Abstract
Epigenetic mechanisms may contribute to the pathogenesis of complex diseases. Early or late environmental influences such as intrauterine malnutrition or sedentary lifestyle have been shown to lead to an increased risk of diabetes. Recently, epigenetic mechanisms were shown to be involved in endocrine cell differentiation and islet function. Genomic profiling of pancreatic islets in non-diabetic and diabetic states is needed in order to dissect the contribution of epigenetic mechanisms to the declining proliferation potential of β cells that we see with aging or the β-cell failure observed in diabetes. In-depth understanding of epigenetic landscapes can help to improve protocols for in vitro differentiation towards the β-cell fate, enhance β-cell proliferation, and lead to the discovery of novel therapeutic targets.
Tackling diabetes: where we are today
Diabetes mellitus is a chronic metabolic disease due to an insufficient insulin response to elevated blood glucose levels, and is caused by autoimmune destruction of the insulin producing pancreatic β cells in type 1 diabetes (T1DM - absolute insulin deficiency) [1], or by impaired sensitivity to insulin of insulin-responsive tissues such as liver and skeletal muscle, coupled with insufficient β-cell compensation in type 2 diabetes (T2DM) (relative insulin deficiency) [2]. Insulin deficiency results in chronic hyperglycemia, which can lead to many long-term complications, such as renal failure, heart attack, and stroke [3].
Increased family risk for developing T2DM has been recognized for a long time, with a sibling having a risk for developing diabetes of about three times that of the risk in the general population [4]. This has prompted decades of research into the genetic causes of diabetes. Initially, the Maturity Onset Diabetes of the Young (MODY) families, which exhibit a highly penetrant autosomal dominant pattern of inheritance [5], proved to be fertile ground for gene discovery. Among the MODY genes is the enzyme glucokinase, which catalyzes the initial step in utilization of glucose by the β-cell and liver and provides the set-point for insulin secretion in response to glucose [6]. DNA binding proteins, or transcriptional regulators, thought to function primarily in β cells are among the MODY identified genes as well [6]. It should be noted, however, that while MODY mutations are highly penetrant and thus of utmost importance for the affected families, they are also quite rare, contributing to only about 2–5% of diabetes incidence [7].
The increased incidence of diabetes and the concomitant increase of its long-term complications call for the development of new therapeutic strategies. Currently, diabetes treatment options include subcutaneous insulin injections for type 1 diabetics [1], and different classes of oral anti-diabetic medication that increase insulin secretion or peripheral insulin sensitivity, or decrease intestinal glucose absorption, for T2DM [8]. T1DM patients have also benefited from the development of the so-called Edmonton protocol for islet transplantation [9], in which cadaveric islets are transplanted into a recipient’s liver via the portal vein, allowing for transient or even long-term normalization of blood glucose levels and insulin independence. Unfortunately, the increasing demand of islet donor material far outstrips the supply of available organs. Novel sources of functional insulin-secreting β cells and glucagon-secreting α cells are the subject of major investigations. Along these lines, many research groups focus on the development of protocols for the directed differentiation of human embryonic stem cells (hESC). Another hotly pursued avenue is the reprogramming of other cell types, such as exocrine cells, to functional β and α cells for diabetes treatment. A third promising therapeutic approach, which might be achieved by the manipulation of the epigenetic landscape, is the improvement of β-cell function in vivo, in type 2 diabetic patients, or an increase in β-cell number in vitro prior to transplantation of donor islets into a T1DM patient.
In another attempt to understand the genes linked to diabetes, investigators have recently taken advantage of the relatively cheap and high coverage platforms used for the genotyping of hundreds of thousands of common single nucleotide polymorphisms (SNP’s), and have performed genome-wide association studies (GWAS) of large cohorts of cases and controls. In rapid succession, several dozen new diabetes risk loci were identified [10], although in most cases the cellular pathway affected or the causative mutation responsible for the altered risk remain to be determined. Despite these breakthroughs and hundreds of millions of dollars spent, all GWAS loci combined explain only about 10% of the familial risk for T2DM [11]. Given that the incidence of T2DM has increased dramatically over the past decades [12], a time period too short to allow for appreciable changes in the genetic makeup of human populations, it is likely that environmental factors, such as diet and sedentary lifestyle, and gene-environment interactions, might play a significant role in the development of the disease. These environmental factors are thought to affect the epigenetic state of the relevant metabolic organs and influence diabetes development [13].
The epigenome in phenotype transmission and development
The word ‘epi’ is of Greek origin (επι) and means ‘above, over’, thus defining that epigenetic modifications regulate various biological processes, from gene transcription to complex metabolic phenotypes, without changing the DNA sequence itself. Recently, this definition was refined to “an operational definition of epigenetics”, to emphasize the impact of environmental influences on epigenetic modifications and the dynamic aspect of the epigenome [14]. This definition of epigenetics proposes three steps of epigenetic signaling; an environmental signal which triggers a second intracellular signal to establish the exact chromatin location where the modification will take place, and a third sustaining signal that helps maintain this modification [14]. More specifically, epigenetics refers to the study of DNA methylation, histone modifications and non-coding RNAs (ncRNAs), and the mechanisms by which these modifications affect phenotype [15]. The functional significance of epigenetic alterations became apparent with the discovery of epigenetic silencing mechanisms of tumor suppressor genes, e.g. by DNA methylation, in cancer [16]. These breakthroughs have prompted epigenetic studies of many tissues in different disease states and developmental processes, have led to the characterization of DNA methylation patterns and the discovery of a diverse set of histone modifications. DNA methylation, which refers to the methylation of cytosine at the 5 position to create 5-methylcytosine, is a highly dynamic process, which is found to be globally decreased during development, with some tissue-specific exceptions. DNA methylation frequently leads to gene silencing and decreased gene expression either by promoting binding of transcriptional repressors or by preventing binding of other DNA-binding proteins [15].
Histones are subject to a wide variety of modifications including lysine (K) acetylation, lysine and arginine (R) methylation, serine (S) and threonine (T) phosphorylation, and lysine ubiquitination and sumoylation. These modifications, which occur primarily within the amino-terminal tails of the histones, greatly affect their function [17]. For example, the trimethylation of the fourth lysine on histone 3 (H3K4me3) is associated with active promoters at genes enriched in CpG islands (genomic regions highly enriched in guanine and cytosine nucleotides), whereas the trimethylation of the 27th lysine on histone 3 (H3K27me3) is linked to repression of transcription [18]. Due to their regulatory role in transcription and their influence on cell fate determination, these two histone marks have been the focus of multiple studies in different tissues and disease states. In embryonic stem cells and early stages of development, both the “activating” H3K4me3 and the “repressive” H3K27me3 mark can be present at the same gene promoter region [19]. The presence of both modifications is referred to as a ‘bivalent’ mark. During the differentiation process, either the H3K4me3 or the H3K27me3 mark is removed from most promoters, repressing or activating the corresponding genes and defining the distinct epigenetic landscape of differentiated cells.
This review will highlight the present and future impact of epigenetics on diabetes research, give an overview of important epigenetic findings, and discuss how these data can be utilized to identify targets for the development of new diabetes treatments.
Diabetes – why focus on epigenetics?
The epigenome can be thought of as an archive that stores information from exposure to environmental influences during embryogenesis and early development, affecting health and disease states in adulthood. At the same time, environmental influences such as increased food intake that leads to obesity (body mass index (BMI) > 30), play a major role in the pathogenesis several diseases, including T2DM [20]. Studies following the Dutch hunger famine have demonstrated that maternal and therefore intrauterine malnutrition and low birth weight followed by an increase of BMI during childhood, lead to an increased likelihood for developing metabolic syndrome [21], a complex disease defined by a group of factors that lead to an increased risk of cardiovascular artherosclerotic diseases and T2DM [22]. This observation, also referred to as the “thrifty phenotype hypothesis”, has been validated in multiple animal models, in which restriction of nutrient supply to the fetus has been shown to lead to a T2DM phenotype in the offspring, which is discussed in more detail below [23, 24]. These well-controlled studies clearly validate the importance of environmental influences in the pathogenesis of T2DM [25], and not only suggest that aberrant chromatin modifications play a role in the pathogenesis of T2DM, but also that epigenetic studies can be used to explore new therapeutic avenues.
The effect of diet on the islet epigenome
As mentioned above, intrauterine malnutrition leads to an increased risk for developing T2DM in adulthood. The environmental influence of malnutrition during embryonic development and the adult phenotype suggest the involvement of epigenetic mechanisms in T2DM pathogenesis. The use of different animal models of diet-induced manipulations of the epigenome, such as (i) supplementation with methyl-donors resulting in increased DNA methylation, (ii) restriction of protein intake in the diet, and (iii) limited nutrient supply by bilateral uterine artery ligation, have confirmed that maternal diet and intrauterine nutrition affect the epigenome of the offspring [25]. Intrauterine growth retardation (IUGR) caused by bilateral uteroplacental insufficiency results in animals with reduced β-cell mass, decreased expression of the essential β-cell transcription factor pancreatic and duodenal homeobox 1 (Pdx1), and a T2DM phenotype, in adulthood [24]. To elucidate the contribution of epigenetic mechanisms to the pathogenesis of T2DM, Park and colleagues analyzed the epigenetic landscape of the Pdx1 gene locus using islets from the IUGR rat model [23]. Chromatin immunoprecipitation (ChIP) analysis revealed that acetylation of both histone H3 and H4 at the Pdx1 locus is reduced in fetal, juvenile (2 weeks), and adult (6 months) islets, and this change is accompanied by progressively lower H3K4me3 levels at the Pdx1 locus and decreased PDX1 expression. In addition, the transcription factor USF-1, which regulates Pdx1 transcription [26], is no longer able to bind to the Pdx1 promoter in IUGR animals, implying that the change in chromatin-composition prevents USF-1 binding. Treatment of isolated 2 week-old islets with the histone deacetylase inhibitor Trichostatin A in vitro results in increased H3 acetylation and H3K4me3 enrichment, and allows for partial restoration of Pdx1 expression in the IUGR islets [23]. Together, these results suggest that epigenetic changes of the Pdx1 locus could be indicative of development of T2DM later in life and that reversal of epigenetic silencing at the Pdx1 locus, for example with a specific histone deacetylase inhibitor, might prevent late onset of the disease.
The transcription factor hepatocyte nuclear factor 4-α (Hnf4α) is important for rodent β-cell replication, in response to metabolic stress and glucose homeostasis, and mutations in HNF4α cause MODY1 in human [27–29]. It has been observed that maternal low protein diet and aging lead to epigenetic changes in the enhancer and the islet-specific P2 promoter region of the Hnf4α gene and in decreased Hnf4α expression levels in the offspring [30]. ChIP analysis showed that maternal diet and/or age decrease the presence of the activating H3Ac and H3K4me1 marks, and increase enrichment of the repressing H3K9me2 and H3K27me3 marks, in the enhancer and P2 promoter regions. These findings suggest that environmental influences lead to epigenetic changes that result in modified Hnf4α enhancer and P2 promoter regions, in altered promoter-enhancer interactions and reduced gene expression which might promote the development of T2DM [30]. In addition, Carone and colleagues have shown in mice that paternal low-protein diet affects cytosine methylation and metabolic gene expression in their offspring [31]. The study demonstrates the transgenerational transmission of epigenetic traits and highlights the importance of environmental influences on the epigenome in disease pathogenesis.
In summary, these studies show that the epigenetic landscape changes with age and in response to altered fetal nutrient supply, and that epigenetic mechanisms can contribute to the pathogenesis of T2DM. However, thus far these studies have been limited to specific candidate loci such as Pdx1 and Hnf4α. Therefore, a genome-wide dissection of the epigenetic landscape of the young, the old and the IUGR endocrine pancreas, and more specifically of pancreatic α and β cells as important regulators of glucose homeostasis, is needed. For example, ChIP-Seq analysis for the histone modification landscape and bisulfite sequencing analysis for the DNA cytosine methylation profile will help to elucidate which gene sets are affected by diet and aging. These investigations will establish if epigenetic markers could serve as biomarkers for T2DM prior to disease onset, and if specific epigenetic manipulation can be utilized for the prevention or treatment of the disease.
Epigenetic profiling of pancreatic islets
Over the past twenty years, developmental geneticists have made great strides in elucidating the transcriptional regulators and signaling pathways that direct the differentiation of foregut endoderm into pancreatic precursors and eventually in the mature endocrine cells [32]. This detailed knowledge was instrumental for the development and improvement of in vitro protocols used for the differentiation of embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) towards the β-cell fate [33, 34]. Despite these successes, the in vitro differentiation protocols only succeed in generating polyhormonal cells that are not glucose-responsive in vitro. At present, only pancreatic precursors derived in vitro which are transplanted into mice for at least three months, to allow for “in vivo maturation”, can differentiate to become more functional endocrine cells [34]. Most likely, some key epigenetic modifications fail to be established in the current in vitro protocols, a serious obstacle that needs to be overcome if cell replacement therapy with ESC or iPSC derived endocrine cells is to become a reality. Epigenetic profiling of the islets of Langerhans and its major cell types α and β cells promises to provide at least part of the missing clues.
Identification of targets for epigenetic therapy requires the detailed analysis of the epigenome in the tissue of interest. The first genome-wide epigenetic studies of human pancreatic islets utilized the techniques of formaldehyde-assisted isolation of regulatory elements coupled with high-throughput sequencing (FAIRE-Seq analysis) and chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq analysis). These studies were able to characterize open chromatin sites [35], the mono-, di- and tri-methylation profile of histone 3 (including the H3K4me1, H3K4me2, H3K4me3, H3K27me3, and H3K79me2 marks), and the binding sites of CTCF (CCCTC-binding factor), an important epigenetic regulator [36], in human islets [37, 38]. These first analyses led to the notion that a small number of bivalent marks are still present in developmental regulatory genes of differentiated human islets, and that histone 3 tri-methylation (H3K4me3) enrichment levels do not correlate with gene expression levels in genes with promoters that are poor in CpG islands [37, 38]. However, β cells represent only a fraction (~35%) of the cells composing the human islet, and follow-up studies analyzing sorted cell populations enriched for α and β cells specifically need to be pursued, in order to validate the previous findings. In addition, this analysis will help assess the cell-type specific differences in the epigenome, and will offer a precise definition of the epigenetic landscape of human α and β cells. Interestingly, characterization of histone modification profiles of murine neural tissues, pancreatic progenitor cells and differentiated β and acinar cells, revealed the active H3K4me3 signature of β cells clusters with neural tissues, but not acinar cells, while the repressive H3K27me3 profile of β cells shows high similarity to acinar cells [39]. These results suggest that (i) the H3K4me3 mark reflects the functional state of the cell type (e.g. stimulus-coupled secretion of vesicles, which is common to β cells and neurons), (ii) indicate that the H3K27me3 mark is reflective of the developmental origin of the cell, and (iii) emphasize the necessity of cell-type specific epigenetic analysis.
Epigenetic regulation of β-cell differentiation and proliferation
Epigenetic modifications play pivotal roles in the regulation of transcription and are important for cell specification and proliferation [15]. Acetylation of histone residues is associated with the regulation of gene expression and cell differentiation [40]. Many enzymes such as histone acetyltransferases (HATs) and histone deacetylases (HDACs) are involved in the process of adding or removing different histone modifications, thereby establishing tissue- and cell- type specific histone modification patterns. The discovery of different classes and subtypes of histone modifying enzymes, the evaluation of their expression patterns, and the effect of their inhibition or deletion in cell function, provides information about the importance of these enzymes and their effect on the development and/or functional maintenance of different tissues.
The large family of HDACs is divided into the classes and subclasses I, IIa, IIb and III, all of which can repress gene expression and regulate differentiation and proliferation [40]. The effect of HDACs on cell proliferation and their potential use as drug targets in cancer therapy drove the recent development of histone deacetylase inhibitors, which are currently in clinical trials, or have been approved for treatment of multiple cancer types [16, 41]. Treatment of cultured embryonic rat pancreata explanted at embryonic day 13.5 (E13.5) with different histone deacetylase inhibitors, results in a decrease in exocrine and an increase in endocrine cell types [42]. Inhibition of class I and class II HDACs with trichostatin A, leads to an increase in endocrine progenitor cells and β cells, while inhibition of class I HDACs with valproic acid enhances the endocrine progenitor and α-cell pool [42]. Collectively, these observations suggest that HDACs play an important role in the early determination of pancreatic cell types.
Lenoir and colleagues determined the expression patterns of class IIa HDACs (HDAC-4, -5, and -9) and the effect of their deletion and forced expression on pancreatic development in mice [43]. Interestingly, expression of class IIa HDACs is only observed in β and δ cells, but not α and acinar cells. The deletion of HDAC5- and -9 (Hdac5−/− and Hdac9−/−) results in an increase in the number of β and δ cells, likely due to increased cell differentiation from endocrine precursors towards these lineages. Treatment of pancreatic explants with MC1568, a specific class IIa histone deacetylase inhibitor, which acts downstream of ‘neurogenin 3’ (Ngn3) and upstream of ‘paired box gene 4’ (Pax4), causes an increase of β and δ-cells. Collectively, these results highlight the role of epigenetic regulation in pancreatic cell fate determination and suggest a potential application of HDAC inhibitors in directing in vitro differentiation towards the β-cell fate, which could be considered a form of epigenetic therapy.
Histone modifying enzymes do not act in isolation, but are directed to their target genes by specific DNA-binding transcription factors. A recent study discovered that the homeodomain transcription factor Nkx2.2 regulates expression of the ‘aristaless homebox’ transcription factor Arx, by binding to its promoter in a repressive complex with HDAC1, the de novo DNA methyltransferase 3a (Dnmt3a), and the co-repressor Groucho-3 (Grg3) [44]. In β cells, this complex binds to the hypermethylated Arx promoter, thereby preserving the β-cell specific state, and blocking β- to α-cell conversion. In addition, the “maintenance” DNA methyltransferase 1 (Dnmt1) is important for preserving β-cell identity. It achieves that by methylating the Arx locus and repressing this α-cell specific transcription factor through binding of the methyl-binding protein MeCP2 and recruitment of the enzyme ‘protein arginine methyltransferase 6′ (PRMT6) [45]. Indeed, deletion of Dnmt1 in β cells leads to loss of Arx repression and gradual conversion of β to α cells [45]. These findings demonstrate that interactions between transcription factors and epigenetic components are important for proper cell differentiation and for maintenance of the differentiated cell state.
The two polycomb repressive complexes 1 and 2 (PRC1 and PRC2), formed by polycomb group (PcG) proteins, are chromatin modifiers, they repress gene transcription, and are critical for cell specification [46]. The PcG protein Bmi-1 is crucial for the function of PRC1, which mono-ubiquitylates histone H2A on lysine 119. Also, ‘enhancer of zeste homolog 2’ (Ezh2) is a histone methyltransferase and part of PRC2 that maintains the repressive H3K27me3 mark [46]. Thus, Bmi-1 and Ezh2 influence the proliferative potential of β cells in the mouse [47, 48]. Dhawan and colleagues reported that Bmi-1 regulates the Ink4/Arf locus, that a decline in Bmi-1 results in reduced β-cell proliferation, and that Bmi-1 plays a role in β-cell regeneration after β-cell ablation by streptozocin treatment [48]. Conditional deletion of Ezh2 in β cells leads to reduced H3K27me3 levels and increased mRNA levels of the two cell cycle inhibitors p16Ink4 and p19Arf, and reduced β-cell proliferation [47]. Consequently, Ezh2-deficient mice exhibit impaired glucose tolerance and an inability to recover from streptozocin-induced β-cell ablation [47]. These results stress the importance of epigenetic mechanisms in the regulation of β-cell proliferation, and suggest that epigenetic manipulation of PcG proteins and their respective histone modification marks in β cells might be a useful tool for promoting β-cell proliferation in vitro and in vivo.
Concluding remarks
Epigenetic diabetes therapy – promises for the future?
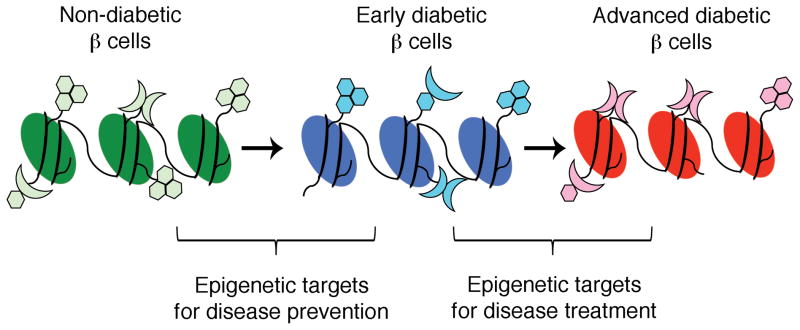
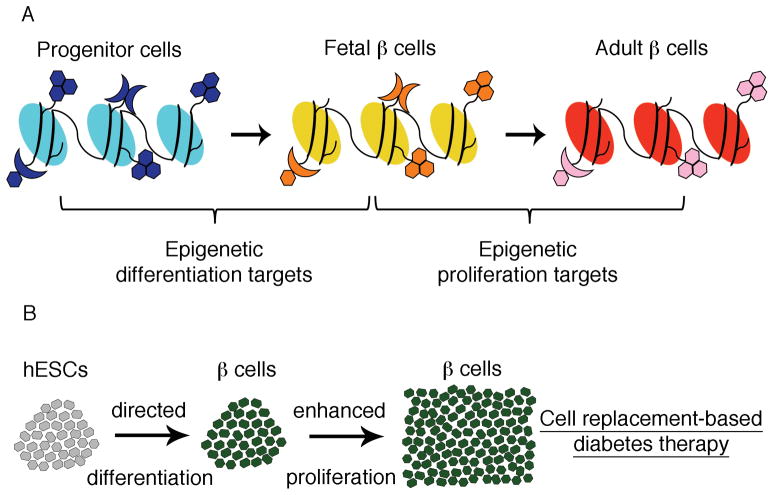
Epigenetic modifications are implicated in the pathogenesis of complex diseases, such as asthma, cancer, and T2DM [13, 49, 50]. The development of epigenetic drugs presents both a major challenge and a promising opportunity. Epigenetic abnormalities are thought to occur before disease onset and might contribute to disease pathogenesis [51]. Hence, discovering markers that might be associated with increased risk for disease development is of great importance. However, the major challenges of epigenetic research are to distinguish between epigenetic modifications that cause the onset of disease and ones that do not, and to evaluate the significant individual variability [51]. In order to determine diabetes-specific epigenetic profiles, precise assessment of the epigenome of endocrine pancreatic cells, e.g. of insulin-secreting β cells, in the non-diabetic state and at early and late diabetic states is necessary, as this approach will help to distinguish between causal and consequential epigenetic modifications. This genome-wide comparison might also uncover disease-specific epigenetic modification patterns, and provide an opportunity for discovering markers for disease onset, and new potential therapeutic targets for diabetes prevention and treatment (Fig. 1). In addition, the profiling of different developmental stages and of proliferative versus non-proliferative β cells will elucidate important epigenetic mechanisms involved in the differentiation and proliferation of β cells, and will help improve the currently used in vitro differentiation protocols (Fig. 2). Most histone modifying enzymes or DNA methyltransferases have a broad target range and are expressed in multiple tissues. This increases the likelihood of “off target” effects, and will present a significant challenge for drug development. Therefore, it is likely that the first applications will be developed for ex vivo use, such as improvement of in vitro protocols for directed differentiation of embryonic stem cells towards the β-cell fate, and the enhancement of β-cell proliferation in donor islets for cell replacement-based diabetes therapy.
Figure 1. Application of epigenetic profiling for the development of novel diabetes treatment strategies.
Precise profiling of the epigenome of insulin-secreting β cells, in the non-diabetic state, and at early and advanced stages of diabetes will lead to the better understanding of diabetes pathogenesis. The discovery of epigenetic markers for disease onset can be used as future targets for disease prevention, and comparison of the epigenetic landscapes of early and advanced diabetic b cells will allow for disclosure of novel epigenetic targets for diabetes treatment.
Figure 2. Use of epigenetic profiling of progenitor cells and mature β cells in diabetes research.
(A) In-depth understanding of the epigenetic landscape of undifferentiated progenitor cells and differentiated β cells will help identify important epigenetic differentiation targets. Comparison of the epigenetic make-up of proliferative, fetal β cells and non-proliferative, adult β cells will elucidate the epigenetic mechanisms contributing to the proliferative capacity of β cells.
(B) The knowledge of epigenetic differentiation markers and the development of epigenetic drugs targeting these markers will provide the opportunity to improve in vitro differentiation protocols of human embryonic stem cells (hESCs) towards the β-cell fate. Furthermore, targeting of epigenetic proliferation markers can enhance in vitro proliferation of b cells for cell replacement-based diabetes therapy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van Belle TL, et al. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 2.Kahn SE, et al. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 3.Kannel WB, McGee DL. Diabetes and cardiovascular disease. The Framingham study. Jama. 1979;241:2035–2038. doi: 10.1001/jama.241.19.2035. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki K, et al. Familial risks for type 2 diabetes in Sweden. Diabetes Care. 2010;33:293–297. doi: 10.2337/dc09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tattersall RB, Fajans SS. A difference between the inheritance of classical juvenile-onset and maturity-onset type diabetes of young people. Diabetes. 1975;24:44–53. doi: 10.2337/diab.24.1.44. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefond A, et al. The emerging genetics of type 2 diabetes. Trends Mol Med. 2010;16:407–416. doi: 10.1016/j.molmed.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Winckler W, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 8.Charbonnel B, Cariou B. Pharmacological management of type 2 diabetes: the potential of incretin-based therapies. Diabetes Obes Metab. 2011;13:99–117. doi: 10.1111/j.1463-1326.2010.01317.x. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro AM, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343:230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- 10.Nolan CJ, et al. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet. 2011;378:169–181. doi: 10.1016/S0140-6736(11)60614-4. [DOI] [PubMed] [Google Scholar]
- 11.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmet P, et al. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 13.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger SL, et al. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21:502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 21.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 22.Kassi E, et al. Metabolic syndrome: definitions and controversies. BMC Med. 2011;9:48. doi: 10.1186/1741-7015-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JH, et al. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoffers DA, et al. Neonatal exendin-4 prevents the development of diabetes in the intrauterine growth retarded rat. Diabetes. 2003;52:734–740. doi: 10.2337/diabetes.52.3.734. [DOI] [PubMed] [Google Scholar]
- 25.Simmons R. Epigenetics and maternal nutrition: nature v. nurture. The Proceedings of the Nutrition Society. 2011;70:73–81. doi: 10.1017/S0029665110003988. [DOI] [PubMed] [Google Scholar]
- 26.Qian J, et al. Upstream stimulatory factor regulates Pdx-1 gene expression in differentiated pancreatic beta-cells. Biochem J. 1999;341 ( Pt 2):315–322. [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta RK, et al. Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev. 2007;21:756–769. doi: 10.1101/gad.1535507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta RK, et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 30.Sandovici I, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 33.D’Amour KA, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 34.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 35.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhandare R, et al. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 2010;20:428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Arensbergen J, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 2010;20:722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M, et al. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 41.Ellis L, et al. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 42.Haumaitre C, et al. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28:6373–6383. doi: 10.1128/MCB.00413-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenoir O, et al. Specific Control of Pancreatic Endocrine {beta}- and {delta}-Cell Mass by Class IIa Histone Deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60:2861–2871. doi: 10.2337/db11-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papizan JB, et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 2011;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhawan S, et al. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20:419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 47.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhawan S, et al. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakyan VK, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]