcinoma and other solid tumors. Breast Cancer Res Treat. 1995;36:169–80. doi: 10.1007/BF00666038. [DOI] [PubMed] [Google Scholar]
- 29.Armellini A, Sarasquete ME, Garcia-Sanz R, et al. Low expression of ZHX2, but not RCBTB2 or RAN, is associated with poor outcome in multiple myeloma. Br J Haematol. 2008;141:212–5. doi: 10.1111/j.1365-2141.2007.06956.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Clement LC, Kanwar YS, et al. ZHX proteins regulate podocyte gene expression during the development of nephrotic syndrome. J Biol Chem. 2006;281:39681–92. doi: 10.1074/jbc.M606664200. [DOI] [PubMed] [Google Scholar]
- 31.Rosenblatt J, Gu Y, Morgan DO. Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proc Natl Acad Sci U S A. 1992;89:2824–8. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoblich JA, Sauer K, Jones L, et al. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–20. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 33.Legartova S, Harnicarova-Horakova A, Bartova E, et al. Expression of RAN, ZHX-2, and CHC1L genes in multiple myeloma patients and in myeloma cell lines treated with HDAC and Dnmts inhibitors. Neoplasma. 2010;57:482–7. doi: 10.4149/neo_2010_05_482. [DOI] [PubMed] [Google Scholar]
- 34.Kramer A, Carstens CP, Wasserman WW, et al. CBP/cycA, a CCAAT-binding protein necessary for adhesion-dependent cyclin A transcription, consists of NF-Y and a novel Mr 115,000 subunit. Cancer Res. 1997;57:5117–21. [PubMed] [Google Scholar]
- 35.Jung YJ, Lee KH, Choi DW, et al. Reciprocal expressions of cyclin E and cyclin D1 in hepatocellular carcinoma. Cancer Lett. 2001;168:57–63. doi: 10.1016/s0304-3835(01)00403-7. [DOI] [PubMed] [Google Scholar]
- 36.Morford LA, Davis C, Jin L, et al. The oncofetal gene glypican 3 is regulated in the postnatal liver by zinc fingers and homeoboxes 2 and in the regenerating liver by alpha-fetoprotein regulator 2. Hepatology. 2007;46:1541–7. doi: 10.1002/hep.21825. [DOI] [PubMed] [Google Scholar]
- 37.Vacher J, Camper SA, Krumlauf R, et al. raf regulates the postnatal repression of the mouse alpha-fetoprotein gene at the posttranscriptional level. Mol Cell Biol. 1992;12:856–64. doi: 10.1128/mcb.12.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.