Abstract
Insulin resistance is a major pathophysiologic abnormality that characterizes metabolic syndrome and type 2 diabetes. A well characterized ethanolic extract of Artemisia dracunculus L., termed PMI 5011, has been shown to improve insulin action in vitro and in vivo, but the cellular mechanisms remain elusive. Using differential proteomics, we have studied mechanisms by which PMI 5011 enhances insulin action in primary human skeletal muscle culture obtained by biopsy from obese, insulin-resistant individuals. Using iTRAQ™ labeling and LC-MS/MS, we have identified over 200 differentially regulated proteins due to treatment with PMI 5011 and insulin stimulation. Bioinformatics analyses determined that several metabolic pathways related to glycolysis, glucose transport and cell signaling were highly represented and differentially regulated in the presence of PMI 5011 indicating that this extract affects several pathways modulating carbohydrate metabolism, including translocation of GLUT4 to the plasma membrane. These findings provide a molecular mechanism by which a botanical extract improves insulin stimulated glucose uptake, transport and metabolism at the cellular level resulting in enhanced whole body insulin sensitivity.
Keywords: botanicals, insulin resistance, iTRAQ, proteomics, skeletal muscle, glucose transporter 4
Introduction
Insulin resistance is a major pathophysiologic parameter that defines metabolic syndrome and type 2 diabetes. Insulin resistance is typically observed 5–10 years prior to the onset and diagnosis of type 2 diabetes and is accompanied by a compensatory increase in insulin secretion [1]. Insulin resistance has been well described to develop with obesity, resulting from increased food intake, sedentary life style and genetic predisposition [1] and is associated with inflammation, dyslipidemia, carbohydrate dysregulation and cardiovascular diseases [2, 3]. Insulin sensitivity can be improved by changes in diet, exercise and use of pharmacological drugs [4]. However, the success rate of maintaining life style changes over prolonged periods of time is low and use of pharmacologic drugs is often accompanied with significant side effects [5]. Thus, nutritional supplementation with naturally occurring products (i.e. botanicals) is a desirable alternative to successfully improve and maintain insulin sensitivity.
Botanical extracts have been widely used for centuries in many cultures in efforts to prevent and treat diseases [6]. Metformin, the most commonly used agent for treatment of type 2 diabetes today, has its origins from a plant source [7]. Due to the complex composition of botanicals, very little is known regarding their exact mode of action. Differential global proteomic technologies provide a broad signature of changes in protein levels which allow identification of key pathways and mechanisms responsible for complex biological effects [8, 9]. Despite advancements in mass spectrometry based proteomic techniques to understand biological processes at the molecular level, only a limited number of studies have used proteomics to study mechanisms by which botanicals induce biological effects [10–13].
Extracts of Artemisia species are widely marketed in over-the-counter dietary supplements. Extracts of Artemisia have also been shown to lower blood glucose levels in rats, and rabbits [14, 15]. We have recently shown that a well characterized ethanolic extract of Artemisia dracunculus L., termed PMI 5011, lowers blood glucose and insulin levels in murine models and improves insulin receptor signaling (e.g. Akt phosphorylation and Phosphatidylinositol 3-kinases (PI3K) activity) [16, 17]. Our studies have also shown that in primary human skeletal muscle culture (HSMC), PMI 5011 improved insulin receptor signaling (Akt phosphorylation and PI3K activity) and increased glucose uptake and glycogen synthesis [18].
Human skeletal muscle culture can be generated from biopsied skeletal muscle tissue from human subjects and retain the metabolic and biochemical properties of skeletal muscle cells noted in the in vivo state [19–23]. Thus, an insulin resistant individual will yield muscle culture that will have diminished insulin signaling and changes in carbohydrate metabolism. Similarly, muscle culture from an insulin sensitive individual will have normal insulin signaling and carbohydrate metabolism. In fact, it has been reported that cultured HSMC from non-diabetic and type 2 diabetic subjects respond to insulin stimulation in a manner consistent with in vivo changes in glucose utilization [19–21, 24, 25]. Thus, HSMC is a good model system to evaluate beneficial effects of botanical extracts under various experimental conditions and to determine molecular mechanisms responsible for improvement in insulin action.
To investigate cellular pathways affected by PMI 5011, we have used two dimensional liquid chromatography-tandem mass spectrometry (2D LC-MS/MS) in conjunction with isobaric tagging for relative and absolute quantification (iTRAQ™ ) of peptides to measure changes in protein expression levels in primary HSMC from obese insulin resistant subjects due to treatment with PMI 5011. We have further utilized immunohistochemistry and western blot analysis to validate results from proteomics experiments and show that PMI 5011 improves actin filament distribution and enhances translocation of glucose transporter 4 (GLUT4) to the plasma membrane resulting in enhanced glucose uptake, transport and metabolism.
Materials and Methods
Extract Preparation
Detailed information about the sourcing, growing conditions, quality control, stability, biochemical characterization and specific preparation of the Artemisia dracunculus L. extract (PMI 5011) tested in this study has been extensively reported [16, 26–29]. Briefly, the Artemisia dracunculus L. extract was produced from plants grown hydroponically in greenhouses maintained under uniform and strictly controlled conditions, thereby standardizing the plants for their phytochemical content. Major compounds identified in the extract have included davidigenin, isomer of demethoxydihydrochalcone and sakuranetin [6].
Primary Human Skeletal Muscle Culture (HSMC)
Primary HSMC were prepared as described in detail previously [11, 18]. Briefly, freshly removed muscle tissue from biopsies of vastus lateralis muscle from five obese diabetic patients was placed in Ham’s F-10 media (HyClone Laboratories, Logan, UT) at 4 °C and dissected, minced, washed, dissociated, centrifuged at 600 x g for 4 min at 37 °C and placed in human skeletal growth medium (SkGM Bullet Kit, Cambrex). Cells were incubated at 37 °C with 95% air and 5% CO2. Media was changed every 2 – 3 days. Myoblasts were sub-cultured and grown to 80 – 90 % confluence. Cells were then differentiated into fused myotubes for seven days by switching to culture media with 2% horse serum. After starvation, cells were treated with 10 μg/mL of PMI 5011 for 16 h. To evaluate effects of PMI 5011 on insulin signaling, cultures were treated with 100 nM insulin for 20 minutes prior to protein extraction. Thus, each experimental set included four HSMC samples: baseline control, PMI 5011 treated, insulin stimulated control and insulin stimulated and PMI 5011 treated. All primary cultured cells used in this study were within five passages.
Sample preparation
Proteins from all samples were extracted by adding 1 mL of lysis buffer (5M Urea, 2M Thiourea, 2% CHAPS, 2% SB3-10, 0.2% Bio-Lyte (pH 3-10), 2% n-dodecyl-b-d-maltoside, 40 mM Tris, 5 mM PMSF, 2 mM TBP and 150U Benzonase) followed by sonication and addition of 50 mM dithiothreitol (DTT) as described previously [11, 30, 31]. The resulting sample mixture was centrifuged for 30 min at 20,800 x g, and the supernatant was acetone precipitated and resolubilized in 0.5 M triethylammonium bicarbonate buffer (TEAB; pH 8.5) and 0.8 M urea. The protein concentration was determined using Bradford Protein Assay (Bio-Rad, Hercules, CA).
iTRAQ labeling
Fifty micrograms of protein from each sample was digested and labeled with iTRAQ™ labeling reagents as per manufacturer’s instructions (AB Sciex, Foster City, CA). Briefly, proteins (2 μg/μL in TEAB) were denatured using 2% SDS and reduced using 50 mM Tris (2-carboxyethyl) phosphine (TCEP). Reduced cysteine residues were blocked using 200 mM methyl methane thiosulfonate (MMTS). Samples were digested using trypsin (Promega, Madison, WI) at an enzyme to substrate ratio of 1:10. All reagents were provided in the iTRAQ™ kit, except for trypsin. Peptides derived from baseline control, PMI 5011 treated, insulin stimulated control and insulin stimulated and PMI 5011 treated samples were labeled with iTRAQ™ tags 114, 115,116 and 117, respectively. After labeling, samples were combined and concentrated under vacuum prior to strong cation exchange chromatography (SCX).
Strong cation exchange chromatography
The iTRAQ™ labeled peptide mixture was fractionated using a PolySulfoethyl A (2.1 mm × 50 mm, 3 μm, 200Å) column as described previously [30]. Peptides were eluted using a linear gradient of 0–40% solvent B (solvent A: 10 mM ammonium formate, 20% acetonitrile, pH 3; solvent B: 600 mM ammonium formate, 20% acetonitrile, pH 3) over 30 min and 40–100% solvent B over 15 min at 0.2 mL/min. Absorbance spectra were recorded at 280 nm. 48 fractions at 1 min intervals were collected in a 96 well plate, and the volume of all fractions was reduced to 5 – 10μL under vacuum. Twenty fractions eluted between 15–34 min were diluted in 0.1% formic acid (2–30 fold) based on SCX chromatogram intensities prior to LC-MS/MS analysis. SCX fractions were analyzed by LC-MS/MS at least three times.
Reverse phase liquid chromatography-tandem mass spectrometry (RPLC-MS/MS)
Reverse phase chromatographic separations of each fraction were carried out using a nanoflow ultraperformance LC directly interfaced to nano-electrospray quadrupole time-of-flight MS (nanoAcquity UPLC-Synapt HDMS, Waters Corp., Milford, MA) [30]. Peptides were loaded on a Symmetry C18 trap column (180 μm × 20mm, 5 μm; Waters Corp.) and washed with 1% solvent B (solvent A: 0.1% formic acid; solvent B: 0.1% formic acid in acetonitrile) for 15 minutes at 5 μL/min. Peptides were separated on a BEH C18 analytical column (1.7 μm, 75 μm ID × 100 mm, Waters Corp.) using a linear gradient of 10–30% Solvent B over 55 minutes. Eluting peptides were introduced into the MS via nano-electrospray ionization source using a fused silica PicoTip emitter (10 μm tip diameter; New Objective, Woburn, MA). The mass spectrometer was operated in positive ion mode, source temperature 150 °C, capillary voltage 3.5 kV, cone voltage 35 V and extraction cone voltage of 2 V. Data were acquired using MassLynx™ 4.1 software (Waters Corp.) in an automatic data dependent acquisition mode. MS-TOF scans were acquired from m/z 400 to 1600, and up to three precursor ions were selected for subsequent MS/MS scans from m/z 50 to 1600 using rolling collision energies to promote fragmentation. Glu1-fibrinogen peptide (GFP) served as lock mass (m/z 785.8426) in the reference sprayer, and its signal was acquired every 30 seconds.
Mass spectrometry data analysis
Mass spectrometry data was processed using ProteinLynx Global Server (PLGS 2.2.5, Waters Corp.) using the following settings: background subtract type, adaptive; smoothing type, Savitzky-Golay; smoothing iterations, 2; smoothing window, 3 channels; deisotoping type, slow with 30 iterations and 3% threshold and centroid top 80%. Lock spray calibration was performed using GFP with 0.1 Da lock mass tolerance. Tandem mass spectra analyzed using PLGS were searched against a UniProt/SwissProt database limited to human proteins (UniProt Release 15.8 with 509,019 entries) [11, 31, 32]. The database search settings included peptide tolerance 100 ppm, fragment-ion tolerance 0.1 Da, estimated calibration error 20 ppm and one missed tryptic cleavage. iTRAQ-labeled lysines and N-termini and MMTS-modified cysteines were set as fixed modifications, and iTRAQ™ labeled tyrosine and methionine oxidation were set as variable modifications. The automodification query was selected to identify peptides with further post-translational modifications in PLGS. Area under the peaks for iTRAQ™ signature peaks at 114.1, 115.1, 116.1 and 117.1 Da was calculated using PLGS Expression 2.0. The top ranking hits with PLGS score >10 were included in the downstream analysis.
Protein identification and iTRAQ™ quantification results for each LC-MS/MS experiment were exported, merged and processed in Microsoft Excel to yield one comprehensive dataset. Bioinformatic redundancy in assignment of identical peptide sequences to different accession numbers was resolved manually following principles of parsimony. For further quantification and statistical analysis, proteins identified with one unique peptide were considered only if that peptide was independently identified at least three times. Relative quantification was performed based on ratios of the areas under the peaks at 114.1, 115.1, 116.1 and 117.1 Da. Peptides containing iTRAQ™ label peaks with area less than 10 were excluded from quantification. Isotopic impurities of iTRAQ™ reporter ions were corrected based on information accompanied with the iTRAQ™ kit. Peak areas of iTRAQ™ reporter ions for peptide sequences unique to each protein were averaged and compared between treatments to determine relative quantification levels of proteins. Peptides shared among two or more proteins were excluded from quantification. The student t-test was applied to determine whether the protein was significantly differentially expressed (Supplementary Table 1).
Pathway analysis
A list of all identified proteins, fold changes and p-values was uploaded to Ingenuity Pathway Analysis (IPA) software to map proteins into biological networks and to retrieve functions and pathways. Only differentially regulated proteins (± 1.2 fold, p-values < 0.05) were considered for network and pathway analyses. The corresponding genes were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base (IPKB). Networks of these focus genes were then algorithmically generated based on their connectivity. Networks are scored based on the number of network eligible molecules within the specific network, size of the network, total number of network eligible molecules in the dataset and total number of molecules in the IPKB that could potentially be included in networks. The significance values for network and pathway analyses were calculated using right-tailed Fisher’s Exact Test.
Immunohistochemistry, actin staining and image analysis
Myoblasts sub-cultured on coverslips were differentiated and treated with insulin in the presence and absence of PMI 5011 as described above. Cells were washed with phosphate buffered saline (PBS, pH 7.4) and fixed in 3.7% formaldehyde at room temperature. Cells were permeabilized with 0.5% Triton X-100 in PBS, blocked with 5% BSA, incubated with anti-GLUT4 antibody (MAB 1262, R&D Systems, 1:250 diluted in 1% BSA) for 1h and rinsed with PBS. Secondary antibody (Alexa-Fluor 594 anti-mouse antibody, A21203, Invitrogen) was applied for 30 min. Cells were washed with PBS and incubated with 200 μL of 3.3 μM Alexa-Fluor 488 phalloidin (A12379, Invitrogen) in PBS for 20 min to stain filamentous actin. Cells were rinsed and mounted on microscope slides. Images were acquired using a FluoView IX81-FV1000 Confocal Laser Scanning Microscope (Olympus America). Images were generated by 10 to 12 optical z-sections with 1.5 μm step-size and analyzed using ImageJ software.
Sub-cellular fractionation and western blot analysis
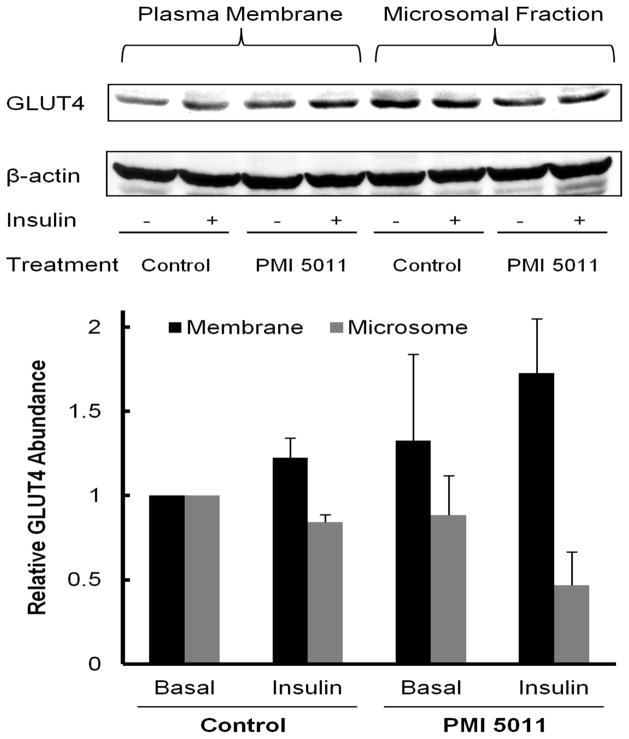
Cells differentiated into fused myotubes for seven days were stimulated with insulin in the presence and absence of PMI 5011 as described above. Myotubes were fractionated as described by Mitsumoto and Klip [33] with minor modifications. Briefly, cells were treated with lysis buffer (250 mM sucrose, 5 mM NaN3, 2 mM EGTA, 200 μM phenylmethysuldony fluoride (PMSF),1 mM pepstatin A, 1 μM aprotinin, and 20 mM HEPES (pH 7.4)) and homogenized for 20 strokes using a PRO200 homogenizer (PRO Scientific, Oxford, CT). The homogenates were centrifuged at 760 x g at 4 °C for 5 min to remove nuclei and unbroken cells. Supernatants were further centrifuged at 4 °C using a Beckman ultracentrifuge TL-100 (Beckman, Fullerton, CA) at 31,000 x g (1h) and 190,000 x g (1h) to pellet crude plasma membrane and microsomes, respectively. Sub-cellular fractions were assayed for expression of GLUT4 using western blot analysis [34]. Briefly, 15 μg of protein from each sample was separated using 10% SDS-PAGE gels and transferred to nitrocellulose membrane. After blocking membranes with 1% BSA, membranes were incubated with anti-GLUT4 monoclonal antibody (1:200 diluted in PBS; R & D systems, Minneapolis, MN) at 4 °C overnight. Membranes were washed and visualized with horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence. Specific bands were quantified by scanning densitometry. Data were normalized to β-actin levels. Protein expression levels in membrane and microsomal fractions were further normalized to levels obtained in their respective baseline control culture (basal). The error bars (Figure 3) are the standard deviation of the mean from results obtained from culture from different obese insulin resistant individuals.
Figure 3.
Western Blot Analysis of GLUT4 in plasma membrane and in microsomal fraction. 15 μg of total protein from both fractions was separated by SDS-PAGE and subjected to western blot analysis. The enhanced GLUT4 expression in membrane compared to microsome after insulin stimulation in the presence of PMI 5011 is consistent with results from 2D-LC-MS/MS.
Results
LC-MS/MS analysis and identification of proteins
Proteins extracted from four treatment groups of HSMC (baseline control, PMI 5011 treated, insulin stimulated control and PMI 5011 treated and insulin stimulated) were subjected to proteolysis and iTRAQ™ labeling followed by 2D-LC-MS/MS for quantitative proteomic analysis. These data allow comparison of protein expression levels at basal and insulin stimulated states with and without PMI 5011 treatment. Based on this analysis, 446 unique proteins were identified and quantified as described in detail in Materials and Methods. 23 of these proteins were identified as viral proteins. Protein expression levels were evaluated using pair-wise comparisons defined as follows: 1) baseline control vs. insulin stimulated control; 2) baseline control vs. PMI 5011 treated; 3) PMI 5011 treated vs. PMI 5011 treated and insulin stimulated; 4) insulin stimulated baseline control vs. PMI 5011 treated and insulin stimulated; and 5) baseline control vs. PMI 5011 treated and insulin stimulated. A summary of the number of differentially regulated proteins in each comparison is provided in Supplementary Table 2. Differentially regulated proteins (± 1.2 fold and p < 0.05) were selected for further analysis. These cut-offs were selected based on our preliminary studies investigating the reproducibility of quantification using iTRAQ™ (unpublished data) and other literature reports [35]. 250 proteins identified in this study were differentially regulated in at least one comparison listed above, and only two proteins were identified to be differentially regulated in all comparisons. Protein accession numbers, number of unique peptides matched to each protein, their molecular weights, percent coverage, score from PLGS software and ratio of iTRAQ™ labels for quantification are listed in Supplementary Table 1.
The biggest change in protein expression levels was observed with insulin stimulation after treatment with PMI 5011, resulting in up-regulation of 210 proteins (Supplementary Table 2). This is highly consistent with the clinical findings for PMI 5011 demonstrating best efficacy for the botanical in the presence of insulin [17]. Interestingly, 41 out of 44 proteins identified to be differentially regulated due to PMI 5011 treatment alone were down-regulated. In contrast, little change in protein expression levels was observed with insulin stimulation in baseline control cells. These results are consistent with the insulin resistant state of the individuals from which the cultures were obtained as it is well known that the HSMC retain the intrinsic properties of skeletal muscle cells noted in the in vivo state [19–23]. The cell cultures used in this study were obtained from biopsied skeletal muscle tissue from obese diabetic individuals and demonstrated to be insulin resistant by gold-standard techniques, i.e. hyperinsulinemic-euglycemic clamps [36].
Pathway analysis
A list containing all 423 identified proteins, fold change and p-values was uploaded to IPA and mapped for functional analysis. IPA software was used to assign cellular location and molecular and cellular functional classes to identified proteins (Supplementary Figure 1). These assignments are based upon underlying biological evidence from the curated IPKB. These results demonstrate that our analysis is not biased towards any one sub-cellular location or functional class. Total numbers of network and pathway analysis eligible differentially regulated proteins in each comparison is listed in Supplementary Table 2. Using these proteins, IPA identified networks and canonical pathways that are most significant to this data set. Several inter-related networks and metabolic pathways related to lipid metabolism, cell signaling, protein synthesis, glycolysis and glucose transport were identified with high statistical significance (Figures 1 and 2 and Supplementary Figures 2 and 3). These metabolic pathways identified based on changes in protein expression levels are consistent with pathways found to be modulated due to PMI 5011 treatment in murine skeletal muscle using gene expression studies [17]. The highest scoring network included NFkB, Akt, insulin, protein kinase C and TNF receptor 17 (Figure 1). Only 6 proteins involved in this network were differentially regulated after insulin stimulation of baseline control samples (Figure 1A), 23 of these proteins were down-regulated due to PMI 5011 treatment (Figure 1B), and 23 proteins were up-regulated after insulin stimulation in the presence of PMI 5011 compared to PMI 5011 alone (Figure 1C).
Figure 1.
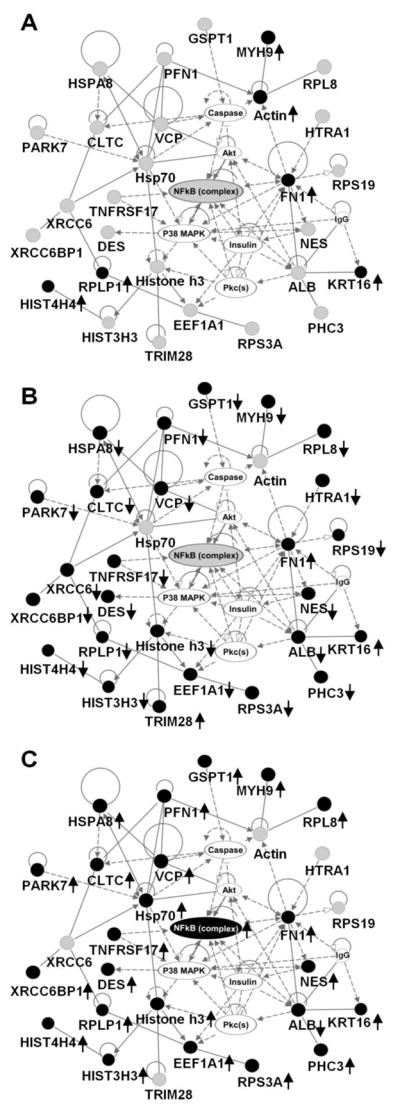
Top connectivity network with p-value 10-65 showing changes in expression of proteins involved in NFκB network in basal and insulin stimulated states with and without PMI 5011 treatment. (A) baseline control vs. insulin stimulated control, (B) baseline control vs. PMI 5011 treated, (C) PMI 5011 treated vs. PMI 5011 treated and insulin stimulated. Proteins labeled with filled circles were identified to be differentially regulated in each comparison. Proteins labeled with gray circles were identified but are not differentially regulated. Proteins labeled with plain circles are imported from the IPA knowledge base. A line indicates interactions, and the dotted lines indicate an inferred or indirect interaction. The abbreviations are: CLTC: clathrin heavy chain, DES: desmin, EEF1A1: eukaryotic translation elongation factor 1 alpha 1, FN1: fibronectin 1, GSPT1: G1 to S phase transition 1, HIST3H3: histone cluster 3 H3, HIST4H4: histone cluster 4 H4, HSPA8: heat shock 70kDa protein 8, HTRA1: HtrA serine peptidase 1, KRT16: keratin, MYH9: myosin heavy chain 9, NES: nestin, PARK7: Protein DJ-1, PFN1: profilin 1, RPL8: ribosomal protein L8, RPLP1: ribosomal protein (large, P1), RPS19: ribosomal protein S19, RPS3A: ribosomal protein S3A, TNFRSF17: tumor necrosis factor receptor 17, TRIM28: cDNA FLJ94025 highly similar to Homo sapiens tripartite motif containing 28, VCP: valosin-containing protein, XRCC6BP1: Mitochondrial inner membrane protease ATP23 homolog.
Figure 2.
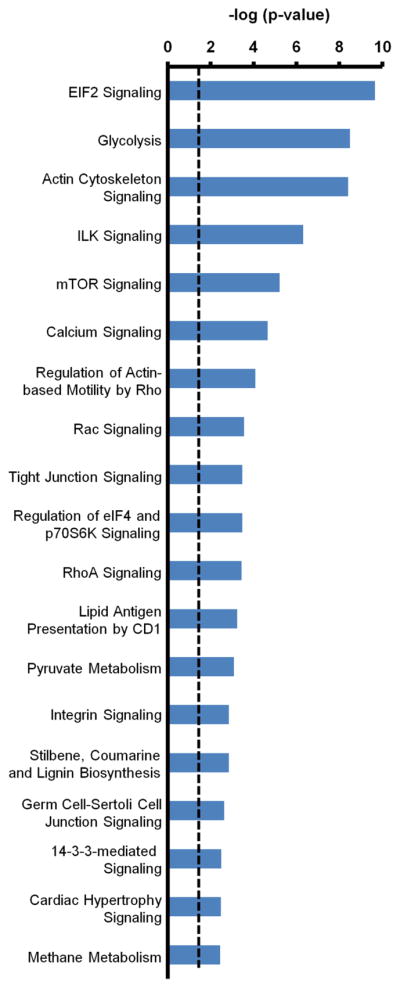
Canonical pathways identified using IPA for proteins differentially regulated due to insulin stimulation after PMI 5011 treatment. The levels of these proteins were unchanged due to PMI 5011 treatment alone. The dotted line represents the threshold of significance (p = 0.05). The length of the bars is a measure of significance (-log of p-value) that identified proteins are part of the specified canonical pathway.
Analysis of proteins differentially regulated due to insulin stimulation in the presence of PMI 5011 but not changing due to PMI 5011 alone showed that various pathways related to protein synthesis were highly represented (Figure 2). Insulin initiates protein synthesis by activating components of the translational system which is mediated by mTOR, eIF2 and eIF4 [37, 38]. After treatment with PMI 5011 and insulin stimulation, levels of several proteins involved in these pathways were elevated compared to all of the other three treatments. In addition, protein synthesis was the most significant biological function identified using pathway analysis (Supplementary Figure 4). Thus, these data clearly demonstrate enhanced insulin-induced protein synthesis due to treatment with PMI 5011.
Several canonical pathways related to glycolysis, pyruvate metabolism, interleukin signaling, actin cytoskeleton signaling and caveolar-mediated endocytosis signaling were also identified using IPA (Supplementary Figure 2). In skeletal muscle, the primary action of insulin is to stimulate glucose uptake and metabolism [39]. Our analysis identified 14 proteins involved in the glycolysis pathway. Upon insulin stimulation of baseline control cells, three of these proteins were moderately up-regulated (Table 1). This is in contrast to insulin stimulation in the presence of PMI 5011 which showed up-regulation of 11 identified proteins (Table 1). Similarly, 13 of the 18 proteins identified in the actin cytoskeleton signaling pathway were up-regulated after insulin stimulation in the presence of PMI 5011 (Table 2).
Table 1.
Proteins Identified in the Glycolysis Pathway
| Protein Accession Number | E.C. Number | Fold Change
|
||||
|---|---|---|---|---|---|---|
| Baseline Control vs. Insulin Stimulated Control | Baseline Control vs. PMI 5011 Treated | PMI 5011 Treated vs. PMI 5011 Treated and Insulin Stimulated | Baseline Control vs. PMI5011 Treated and Insulin Stimulated | |||
| P00352 | Aldehyde dehydrogenase (NAD+) | 1.2.1.3 | 1.08 | −1.18 | 1.36a | 1.15 |
| P04075 | Fructose bisphosphate aldolase A | 4.1.2.13 | 1.13 | −1.16 | 1.37a | 1.18 |
| P06733 | Alpha enolase (phosphopyruvate hydratase) | 4.2.1.11 | 1.12 | −1.03 | 1.24a | 1.2 |
| P04406 | Glyceraldehyde 3 phosphate dehydrogenase | 1.2.1.12 | 1.17 | −1.11 | 1.37a | 1.23a |
| B4DE36 | Glucose 6 phosphate isomerase | 5.3.1.9 | 1.13 | −1.16 | 1.32a | 1.14 |
| P00338 | L lactate dehydrogenase A chain | 1.1.1.27 | 1.26a | −1.00 | 1.32a | 1.32a |
| P07195 | L lactate dehydrogenase B chain | 1.1.1.27 | 1.14 | −1.11 | 1.33a | 1.2 |
| P18669 | 2,3 bis-phosphoglycerate mutase | 5.4.2.4 | 1.26a | 1.07 | 1.22a | 1.3a |
| 3.1.3.13 | ||||||
| 5.4.2.1 | ||||||
| A8K4W6 | Phosphoglycerate kinase | 2.7.2.3 | 1.13 | −1.09 | 1.37a | 1.25a |
| P14618 | Pyruvate kinase | 2.7.1.40 | 1.18 | −1.03 | 1.29a | 1.24a |
| P60174 | Triose-phosphate isomerase | 5.3.1.1 | 1.12 | −1.05 | 1.34a | 1.28a |
| Q658X4 | Acetyl-CoA synthetase | 6.2.1.1 | 1.27a | 1.04 | 1.27 | 1.31 |
| P11766 | Alcohol dehydrogenase | 1.1.1.1 | 1.07 | 1.11 | 1.17 | 1.04 |
| B4DDQ8 | Phosphoglucomutase | 5.4.2.2 | −1.7 | 1.28 | −1.24 | 1.03 |
Significantly differentially regulated proteins with p-value < 0.05.
Table 2.
Proteins Identified in the Actin Cytoskeleton Signaling Pathway
| Protein Accession Number | Protein name | Fold change
|
|||
|---|---|---|---|---|---|
| Baseline Control vs. Insulin Stimulated Control | Baseline Control vs. PMI 5011 Treated | PMI 5011 Treated vs. PMI 5011 Treated and Insulin Stimulated | Baseline Control vs. PMI 5011 Treated and Insulin Stimulated | ||
| Q6ZS32 | Putative uncharacterized protein / FYVE finger containing phosphoinositide kinase | 1.67 | 1.61 | 1.48 | 2.37 |
| Q5CZ99 | fibronectin 1 | 1.28a | 1.26a | 1.28a | 1.62a |
| Q6ZNL4 | FLJ00279 protein fragment/non-muscle myosin heavy chain 9 | 1.08 | −1.21a | 1.42a | 1.17 |
| P07737 | Profilin 1 | −1.02 | −1.36a | 1.51a | 1.11 |
| P05556 | Integrin beta 1 | −1.10 | −1.05 | 1.20 | 1.14 |
| Q53HL1 | Myosin regulatory light chain MRCL3 variant | −1.18 | −1.46 | 1.49 | 1.02 |
| P12814 | Alpha actinin 1 | 1.10 | −1.04 | 1.28a | 1.22a |
| B4DM63 | cDNA FLJ51245 highly similar to Actin related protein 2 3 complex subunit 3 | 1.10 | −1.19 | 1.47a | 1.23a |
| P23528 | Cofilin-1 | 1.15 | −1.08 | 1.43a | 1.32a |
| P06396 | Gelsolin | 1.16 | −1.14 | 1.42a | 1.25a |
| Q5FWG8 | IQGAP1 protein Fragment | −1.17 | −1.46 | 1.39a | −1.05 |
| P26038 | Moesin | 1.12 | −1.16 | 1.39a | 1.21a |
| Q7Z7R0 | Smooth muscle myosin heavy chain | 1.23a | −1.10 | 1.28a | 1.16 |
| P60660 | Myosin light polypeptide 6 | −1.06 | 1.13 | 1.07 | 1.21a |
| Q9BVT0 | ARHA protein aplysia ras-related homolog 12 | 1.24a | −1.01 | 1.29a | 1.28a |
| A2VCK8 | Thymosin beta 4 X linked | 1.12 | −1.13 | 1.39a | 1.23a |
| B4DTM7 | cDNA FLJ53006 highly similar to Vinculin | 1.49a | −2.30 | 3.16a | 1.37 |
| B4DW52 | cDNA FLJ55253 highly similar to Actin cytoplasmic 1 | 1.21a | −1.15 | 1.25 | 1.08 |
Significantly differentially regulated proteins with p-value < 0.05.
GLUT4 immunohistochemistry and translocation
Several of the canonical pathways including glycolysis, actin cytoskeleton signaling and caveolar-mediated endocytosis are interrelated through glucose transporter GLUT4 which is synthesized in the endoplasmic reticulum and transported via intracellular vesicles that form caveolae after fusing with the plasma membrane where it facilitates insulin-stimulated glucose uptake. To evaluate a functional marker that supports our proteomics data, specific GLUT4 translocation studies were performed.
Immunostaining and subsequent fluorescence imaging of myotubes revealed weak GLUT4 and actin filament staining in baseline control cells (Supplementary Figure 5) which increased slightly in the vicinity of the nucleus upon insulin stimulation. The strongest response was observed when myotubes were stimulated with insulin in the presence of PMI 5011, showing increased GLUT4 staining throughout the cytoplasm and increased actin filament distribution (Supplementary Figure 5). These results are consistent with proteomics data showing increased expression levels of proteins (filamin, cofilin, Arp2/3) involved in actin polymerization (Table 2).
GLUT4 translocation studies using western blot analysis are presented in Figure 3. As shown, we evaluated not only total GLUT4 content, but the relative change in GLUT4 expression in microsomes compared to the plasma membrane of HSMC with and without insulin stimulation and PMI 5011 treatment. Insulin stimulation of the baseline control culture resulted in a significant increase in membrane bound GLUT4 content and a corresponding decrease in the microsomal content. In contrast, in PMI 5011 treated culture there was an increase in GLUT4 levels in the plasma membrane before insulin stimulation and a corresponding decrease in the microsomal content as compared to the baseline control culture. Insulin stimulation of the PMI 5011 treated culture resulted in a further increase in GLUT4 expression in the plasma membrane and a decrease in GLUT4 levels in microsomal fraction. These results clearly show that PMI 5011 enhances translocation of GLUT4 to the plasma membrane.
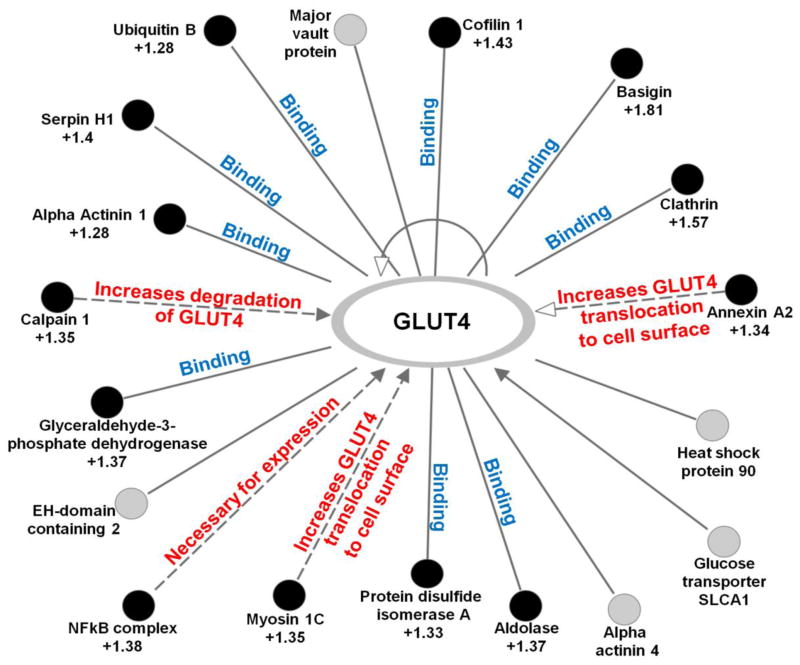
Taken together, proteomics data suggest that PMI 5011 treatment enhances GLUT4 translocation to the plasma membrane which results in increased glucose uptake and insulin sensitivity in HSMC. Using IPA, we generated a GLUT4 interaction network based on our proteomics data (Figure 4). The generated network identified 18 proteins interacting with GLUT4 with 13 of these proteins being significantly up-regulated. Several of the up-regulated proteins (annexin A2, myosin 1C, calpain 2) are known to increase GLUT4 translocation to the plasma membrane and increase GLUT4 degradation/turnover [40–43]. These findings are supported in HSMC using immunohistochemistry and western blot analysis in which a functional measure of GLUT4 translocation was assessed.
Figure 4.
Interaction network of proteins for SLC2A4 (GLUT4) generated using IPA. Proteins identified as significantly differentially regulated (p < 0.05) due to insulin stimulation in the presence of PMI 5011 compared to PMI 5011 alone are shown as filled circles. The fold change for each of these proteins is indicated in the Figure. Proteins labeled with gray circles were identified but are not differentially regulated.
Discussion
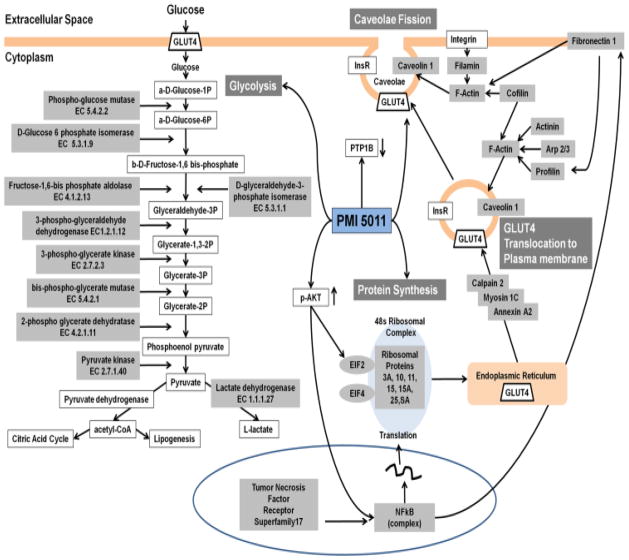
Skeletal muscle accounts for 70–85% of the whole body insulin-stimulated glucose uptake [39]. Insulin initiates its cellular action by first binding to the transmembrane receptor, triggering a cascade of intracellular molecular signaling pathways [44]. Insulin signaling has been shown to be attenuated in insulin resistant states [39, 45]. The differential proteomics data demonstrate that the ethanolic extract of Artemisia dracunculus L. (PMI 5011) modulates cellular pathways that regulate carbohydrate metabolism in HSMC. Specifically, proteomics data show significant changes in expression levels of proteins involved in actin cytoskeleton signaling, inflammation, caveolar-mediated endocytosis and glycolysis. These pathways are interrelated through GLUT4 (as described below) which is transported to the plasma membrane to facilitate glucose uptake. Western blot data confirm that PMI 5011 enhances GLUT4 translocation to the plasma membrane. Thus, collectively, our data support and extend findings reported in previous studies and provide data on additional cellular mechanisms, supporting the role of PMI 5011 to favorably alter glucose uptake and metabolism as summarized in Figure 5.
Figure 5.
Schematic representation of selected differentially regulated proteins and pathways identified using our proteomics analysis (gray) and from previous studies and their role in glucose uptake, glycolysis and GLUT4 turnover.
One of the key aspects of cellular response to insulin is the facilitation of glucose uptake into the cell by GLUT4 and the actin cytoskeleton network. This has been demonstrated to play a major role in the translocation of GLUT4 to the plasma membrane [46]. Insulin also stimulates actin remodeling via PI3K and profilin. Our previous reports have supported the role of PMI 5011 to enhance insulin stimulated PI3K activity in both in vitro and in vivo studies [17, 18]. In addition, in insulin resistant states, filamentous actin (F-actin) is disrupted which impedes GLUT4 transport [47]. In this regard, eighteen proteins involved in the actin cytoskeleton signaling pathway were identified in the data presented here (Table 2). Consistent with the literature, insulin stimulation in PMI 5011 treated cells resulted in an increased expression of proteins such as myosin, gelsolin and fibronectin 1, known to regulate actin cytoskeleton signaling [48, 49] (Figure 5). These results are consistent with increased F-actin enhancing GLUT4 transport to the plasma membrane and are very consistent with the noted efficacy of PMI 5011 to have significant effects after insulin administration in vivo [17].
It is well known that increased activation of inflammatory pathways plays a major role in regulating glucose metabolism and represents a fundamental step in the development of insulin resistance [3, 50]. An increase in inflammation in the insulin resistant state as in our baseline experiments has been shown to be a result of increased free fatty acid (FFA) flux [3, 51–56]. Recent reports also demonstrate that PTP1B overexpression in multiple tissues in obesity may be regulated by inflammation [57]. Our previous work indicates that PMI 5011 modulates PTP1B levels and activity in muscle [17, 18]. Overactivation of NFκB has also been shown to decrease GLUT4 transport [58]. Consistent with our previous studies [11], the current proteomics data show reduced expression of 23 proteins interacting with NFκB due to treatment with PMI 5011 (Figure 1b), suggesting that PMI 5011 improves insulin sensitivity by down-regulating proteins involved in regulation of inflammation in an insulin-independent manner. Thus, the data collected to date, including data in this manuscript showing a decrease in levels of proteins involved in regulation of inflammation, provide an intriguing unifying hypothesis linking inflammation related to FFA flux to insulin signaling, and modulation of this mechanism by PMI 5011.
In addition to the observation that inflammatory pathways were modulated, expression levels of caveolin 1 and other proteins involved in caveolar-mediated endocytosis signaling (Supplementary Figure 6) were increased upon insulin stimulation in the presence of PMI 5011 consistent with enhanced GLUT4 incorporation in the plasma membrane [59]. Furthermore, 18 proteins were identified as interacting with GLUT4 of which 13 were up-regulated upon insulin stimulation in the presence of PMI 5011 (Figure 4). These include proteins known to be involved in expression, translocation and turnover of GLUT4. Specifically, calpain has been shown to be involved in insulin-stimulated GLUT4 translocation, and its levels are decreased in type 2 diabetes. Furthermore, inhibition of calpain results in reduced glucose uptake by 60% [43]. In the presence of PMI 5011, proteomics data show that calpain is upregulated after insulin stimulation. In contrast, in insulin stimulated baseline control cells, none of these proteins were differentially regulated, underscoring the insulin resistant state of these cells.
Our results from proteomics studies suggesting enhanced GLUT4 transport to the plasma membrane were validated using immunohistochemistry and analysis of GLUT4 expression levels in membrane and microsomal fractions from HSMC. Specifically, our results (see Figure 3) demonstrate that the relative amount of GLUT4 is enhanced in plasma membrane due to PMI 5011 in basal states and is further increased due to insulin stimulation. This increase in GLUT4 content in plasma membrane is complemented by a decrease in GLUT4 levels in microsomal fraction due to PMI 5011 treatment and insulin stimulation. Thus, these studies support an effect of PMI 5011 to increase GLUT4 translocation to the plasma membrane (Figure 3).
In summary, we have shown that PMI 5011 modulates mechanisms in skeletal muscle consistent with enhanced carbohydrate metabolism. Specifically, we demonstrate enhanced expression of proteins involved in inflammation, glycolysis, actin cytoskeleton signaling, GLUT4 turnover and transport. The increase in GLUT4 transport results in increased glucose uptake and metabolism; hence, improving whole body insulin sensitivity. Thus, these studies have identified cellular pathways by which PMI 5011 improves insulin stimulated glucose uptake, transport and metabolism.
Supplementary Material
Highlights.
Insulin resistance is a major risk factor for type 2 diabetes and cardiovascular diseases.
Evaluate cellular mechanisms by which a botanical extract improves insulin action.
iTRAQ-based proteomic tools were used to identify differentially regulated proteins.
Proteomics data reveals that pathways modulating carbohydrate metabolism are affected.
Enhanced GLUT4 translocation results in increased glucose uptake and insulin sensitivity.
Acknowledgments
This work was supported by the grant P50AT002776 from the National Center for Complementary and Alternative Medicine and the Office of Dietary Supplements which funds the Botanical Research Center of Pennington Biomedical Research Center and by the T-32 postdoctoral fellowship award (AT004094) to P.S. We especially thank Ginger Ku for support on the nanoAcquity/Synapt HDMS system, Z. Elizabeth Floyd for helpful discussions and Weihong Pan for access to the Olympus FV1000 Laser Scanning Microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldstein BJ. Insulin resistance as the core defect in type 2 diabetes mellitus. Am J Cardiol. 2002;90:3G–10G. doi: 10.1016/s0002-9149(02)02553-5. [DOI] [PubMed] [Google Scholar]
- 2.Bloomgarden ZT. Insulin resistance, dyslipidemia, and cardiovascular disease. Diabetes Care. 2007;30:2164–70. doi: 10.2337/dc07-zb08. [DOI] [PubMed] [Google Scholar]
- 3.de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borghouts LB, Keizer HA. Exercise and insulin sensitivity: a review. Int J Sports Med. 2000;21:1–12. doi: 10.1055/s-2000-8847. [DOI] [PubMed] [Google Scholar]
- 5.Stafylas PC, Sarafidis PA, Lasaridis AN. The controversial effects of thiazolidinediones on cardiovascular morbidity and mortality. Int J Cardiol. 2009;131:298–304. doi: 10.1016/j.ijcard.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt B, Ribnicky DM, Poulev A, Logendra S, Cefalu WT, Raskin I. A natural history of botanical therapeutics. Metabolism. 2008;57:S3–9. doi: 10.1016/j.metabol.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusi K, DeFronzo RA. Metformin: a review of its metabolic effects. Diabetes Rev. 1998;6:89–131. [Google Scholar]
- 8.Wilm M. Quantitative proteomics in biological research. Proteomics. 2009;9:4590–605. doi: 10.1002/pmic.200900299. [DOI] [PubMed] [Google Scholar]
- 9.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen-Khuong T, White MY, Hung TT, Seeto S, Thomas ML, Fitzgerald AM, Martucci CE, Luk S, Pang SF, Russell PJ, Walsh BJ. Alterations to the protein profile of bladder carcinoma cell lines induced by plant extract MINA-05 in vitro. Proteomics. 2009;9:1883–92. doi: 10.1002/pmic.200700839. [DOI] [PubMed] [Google Scholar]
- 11.Kheterpal I, Coleman L, Ku G, Wang ZQ, Ribnicky D, Cefalu WT. Regulation of Insulin Action by an Extract of Artemisia dracunculus L. in Primary Human Skeletal Muscle Culture - A Proteomics Approach. Phytotherapy Res. 2010;24:1278–84. doi: 10.1002/ptr.3093. [DOI] [PubMed] [Google Scholar]
- 12.Lu QY, Yang Y, Jin YS, Zhang ZF, Heber D, Li FP, Dubinett SM, Sondej MA, Loo JA, Rao JY. Effects of green tea extract on lung cancer A549 cells: proteomic identification of proteins associated with cell migration. Proteomics. 2009;9:757–67. doi: 10.1002/pmic.200800019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baiges I, Palmfeldt J, Blade C, Gregersen N, Arola L. Lipogenesis is decreased by grape seed proanthocyanidins according to liver proteomics of rats fed a high fat diet. Mol Cell Proteomics. 2010;9:1499–513. doi: 10.1074/mcp.M000055-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkmaz H, Gurdal A. Effect of Artemisia santonicum L. on blood glucose in normal and alloxan-induced diabetic rabbits. Phytother Res. 2002;16:675–6. doi: 10.1002/ptr.943. [DOI] [PubMed] [Google Scholar]
- 15.Subramoniam A, Pushpangadan P, Rajasekharan S, Evans DA, Latha PG, Valsaraj R. Effects of Artemisia pallens Wall on blood glucose levels in normal and alloxan-induced diabetic rats. J Ethnopharmacol. 1996;50:13–7. doi: 10.1016/0378-8741(95)01329-6. [DOI] [PubMed] [Google Scholar]
- 16.Ribnicky DM, Poulev A, Watford M, Cefalu WT, Raskin I. Antihyperglycemic activity of Tarralin, an ethanolic extract of Artemisia dracunculus L. Phytomedicine. 2006;13:550–7. doi: 10.1016/j.phymed.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZQ, Ribnicky D, Zhang XH, Zuberi A, Raskin I, Yu Y, Cefalu WT. An extract of Artemisia dracunculus L. enhances insulin receptor signaling and modulates gene expression in skeletal muscle in KK-A(y) mice. J Nutr Biochem. 2011;22:71–8. doi: 10.1016/j.jnutbio.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ZQ, Ribnicky D, Zhang XH, Raskin I, Yu Y, Cefalu WT. Bioactives of Artemisia dracunculus L enhance cellular insulin signaling in primary human skeletal muscle culture. Metabolism. 2008;57:S58–64. doi: 10.1016/j.metabol.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cha BS, Ciaraldi TP, Park KS, Carter L, Mudaliar SR, Henry RR. Impaired fatty acid metabolism in type 2 diabetic skeletal muscle cells is reversed by PPARgamma agonists. Am J Physiol Endocrinol Metab. 2005;289:E151–9. doi: 10.1152/ajpendo.00141.2004. [DOI] [PubMed] [Google Scholar]
- 20.Ciaraldi TP, Abrams L, Nikoulina S, Mudaliar S, Henry RR. Glucose transport in cultured human skeletal muscle cells. Regulation by insulin and glucose in nondiabetic and non-insulin-dependent diabetes mellitus subjects. J Clin Invest. 1995;96:2820–7. doi: 10.1172/JCI118352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry RR, Abrams L, Nikoulina S, Ciaraldi TP. Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes. 1995;44:936–46. doi: 10.2337/diab.44.8.936. [DOI] [PubMed] [Google Scholar]
- 22.Ukropcova B, McNeil M, Sereda O, de Jonge L, Xie H, Bray GA, Smith SR. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J Clin Invest. 2005;115:1934–41. doi: 10.1172/JCI24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry RR, Ciaraldi TP, Mudaliar S, Abrams L, Nikoulina SE. Acquired defects of glycogen synthase activity in cultured human skeletal muscle cells: influence of high glucose and insulin levels. Diabetes. 1996;45:400–7. doi: 10.2337/diab.45.4.400. [DOI] [PubMed] [Google Scholar]
- 24.Gaster M, Petersen I, Hojlund K, Poulsen P, Beck-Nielsen H. The diabetic phenotype is conserved in myotubes established from diabetic subjects: evidence for primary defects in glucose transport and glycogen synthase activity. Diabetes. 2002;51:921–7. doi: 10.2337/diabetes.51.4.921. [DOI] [PubMed] [Google Scholar]
- 25.Ortenblad N, Mogensen M, Petersen I, Hojlund K, Levin K, Sahlin K, Beck-Nielsen H, Gaster M. Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: evidence for an intrinsic oxidative enzyme defect. Biochim Biophys Acta. 2005;1741:206–14. doi: 10.1016/j.bbadis.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Logendra S, Ribnicky DM, Yang H, Poulev A, Ma J, Kennelly EJ, Raskin I. Bioassay-guided isolation of aldose reductase inhibitors from Artemisia dracunculus. Phytochemistry. 2006;67:1539–46. doi: 10.1016/j.phytochem.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Ribnicky DM, Kuhn P, Poulev A, Logendra S, Zuberi A, Cefalu WT, Raskin I. Improved absorption and bioactivity of active compounds from an anti-diabetic extract of Artemisia dracunculus L. Int J Pharm. 2009;370:87–92. doi: 10.1016/j.ijpharm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govorko D, Logendra S, Wang Y, Esposito D, Komarnytsky S, Ribnicky D, Poulev A, Wang Z, Cefalu WT, Raskin I. Polyphenolic compounds from Artemisia dracunculus L. inhibit PEPCK gene expression and gluconeogenesis in an H4IIE hepatoma cell line. Am J Physiol Endocrinol Metab. 2007;293:E1503–10. doi: 10.1152/ajpendo.00420.2007. [DOI] [PubMed] [Google Scholar]
- 29.Ribnicky D, Poulev A, Rood J, Raskin I, Cefalu WT. Plasma Abundance of bioactives of Artemisia dracunculus L. and insulin sensitivity in obese, insulin-resistant human subjects: A pilot tria. J Alt Comp Med. 2011 in press. [Google Scholar]
- 30.Scherp P, Ku G, Coleman L, Kheterpal I. Gel-based and gel-free proteomic technologies. Methods Mol Biol. 2010;702:163–90. doi: 10.1007/978-1-61737-960-4_13. [DOI] [PubMed] [Google Scholar]
- 31.Zvonic S, Lefevre M, Kilroy G, Floyd ZE, DeLany JP, Kheterpal I, Gravois A, Dow R, White A, Wu X, Gimble JM. Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6:18–28. doi: 10.1074/mcp.M600217-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Kheterpal I, Ku G, Coleman L, Yu G, Ptitsyn AA, Floyd ZE, Gimble JM. Proteome of human subcutaneous adipose tissue stromal vascular fraction cells versus mature adipocytes based on DIGE. J Proteome Res. 2011;10:1519–27. doi: 10.1021/pr100887r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsumoto Y, Klip A. Development regulation of the subcellular distribution and glycosylation of GLUT1 and GLUT4 glucose transporters during myogenesis of L6 muscle cells. J Biol Chem. 1992;267:4957–62. [PubMed] [Google Scholar]
- 34.Wang ZQ, Bell-Farrow AD, Sonntag W, Cefalu WT. Effect of age and caloric restriction on insulin receptor binding and glucose transporter levels in aging rats. Exp Gerontol. 1997;32:671–84. doi: 10.1016/s0531-5565(97)00054-5. [DOI] [PubMed] [Google Scholar]
- 35.Moulder R, Lonnberg T, Elo LL, Filen JJ, Rainio E, Corthals G, Oresic M, Nyman TA, Aittokallio T, Lahesmaa R. Quantitative proteomics analysis of the nuclear fraction of human CD4+ cells in the early phases of IL-4-induced Th2 differentiation. Mol Cell Proteomics. 2010;9:1937–53. doi: 10.1074/mcp.M900483-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stull AJ, Galgani JE, Johnson WD, Cefalu WT. The contribution of race and diabetes status to metabolic flexibility in humans. Metabolism. 2010;59:1358–64. doi: 10.1016/j.metabol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proud CG. Regulation of protein synthesis by insulin. Biochem Soc Trans. 2006;34:213–6. doi: 10.1042/BST20060213. [DOI] [PubMed] [Google Scholar]
- 38.Proud CG, Denton RM. Molecular mechanisms for the control of translation by insulin. Biochem J. 1997;328 ( Pt 2):329–41. doi: 10.1042/bj3280329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdul-Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010:476279. doi: 10.1155/2010/476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Hsia SH, Imamura T, Usui I, Olefsky JM. Annexin II is a thiazolidinedione-responsive gene involved in insulin-induced glucose transporter isoform 4 translocation in 3T3-L1 adipocytes. Endocrinology. 2004;145:1579–86. doi: 10.1210/en.2003-1197. [DOI] [PubMed] [Google Scholar]
- 41.Bose A, Robida S, Furcinitti PS, Chawla A, Fogarty K, Corvera S, Czech MP. Unconventional myosin Myo1c promotes membrane fusion in a regulated exocytic pathway. Mol Cell Biol. 2004;24:5447–58. doi: 10.1128/MCB.24.12.5447-5458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otani K, Han DH, Ford EL, Garcia-Roves PM, Ye H, Horikawa Y, Bell GI, Holloszy JO, Polonsky KS. Calpain system regulates muscle mass and glucose transporter GLUT4 turnover. J Biol Chem. 2004;279:20915–20. doi: 10.1074/jbc.M400213200. [DOI] [PubMed] [Google Scholar]
- 43.Paul DS, Harmon AW, Winston CP, Patel YM. Calpain facilitates GLUT4 vesicle translocation during insulin-stimulated glucose uptake in adipocytes. Biochem J. 2003;376:625–32. doi: 10.1042/BJ20030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White MF. The insulin signalling system and the IRS proteins. Diabetologia. 1997;40 (Suppl 2):S2–17. doi: 10.1007/s001250051387. [DOI] [PubMed] [Google Scholar]
- 45.Olefsky JM. Insulin resistance and the pathogenesis of non-insulin dependent diabetes mellitus: cellular and molecular mechanisms. Adv Exp Med Biol. 1993;334:129–50. doi: 10.1007/978-1-4615-2910-1_10. [DOI] [PubMed] [Google Scholar]
- 46.Brozinick JT, Jr, Berkemeier BA, Elmendorf JS. “Actin”g on GLUT4: membrane & cytoskeletal components of insulin action. Curr Diabetes Rev. 2007;3:111–22. doi: 10.2174/157339907780598199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarthy AM, Spisak KO, Brozinick JT, Elmendorf JS. Loss of cortical actin filaments in insulin-resistant skeletal muscle cells impairs GLUT4 vesicle trafficking and glucose transport. Am J Physiol Cell Physiol. 2006;291:C860–8. doi: 10.1152/ajpcell.00107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom L, Ingham KC, Hynes RO. Fibronectin regulates assembly of actin filaments and focal contacts in cultured cells via the heparin-binding site in repeat III13. Mol Biol Cell. 1999;10:1521–36. doi: 10.1091/mbc.10.5.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–82. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 50.Perseghin G, Petersen K, Shulman GI. Cellular mechanism of insulin resistance: potential links with inflammation. Int J Obes Relat Metab Disord. 2003;27 (Suppl 3):S6–11. doi: 10.1038/sj.ijo.0802491. [DOI] [PubMed] [Google Scholar]
- 51.Lara-Castro C, Garvey WT. Intracellular lipid accumulation in liver and muscle and the insulin resistance syndrome. Endocrinol Metab Clin North Am. 2008;37:841–56. doi: 10.1016/j.ecl.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD, Muoio DM. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 53.Koh EH, Lee WJ, Kim MS, Park JY, Lee IK, Lee KU. Intracellular fatty acid metabolism in skeletal muscle and insulin resistance. Curr Diabetes Rev. 2005;1:331–6. doi: 10.2174/157339905774574347. [DOI] [PubMed] [Google Scholar]
- 54.Kelley DE, Goodpaster BH, Storlien L. Muscle triglyceride and insulin resistance. Annu Rev Nutr. 2002;22:325–46. doi: 10.1146/annurev.nutr.22.010402.102912. [DOI] [PubMed] [Google Scholar]
- 55.McGarry JD. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- 56.Boden G. Obesity and free fatty acids. Endocrinol Metab Clin North Am. 2008;37:635–46. viii–ix. doi: 10.1016/j.ecl.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem. 2008;283:14230–41. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Wu W, Li D, Guo Y, Ding H. Overactivation of NF-kappaB impairs insulin sensitivity and mediates palmitate-induced insulin resistance in C2C12 skeletal muscle cells. Endocrine. 2010;37:157–66. doi: 10.1007/s12020-009-9283-y. [DOI] [PubMed] [Google Scholar]
- 59.Oh YS, Khil LY, Cho KA, Ryu SJ, Ha MK, Cheon GJ, Lee TS, Yoon JW, Jun HS, Park SC. A potential role for skeletal muscle caveolin-1 as an insulin sensitivity modulator in ageing-dependent non-obese type 2 diabetes: studies in a new mouse model. Diabetologia. 2008;51:1025–34. doi: 10.1007/s00125-008-0993-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.