Abstract
With the recent growth of the global monoclonal antibody market, ultrasensitive techniques are required for rapid analysis of possible immunogenic residues, such as galactose-α-1,3-galactose (α-1,3-Gal) on therapeutic proteins expressed in murine or CHO cell lines. We report a capillary electrophoretic approach in conjunction with exoglycosidase digestion for structural elucidation of N-linked IgG glycans containing the above immunogenic epitope. The method uses commercially available reagents and instrumentation, thus making the described methodology readily available for implementation and validation within the biotechnology industry. The method was first evaluated using polyclonal mouse IgG N-glycans which are known to contain α-1,3-Gal containing epitopes. High reproducibility in migration time enabled determination of GU values for five α-1,3-Gal containing structures. The method was successfully applied to the analysis of a NCI reference standard monoclonal antibody and two development phase monoclonal antibodies. The limit of detection and limit of quantitation were 1 and 2 µg of intact protein IgG starting material, respectively, further indicating the high sensitivity of the described method.
Keywords: therapeutic antibody, monoclonal antibody, alpha galactose, immunogenic epitopes, capillary electrophoresis, exoglycosidase digestion, glycomics
1.0 INTRODUCTION
Characterization of the oligosaccharides present on therapeutic glycoproteins is an important regulatory requirement during product development and lot release. One of the most important aspects of glycomic analysis is the determination of the presence or absence of potential immunogenic epitopes, even at trace levels.1 Non-human oligosaccharide motifs, including galactose-α-1,3-galactose (α-1,3-Gal) and N-glycolyl neuraminic acid (NGNA), are known to have the potential to elicit an immunogenic response.1
Immunoglobulin-1 (IgG1) isotypes, which represent the major number of current antibody therapeutics, contain ~2–3% carbohydrate by mass, primarily attached to the highly conserved N-glycosylation sites at asparagine 297 in the CH2 domain of the Fc region of each heavy chain. Additional glycosylation sites may be found in the hypervariable regions of the Fab portion.2 In species that express the Ggta1 gene, the non-reducing ends of IgG Fc and Fab N-glycans can potentially be capped by galactose-α-1,3-galactose structures, as opposed to sialic acids during competitive processing by the glycosyltransferase enzymes within the Golgi apparatus. Such epitopes may adversely affect the safety of biotherapeutics products.3
Humans have high circulating levels of anti-α-1,3-Gal antibodies (approximately 1% of total serum antibodies) due to the evolutionary loss of the N-acetyllactosaminide-3-α-galactosyltransferase-1 enzyme associated with the construction of these α-1,3-Gal containing glycoforms making such sugars foreign to our immune system.4 Minimization of the level of α-1,3-Gal epitopes during biotherapeutic development and production is an important issue.5 For example, Cetuximab, an anti-epidermal growth factor receptor monoclonal antibody used in the treatment of metastatic cancer and produced in murine SP2/0 cells is associated with the generation of an IgE-mediated anaphylactic response following administration of the drug in certain patients.1 The actual amount of α-1,3-Gal that can induce such hypersensitivity reactions is not known.6 Therefore, analysis of the glycosylation profile and identification of this and potentially other immunogenic epitopes of therapeutic antibodies is of high importance in biotherapeutics production and release, as already advised by regulatory agencies.7 As glycosylation is subject to the cell culture conditions, alterations in the process may result in different levels of α-1,3-galactosylation.8 Chinese hamster ovary (CHO) cells, commonly used as host cells for therapeutic monoclonal antibody production, have been assumed until recently, not to synthesize galactose-α-1,3-galactose epitopes.5 Recent studies revealed that CHO cells can also produce α-1,3-Gal residues, therefore making it essential to screen for these immunogenic structural motifs in therapeutic monoclonal antibodies made from CHO cells.5
The high structural diversity of protein glycosylation makes carbohydrate analysis challenging as glycans lack chromophoric/fluorophoric moieties and easily ionizable groups. High performance analytical techniques such as hydrophilic interaction chromatography (HILIC), graphitized carbon chromatography, microfluidics, ultra-performance liquid chromatography and mass spectrometry (MS) are usually employed, in most cases following analyte derivatization.9–13 However, in the case of MS, problems with quantitation and qualitative confirmation due to the loss of labile terminal monosaccharides during analyte transfer through the mass spectrometer have been reported.14 Among the electric field mediated separation techniques, capillary electrophoresis (CE) is one of the most powerful methods to analyze complex glycans.15–16 This method is today a widely accepted glycan analysis procedure by regulatory agencies.
LC-, CE- and MS-based methods are often performed in combination with exoglycosidase digestion for complete oligosaccharide characterization.17 Structural elucidation of glycans by consecutive enzymatic digestion of carbohydrates using exoglycosidase combinations, followed by orthogonal separation methods of capillary electrophoresis and/or LC separation, or mass spectrometric analysis of the digestion products are well-established tools for such analysis.18–19 Comparison of the retention/migration times of the exoglycosidase digestion products to an oligosaccharide ladder enables calculation of migration shifts due to cleavage based on the exoglycosidase used. Thus, the particular sequence of each oligosaccharide in a glycan pool can be predicted with high confidence.19 Often so called annotated and curated GU libraries (under construction) can be used and the exoglycosidase digestion approach serves for qualitative confirmation of the annotated structural predictions.
In this paper, we focus on developing a CE-LIF approach for the ultrasensitive analysis of potentially immunogenic α-1,3-Gal containing structures in antibody therapeutics using commercially available reagents and instrumentation. The method can be easily adapted and validated by the biopharmaceutical industry. For our studies, we selected the α-1,3-Gal containing structures derived from polyclonal murine serum IgG as a representative reference sample for this challenging problem. After successful identification of the α-1,3-Gal containing glycans in mouse polyclonal IgG, we applied the method to the comprehensive structural annotation of an NCI monoclonal antibody reference standard and two developmental phase therapeutic antibodies. Five oligosaccharides containing the α-1,3-Gal moiety were identified. In addition to the identification of α-1,3-Gal structures, the approach also enables the determination of the antennary position of the α-1,3-Gal residues with high sensitivity due to the excellent selectivity of the CE separation.
2.0 MATERIALS AND METHODS
2.1 Chemicals and reagents
Citric acid, 8-aminopyrene-1,3,6-trisulfonic-acid trisodium salt (APTS), 2-aminoacridone (AMAC), ammonium acetate, acetic acid, sodium cyanoborohydrate (1 M solution in tetrahydrofuran), polyclonal IgG from mouse serum, maltose and glucuronic acid were from Sigma-Aldrich (St. Louis, MO). The maltodextrin (M040) ladder was provided by Grain Processing Corporation (Muscatine, IA). α(2–3,6,8,9)-Neuraminidase from Arthrobacter ureafaciens (ABS), α(1–2,3,4,6)-fucosidase from bovine kidney (BKF), β(1–3,4)-galactosidase from bovine testis (BTG), α(1–3,4,6)-galactosidase from green coffee bean (CBG) and β(1–2,3,4,6)-N-acetylhexosaminidase from jack bean (GUH) were all obtained from Prozyme (Hayward, CA). α(1–2,3)-Mannosidase from Chryseobacterium meningosepticum (Mα1–2,3) for antennary position determination was purchased from New England Biolabs (Ipswitch, MA). The chimeric monoclonal antibody reference material (NSC#623408) was from NCI (Frederick, MD). Two in-house development stage monoclonal antibodies (UTC1 and UTC2) were also analyzed during the study (United Therapeutics Corporation). Acetonitrile and water, both HPLC-MS grade, were purchased from Thermo Fisher Scientific (Fair Lawn, NJ).
2.2 Release of N-linked glycans from monoclonal antibodies
Monoclonal antibody N-glycans were released using the GlykoPrep Digestion Module (Prozyme), according to the manufacturer’s protocol. Briefly, 50 µg of denatured glycoprotein sample were loaded onto the equilibrated cartridges and following immobilization of the monoclonal antibodies 25 mU PNGase F was added to each sample. The solution was incubated at 55°C for 1 hour and released glycans were eluted from the cartridges with 100 µL of water, followed by drying in a centrifugal vacuum evaporator (Centrivap, Labconco, Kansas City, MO). The Fab portions of the IgG molecules for fragment specific glycosylation studies were prepared as previously described.20
2.3 APTS and AMAC labeling
Oligosaccharide standards and released glycans were labeled according to our recently published protocol.21 Briefly, 2 µL of APTS solution (50 mM in 1.2 M citric acid) and 2 µL of NaBH3CN (1 M in THF) were added to the dried samples, followed by incubation of the reaction mixture at 55°C for 60 minutes. The reactions were quenched by the addition of 50 µL of HPLC water. The glucuronic acid (upper bracketing standard for run to run normalization) was labeled with AMAC as follows: 2.5 µL of AMAC solution (1 M in DMSO) and 2.5 µL of NaBH3CN (1 M in DMSO) were added to 20 µg of glucuronic acid, and the mixture was incubated overnight at 37°C. The reaction was stopped by the addition of 50 µL of HPLC water.
2.4 Purification of the labeled glycans
To remove unconjugated APTS, 450 µL of acetonitrile was added to the reaction mixture and the solution was loaded onto polyamide resin filled pipette tips (10 µL bed volume, PhyNexus, San Jose, CA) using a semi-automated 12-channel pipettor (Phynexus). The purification process was controlled by the PhyTip operating software (Phynexus). After glycan capture, the tips were washed four times with 500 µL of 95 % acetonitrile by applying 8 in take/expel cycles. The captured APTS labeled glycans were then eluted with 50 µL of HPLC water and analyzed by CE-LIF.
2.5 Exoglycosidase structural elucidation of the labeled glycans
To obtain the necessary structural information of the glycans of interest, APTS labeled, purified glycan samples released from 50 µg of each antibody (6 parallels) were subject to exoglycosidase digestion, as previously described.22 Aliquots were processed using different exoglycosidase combinations in 50 mM ammonium acetate (pH 5.5) digestion buffer. After digestion, the aliquots were evaporated to dryness, dissolved in 50 µL of water and analyzed by CE-LIF.
2.6 CE-LIF analysis
All capillary electrophoresis experiments were implemented on a PA 800 Plus Pharmaceutical Analysis System (Beckman Coulter, Inc., Brea, CA) equipped with laser-induced fluorescence detection (488 nm excitation / 520 nm emission). eCAP NCHO coated capillaries (50 µm i.d., 60 cm total length, 50 cm effective length) were used for the separations, with 500 V/cm applied electric field strength. The Carbohydrate Separation Gel Buffer-N (Beckman Coulter) was used as background electrolyte in each experiment. All samples were pressure injected (0.5 psi for 5 seconds).
3.0 RESULTS
The application of an ultrasensitive method for the analysis of N-linked oligosaccharides released from therapeutic antibodies for their galactose α-1–3 linked galactose epitope content using capillary electrophoresis with laser induced fluorescence detection and exoglycosidase digestions is described. The method was applied to the comprehensive analysis of a NCI reference standard and two development phase therapeutic monoclonal antibodies. The high resolving power and ultrahigh sensitivity of the CE-LIF/exoglycosidase digestion method are demonstrated. The method uses standard reagents and instrumentation, thus making the approach readily available for direct implementation and further validation within the biopharmaceutical industry.
3.1 Exoglycosidase digestion-based structural annotation of α-1,3-Gal containing mouse IgG glycans
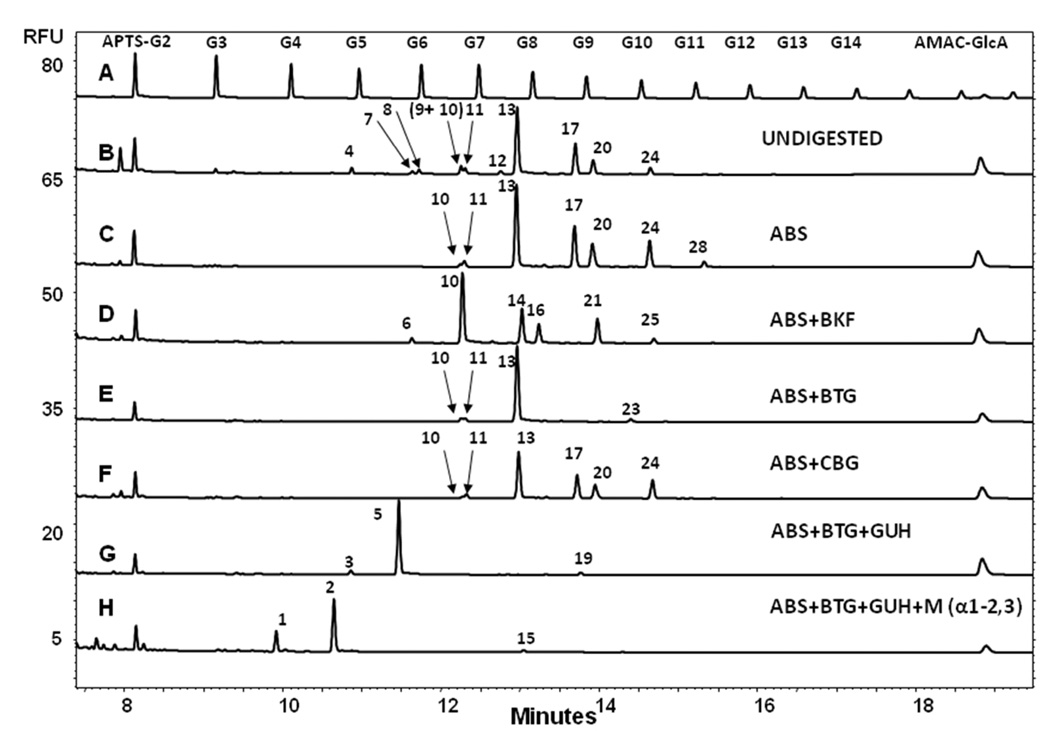
Polyclonal mouse serum IgG was used as a source of galactose α-1–3 linked galactose epitope-containing oligosaccharides for initial investigations in order to elucidate the migration position of the glycans of interest. Figure 1 shows the CE-LIF profile (trace B) and the exoglycosidase digestion-based structural annotation (traces C-H) of mouse IgG N-glycans compared to the malto-oligosaccharide ladder (trace A). APTS labeled maltose (APTS-G2) and AMAC labeled glucuronic acid (AMAC-GlcA) were used as lower and upper bracketing standards, respectively, to increase the precision of peak assignment.19
Figure 1.
Exoglycosidase digestion-based structural annotation of mouse IgG N-glycans. Trace A: malto-oligosaccharide ladder, Trace B: APTS labeled undigested mouse IgG N-glycans, Trace C-H: ABS, ABS+BKF, ABS+BTG, ABS+CBG, ABS+BTG+GUH and ABS+BTG+GUH+Mα-1–2,3 digests. Conditions: eCAP NCHO coated capillary (50 µm i.d., 60 cm total length, 50 cm effective length); Applied electric field strength: 500 V/cm; Background electrolyte: Carbohydrate Separation Gel Buffer-N; Separation temperature: 25°C; Injection: 0.5 psi for 5 seconds. APTS-G2: APTS labeled maltose lower bracketing standard. AMAC-GlcA: AMAC labeled glucuronic acid upper bracketing standard.
Trace B presents 10 peaks corresponding to 11 glycan structures as shown in Table 1. CE-LIF / exoglycosidase digestion was applied in conjunction with an earlier established partial database for full structural characterization in order to accurately assign glycan compositions to the individual glycoforms.19 We anticipated that peaks 4–12 could contain sialic acid residues at their non-reducing ends. First α(2–3,6,8,9)-neuraminidase (ABS) was applied for enzymatic desialylation. Trace C depicts the resulting electropherogram showing shifts for peaks 4, 7, 8, 9 and 12 towards the later migrating, less charged asialo glycan regime. These shifts increased the relative peak areas for peaks 17, 20 and 24 and resulted in the appearance of peak 28. The long migration time of this latter species suggested the presence of additional monosaccharide residues over the commonly reported bi-antennary digalactosyl oligosaccharide structure (see Table 1), which was suspected to be α-1,3-Gal.
Table 1.
Structure abbreviated names of all glycans determined in this study using CE-LIF and UPLC-fluorescence with glucose unit (GU) values.
| Peak | Structure | CE-LIF GU values |
HILIC GU Values |
|---|---|---|---|
| 1 | M2 | 3.93 | - |
| 2 | FM2 | 4.64 | - |
| 3 | M3 | 4.89 | - |
| 4 | FA2G2S2 | 4.91 | 10.12 |
| 5 | FM3 | 5.66 | - |
| 6 | A1 | 5.86 | 4.98 |
| 7 | FA2[6]G1S1 | 5.88 | 8.35 |
| 8 | FA2[3]G1S1 | 5.99 | 8.48 |
| 9 | FA2G2S1 | 6.75 | 9.26 |
| 10 | A2 | 6.75 | 5.37 |
| 11 | FA1 | 6.82 | 5.44 |
| 12 | FA2G2Gal[6]1S1 | 7.49 | 10.12 |
| 13 | FA2 | 7.75 | 5.91 |
| 14 | A2[6]G1 | 7.82 | 6.28 |
| 15 | FM2A1G1Gal[3]1 | 7.86 | - |
| 16 | A2[3]G1 | 8.13 | 6.39 |
| 17 | FA2[6]G1 | 8.85 | 6.75 |
| 18 | A2G1Gal[6]1 | 8.88 | 7.12 |
| 19 | FA1G1Gal[6]1 | 8.92 | 6.81 |
| 20 | FA2[3]G1 | 9.17 | 6.87 |
| 21 | A2G2 | 9.23 | 7.23 |
| 22 | FA1G1Gal[3]1 | 9.67 | 6.89 |
| 23 | FA2G1Gal[6]1 | 9.87 | 7.65 |
| 24 | FA2G2 | 10.24 | 7.66 |
| 25 | A2G2Gal[6]1 | 10.26 | 8.02 |
| 26 | FA2[3]G1Gal1 | 10.50 | 7.79 |
| 27 | A2G2Gal[3]1 | 10.53 | 8.07 |
| 28 | FA2G2Gal[6]1 | 11.25 | 8.47 |
| 29 | FA2G2Gal[3]1 | 11.54 | 8.48 |
| 30 | A2G2Gal2 | 11.54 | 8.85 |
| 31 | FA2G2Gal2 | 12.53 | 9.63 |
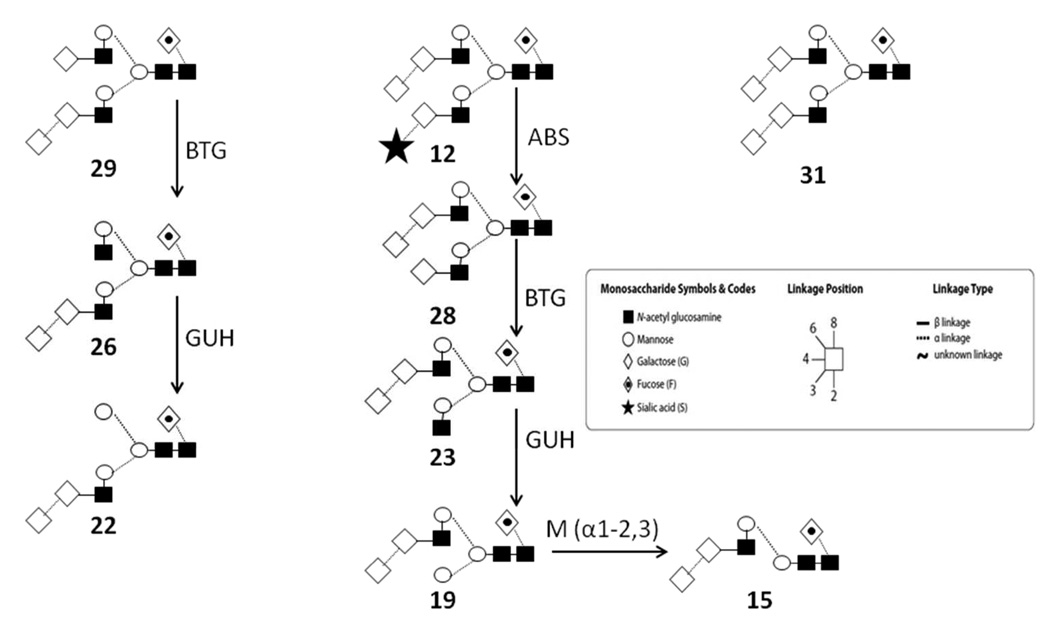
Further exoglycosidase digestion was applied to the APTS labeled mouse N-glycan pool to investigate the potential presence of galactose α-1–3 linked galactose epitopes. Simultaneous digestion with neuraminidase (ABS) and α-fucosidase (BKF) (trace D) resulted in a shift of all peaks (except peak 10) to lower migration times, including peak 28 (to position 25), suggesting that all of these glycans (except peak 10) possessed core fucosylation. Digestion of the glycan pool with a mixture of neuraminidase (ABS) and β-galactosidase (BTG) (trace E) resulted in the shift of all β-galactose containing structures, affecting peaks 17, 20 and 24, but not peaks 10, 11 and 13; however, the digestion with ABS and BTG caused the appearance of peak 23, suggesting that one antenna of the structure present in peak 28 terminates with a β-galactose residue. Due to its long migration time relative to the other peaks, the other antenna is potentially capped with α-1,3-Gal.
To further investigate the above interpretation, neuraminidase (ABS) and α-galactosidase (CBG) were simultaneously employed for the digestion of the mouse polyclonal IgG N-glycan pool (trace F). Following the digestion step, an increase of the peak area of 24 was observed, indicating that the structure of peak 12 contained both sialic acid and α-1,3-Gal caps, resulting in the shift to peak 24. The final electropherogram, following digestion of the total glycan pool with a mixture of neuraminidase, β-galactosidase and β-N-acetylhexosaminidase (GUH) (trace G), contained only three peaks (3, 5 and 19), suggesting that the structure corresponding to peak 19 still contained an α-1,3-Gal residue on one antenna of the biantennary structure, with complete digestion of the other arm to the antennary mannose residue (Scheme 1). The antenna displaying α-1,3-Gal was identified by the addition of α(1–2,3)-mannosidase (Mα1–2,3) to the above reaction mixture (ABS+BTG+GUH) (trace H), revealing that the α-1,3-Gal residue was present in a terminal position of the 1–6 antenna, as shown by the glycan 19 to glycan 15 transition in Scheme 1.
Scheme 1.
Characteristic α-1,3-Gal containing intact and exoglycosidase digested structures. BTG: β(1–3,4)-galactosidase; GUH: β(1–2,3,4,6)-N-acetylhexosaminidase; Mα-1–2,3: α(1–2,3)-Mannosidase. Numbers correspond to the abbreviated names in Table 1.
Table 1 lists all IgG glycans found during this study with their abbreviated structure names23 and their CE-LIF glucose unit values.19 The characteristic positionally specific GU value shifts of the various sugar residues are listed in Table 2. This data will be deposited in the CE stream of GlycoBase, the HPLC relational GU database, which is currently under final development prior to public release, (http://glycobase.nibrt.ie). In addition, UPLC-fluorescence analysis, as previously described was also performed for confirmatory purposes and the experimentally determined GU values for each oligosaccharide are presented in Table 1.19
Table 2.
Characteristic CE-LIF GU value shifts of the sugar residues.
| Sugar residue | ΔGU |
|---|---|
| 2 × (α-2,3) sialic acids on FA2G2 | −5.33 ± 0.03 |
| 1 × (α-2,3) sialic acid on FA2[6]G1, α-1,6 antenna | −2.97 ± 0.02 |
| 1 × (α-2,3) sialic acid on FA2[3]G1, α-1–3 antenna | −3.18 ± 0.02 |
| 1 × (α-1,6) fucose on trimannosyl core (M3) | +1.01 ± 0.01 |
| 1 × (β-1,4) galactose on FA2, α-1,6 antenna | +1.10 ± 0.02 |
| 1 × (β-1,4) galactose on FA2, α-1,3 antenna | +1.42 ± 0.02 |
| 1 × (α-1,3) galactose on FA2G2, α-1,6 antenna | +1.01 ± 0.01 |
| 1 × (α-1,3) galactose on FA2G2, α-1,3 antenna | +1.30 ± 0.01 |
| 1 × (β-1,2) GlcNAc on trimannosyl core, α-1,6 antenna | +0.83 ± 0.03 |
| 1 × (β-1,2) GlcNAc on trimannosyl core , α-1,3 antenna | +0.95 ± 0.02 |
| 1 × (α-1,3) Mannose (1–3) on FM2 | +1.10 ± 0.01 |
3.2 Determination of α-1,3-Gal containing structures in the NCI reference standard monoclonal antibody and two development-phase monoclonal antibody therapeutics
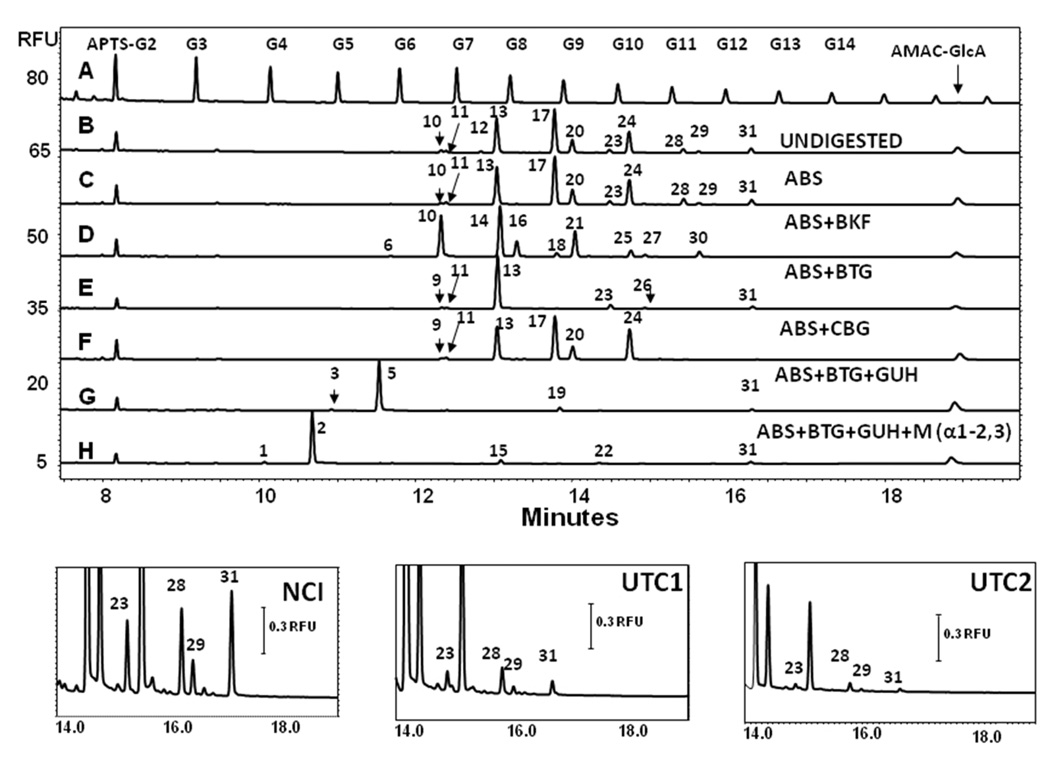
The combined CE-LIF exoglycosidase platform for the analysis of α-1,3-Gal containing structures was next applied to the N-glycan pool released from the NCI reference monoclonal antibody. The upper panel of Figure 2 presents the CE-LIF traces of APTS-labeled NCI reference standard glycans (trace B), along with the ABS, ABS+BKF, ABS+BTG, ABS+CBG, ABS+BTG+GUH and ABS+BTG+GUH+Mα1–2,3 digestion traces (B–H, respectively), compared to the malto-oligosaccharide ladder (trace A). The CE-LIF trace of the APTS-labeled intact glycan pool of this mAB revealed 11 well separated peaks, but none in the heavily sialylated region of the electropherogram, i.e. section G5-G7 of the malto-oligosaccharide ladder trace. Indeed, the results after neuraminidase (ABS) digestion resulted in a shift only for peaks 12 to 28, suggesting low quantities of sialylated structures in this particular monoclonal antibody.
Figure 2.
CE-LIF traces of the exoglycosidase digestion based structural annotation of N-glycans present in the NCI reference standard monoclonal antibody (upper panel). The three lower panels show the relevant sections of CE-LIF traces showing the peaks corresponding the structures containing the α-1,3-Gal moieties of the N-glycans of the NCI reference standard monoclonal antibody (NCI) and two development-phase monoclonal antibody therapeutics (UTC 1 and UTC2). Exoglycosidase digestion enzyme combinations, separation conditions and bracketing standards were the same as of in Figure 1.
Further digestion with the above enzyme combinations revealed several α-1,3-Gal residue related peaks (23, 28, 29 and 31, see structures in Table 1). These peaks did not shift following ABS treatment (trace C), but all shifted by approximately 1 GU after fucosidase treatment (trace D). Peaks 28 and 29 were altered by one galactose ΔGU value, suggesting a single β-galactose residue in one of the arms (trace E). Most importantly, all four glycans shifted by the expected ΔGU value (1.01–1.30) after the α-Gal specific CBG digestion (trace F). Scheme 1 shows the structural interpretation of the changes after treatment with the various exoglycosidase enzymes. Based on calculations, glycan 29 is assumed to contain one α-1,3-Gal residue on the 1–3 arm of the biantennary structure and interestingly, after BTG and GUH digestion, the resulting products of 26 and 22 still contained the α-1,3-Gal residue. Similar observations were noted for the oligosaccharide present in peak 28 with the α-1,3-Gal residue remaining on its 1–6 arm, following BTG and GUH digestion. Treating glycan 19 with Mα-1–2,3 revealed the location of the α-1,3-Gal residue to be on the 1–6 arm. Glycan 31 holds two α-1,3-Gal residues on both arms of the biantennary structure. Thus, the important immunogenic epitope of terminal α-1,3-Gal residues in this mAb was found in structures corresponding to peaks 12, 23, 28, 29 and 31. This ability to accurately determine the position of the α-1,3-Gal residues is of importance as the immunogenic potential of glycans bearing the alpha gal on the antenna extending from the α-1,3 mannose residue may be higher due to the accessibility of the epitope, based on its position, to circulating anti-α-galactose antibodies.
The three lower panels of Figure 2 show the relevant sections of CE-LIF traces of the APTS-labeled intact N-glycans from the NCI reference standard monoclonal antibody (NCI) and the two development-phase monoclonal antibody therapeutics (UTC 1 and UTC2). In addition to the structure corresponding to peak 12 (Table 1), the same α-1,3-Gal containing structures were found in all analyzed samples, i.e. peaks 23, 28, 29 and 31 corresponding to FA2G1Gal[6]1, FA2G2Gal[6]1, FA2G2Gal[3]1 and FA2G2Gal2, respectively. The total amounts of α-1,3-Gal containing structures were found to be 9.72 ± 0.02%, 1.50 ± 0.02% and 1.11 ± 0.02%, all n = 5, for the NCI reference material and the UTC 1 and UTC 2 development phase antibodies, respectively, see Table 3. No glycans were found in the Fab fractions of the investigated samples.
Table 3.
Corrected peak areas and the calculation %RSD for oligosaccharides containing α-1,3-Gal epitopes in the N-glycan pool of the NCI Reference Standard.
| NCI | Peak Number | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Releas e |
10 | 11 | 12 | 13 | 17 | 20 | 23 | 24 | 28 | 29 | 31 | α-1,3-Gal content, % |
|
| 1 | Corrected peak area Corrected peak area % |
12317 2.09 |
9404 1.59 |
5394 0.91 |
175374 29.71 |
193566 32.79 |
55868 9.46 |
12349 2.09 |
85985 14.57 |
15424 2.61 |
6013 1.02 |
18650 3.16 |
9.80 |
| 2 | Corrected peak area Corrected peak area % |
12885 2.06 |
10602 1.70 |
5545 0.89 |
186217 29.78 |
204462 32.70 |
59587 9.53 |
12865 2.06 |
90697 14.50 |
16748 2.68 |
6330 1.01 |
19368 3.10 |
9.73 |
| 3 | Corrected peak area Corrected peak area % |
9691 2.02 |
7169 1.49 |
4370 0.91 |
142603 29.71 |
157837 32.88 |
45510 9.48 |
10099 2.10 |
70280 14.64 |
12405 2.58 |
4933 1.03 |
15103 3.15 |
9.77 |
| 4 | Corrected peak area Corrected peak area % |
10532 2.00 |
8431 1.60 |
4685 0.89 |
157444 29.84 |
173171 32.82 |
50289 9.53 |
10947 2.08 |
76785 14.55 |
13620 2.58 |
5333 1.01 |
16345 3.10 |
9.65 |
| 5 | Corrected peak area Corrected peak area % |
14333 2.24 |
10419 1.63 |
5600 0.87 |
176665 27.56 |
219307 34.22 |
61205 9.55 |
13207 2.06 |
97335 15.19 |
15999 2.50 |
6594 1.03 |
20299 3.17 |
9.63 |
3.3 Limit of detection and limit of quantitation (LOQ) for α-1,3-Gal containing structures
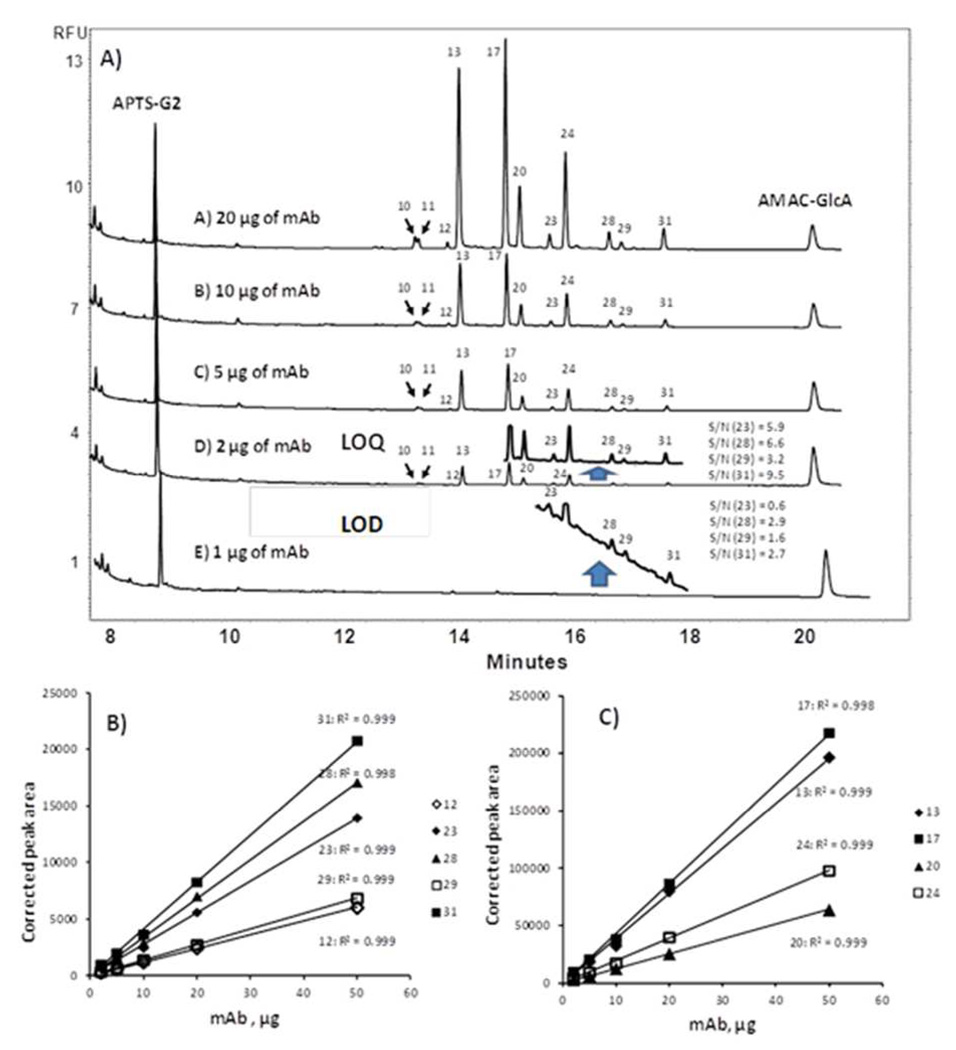
High analytical sensitivity is required to ensure the detection of even trace levels of the α-1,3-Gal residues to guarantee product safety. The NCI reference standard monoclonal antibody was used for the limit of detection and limit of quantitation (LOQ) determination of α-1,3-Gal containing structures. This standard contained approximately 10% of α-1,3-Gal residue bearing glycans. Experimentally, 20, 10, 5, 2 and 1 µg of the NCI reference standard monoclonal antibody were N-deglycosylated; the corresponding CE-LIF traces are compared in Figure 3. At the lower IgG levels the insets show expansion of the relevant section of the electropherogram spanning the migration window of most α-1,3-Gal residue containing structures (peaks 2–31). As noted above, an average of 9.72 ± 0.02% of α-1,3-Gal containing structures were found from the release reactions of 20, 10, 5, and 2 µg of the NCI reference standard suggesting detection linearity in this range. Analysis of 1 µg reference standard material resulted in 10.92% α-1,3-Gal structures, almost 40% different from the linear quantitation range. Based on the detection linearity data and the signal to noise ratio in these traces, the LOQ was determined as 2 µg mAb (average signal to noise ratio of 6.3) and 1 µg as LOD (average signal to noise ratio of 2.0). These values represent ultrahigh sensitivity detection for the CE-LIF methodology.
Figure 3.
(A) Limit of detection and limit of quantitation (LOQ) for α-1,3-Gal containing structures. Separation conditions and bracketing standards are the same as in Figure 1. Estimation of linearity for (B) α-1,3-Gal containing structures and (C) the main oligosaccharides present, numbering in the legend refers to peak identity in Table 1.
DISCUSSION
In this paper, we have presented a rapid and sensitive method for the identification and quantitation of N-linked oligosaccharides containing potentially immunogenic α-1,3-Gal epitopes using a combination of CE-LIF and exoglycosidase digestion. The methodology is conceptually simple and employs commercially available consumables and kits, thereby facilitating easy implementation within the biopharmaceutical arena. As noted previously, using the current CE-LIF separation parameters, an incomplete separation of fucosylated and afucosylated glycans was achieved.19 However, we recently reported an alternative, complementary and rapid high performance CE based separation strategy for complete resolution of these functionally important oligosaccharides.24 Using exoglycosidase digestion, we confidently identified α-1,3-Gal epitopes on both fucosylated and afucosylated neutral and monosialylated oligosaccharides. The use of upper and lower bracketing standards aided in improving the annotation of analytical peaks with GU values; indeed, the GU data obtained in the present study was in close agreement with that previously presented,19 wherein the characterization of polyclonal IgG extracted from the serum of healthy donors was performed.19 GU values identified for α-1,3-Gal containing oligosaccharides will be deposited into the CE-LIF stream of GlycoBase, thereby facilitating easy application of the technology by the wider research and regulatory community.25 The high precision offered by the platform allows users to confidently identify or confirm the presence or absence of α-1,3-Gal containing N-glycans on their therapeutic proteins with exoglycosidase digestion only required for further confirmation. Additionally, the Δ GU values presented in Table 2 enable the prediction of the migration time in a linkage and positional specific manner due to the excellent selectivity and reproducibility of the CE separation, a feature that can also be used for tentative structural assignment.
A distinct advantage of the current approach is the ability to characterize the antennary position of attachment of the α-1,3-Gal epitope. Interestingly, the majority of α-1,3-Gal epitopes identified were noted to be on the antenna extending from the α-1–6 linked core mannose residue with smaller quantities of α-1,3-Gal epitopes present on the antenna extending from the α-1–3 linked core mannose residue. Therefore, the question regarding antennary position and antigenic potential of the α-1,3-Gal epitope can be considered. The antenna extending from the α-1–6 linked core mannose residue is known to make multiple interactions with the polypeptide chain of the CH2 domain; such interactions are known to be crucial for conformational stability. On the other hand, the antenna extending from the α-1–3 linked core mannose residue is positioned in the interstitial space between the CH2 domains.26 The addition of sialic acid appears to be more favorable on the α-1–3 antenna in contrast to the α-1–6 antenna, where the presence of negatively charged amino acids in the vicinity hinders the addition of sialic acid to the α-1–6 antennary galactose.27–28 The observation of increased levels of α-1,3-Gal epitopes attached to the α-1–6 antennary galactose thus suggest that further processing of this particular residue is not governed by steric factors but rather by the nature of the polypeptide sequence present in the local environment. Furthermore, as the α-1–6 antenna is in close and multiple contact with the polypeptide chain, the antigenic potential of an α-1,3-Gal epitope present on this arm of the oligosaccharide is likely lower than that of an α-1,3-Gal epitope linked to the galactose of the α-1–3 antenna due to the more direct accessibility for recognition by anti-α-1,3-Gal antibodies. However, possible alteration of the local polypeptide structure following the addition of an additional monosaccharide to the α-1–6 antenna thereby creating confirmations that may potentially also elicit an immune response or alter the ability of the antibody to evoke Fc effector functions should also be considered. As was previously noted, the antigenic potential of α-1,3-Gal residues on the oligosaccharides present in the Fc region of the molecule are lower than those in the Fab region, likely due to the accessibility of Fab oligosaccharides, if present, for recognition by circulating anti-α-1,3-Gal antibodies.29 Although further research is required to evaluate the role of positional attachment and antigenic potential, the current methodology offers drug developers a platform to perform in depth characterization of their molecules, thereby facilitating more complete structural information to assess immunogenic potential.
CONCLUSIONS
A rapid and reproducible method for ultrasensitive analysis of therapeutic antibody glycosylation containing immunogenic α-1,3-Gal epitopes is described. Our studies demonstrate the high resolution and high precision of CE-LIF with the specificity of exoglycosidase digestion sequence (ABS, ABS+BKF, ABS+BTG, ABS+CBG, ABS+BTG+GUH and ABS+BTG+GUH+Mα-1–2,3) are an information rich methods capable of providing direct structural annotation of N-linked glycans of therapeutic antibodies. Our approach has established the GU values of five important α-1,3-Gal containing structures, which can be directly used in the future for data analysis without the necessity for exoglycosidase digestions. The combined technique was evaluated on a polyclonal mouse IgG model and then applied to the ultrasensitive α-1,3-Gal structural analysis of an NCI reference standard monoclonal antibody (expressed in SP2/0) and two development-phase monoclonal antibody therapeutics (also expressed in SP2/0). The focus of our work was to determine possible immunogenic epitopes in therapeutic monoclonal antibodies with ultrahigh sensitivity through the use of commercially available instrumentation and reagents, thereby allowing the method to be readily adaptable within the scientific community. The ultrasensitive and high resolution technique described in this paper may also play an important role in the evaluation of comparability of innovator products and biosimilars, revealing the actual structures of glycans of interest. As a continuation of this work, a highly sensitive method will be described in a separate paper on the analysis of N-glycolylneuraminic acid, another possible source of immunogenicity in therapeutic antibodies.
ACKNOWLEDGEMENT
This research was supported by NIH GM 15847. The authors also thank Beckman Coulter, Prozyme and PhyNexus for their generous gifts of products to support this study. Contribution No.1000 from the Barnett Institute.
Abbreviations
- APTS
8-aminopyrene-1,3,6-trisulfonic-acid trisodium salt
- AMAC
2-aminoacridone
- ABS
α(2–3,6,8,9)-neuraminidase from Arthrobacter ureafaciens
- BKF
α(1–2,3,4,6)-fucosidase from bovine kidney
- BTG
β(1–3,4)-galactosidase from bovine testis
- CBG
α(1–3,4,6)-galactosidase from green coffee bean
- GU
Glucose Unit
- GUH
β(1–2,3,4,6)-N-acetylhexosaminidase from jack bean
- Mα1–2,3
α(1–2,3)-mannosidase from Chryseobacterium meningosepticum
REFERENCES
- 1.Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, Slebos RJ, Zhou Q, Gold D, Hatley T, Hicklin DJ, Platts-Mills TA. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. 2008;358(11):1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobata A. The N-linked sugar chains of human immunoglobulin G: their unique pattern, and their functional roles. Biochim Biophys Acta. 2008;1780(3):472–478. doi: 10.1016/j.bbagen.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Thakur D, Rejtar T, Karger BL, Washburn NJ, Bosques CJ, Gunay NS, Shriver Z, Venkataraman G. Profiling the glycoforms of the intact alpha subunit of recombinant human chorionic gonadotropin by high-resolution capillary electrophoresis-mass spectrometry. Anal Chem. 2009;81(21):8900–8907. doi: 10.1021/ac901506p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koike C, Uddin M, Wildman DE, Gray EA, Trucco M, Starzl TE, Goodman M. Functionally important glycosyltransferase gain and loss during catarrhine primate emergence. Proc Natl Acad Sci U S A. 2007;104(2):559–564. doi: 10.1073/pnas.0610012104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosques CJ, Collins BE, Meador JW, 3rd, Sarvaiya H, Murphy JL, Dellorusso G, Bulik DA, Hsu IH, Washburn N, Sipsey SF, Myette JR, Raman R, Shriver Z, Sasisekharan R, Venkataraman G. Chinese hamster ovary cells can produce galactose-alpha-1,3-galactose antigens on proteins. Nat Biotechnol. 2010;28(11):1153–1156. doi: 10.1038/nbt1110-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian J, Liu T, Yang L, Daus A, Crowley R, Zhou Q. Structural characterization of N-linked oligosaccharides on monoclonal antibody cetuximab by the combination of orthogonal matrix-assisted laser desorption/ionization hybrid quadrupole-quadrupole time-of-flight tandem mass spectrometry and sequential enzymatic digestion. Anal Biochem. 2007;364(1):8–18. doi: 10.1016/j.ab.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 7.EMEA. Guideline on development, production, characterisation and specifications for monoclonal antibodies and related products. 2009. [Google Scholar]
- 8.Smith DF, Larsen RD, Mattox S, Lowe JB, Cummings RD. Transfer and expression of a murine UDP-Gal:beta-D-Gal-alpha 1,3-galactosyltransferase gene in transfected Chinese hamster ovary cells. Competition reactions between the alpha 1,3-galactosyltransferase and the endogenous alpha 2,3-sialyltransferase. J Biol Chem. 1990;265(11):6225–6234. [PubMed] [Google Scholar]
- 9.Bynum MA, Yin H, Felts K, Lee YM, Monell CR, Killeen K. Characterization of IgG N-glycans employing a microfluidic chip that integrates glycan cleavage, sample purification, LC separation, and MS detection. Anal Chem. 2009;81(21):8818–8825. doi: 10.1021/ac901326u. [DOI] [PubMed] [Google Scholar]
- 10.Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(5):721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 11.Kurihara T, Min JZ, Hirata A, Toyo'oka T, Inagaki S. Rapid analysis of N-linked oligosaccharides in glycoproteins (ovalbumin, ribonuclease B and fetuin) by reversed-phase ultra-performance liquid chromatography with fluorescence detection and electrospray ionization time-of-flight mass spectrometry. Biomed Chromatogr. 2009;23(5):516–523. doi: 10.1002/bmc.1147. [DOI] [PubMed] [Google Scholar]
- 12.Ruhaak LR, Deelder AM, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2009;394(1):163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi N. Three-dimensional mapping of N-linked oligosaccharides using anion-exchange, hydrophobic and hydrophilic interaction modes of high-performance liquid chromatography. J Chromatogr A. 1996;720(1–2):217–225. doi: 10.1016/0021-9673(95)00328-2. [DOI] [PubMed] [Google Scholar]
- 14.Chelius D, editor. Well Characterized Biologicals. Washington DC: 2011. Fully Automated N-glycosylation analysis of monoclonal antibodies. 'Ed. '^'Eds.' 'Vol.' p^pp. [Google Scholar]
- 15.Guttman A. High-resolution carbohydrate profiling by capillary gel electrophoresis. Nature (London) 1996;380(6573):461–462. doi: 10.1038/380461a0. [DOI] [PubMed] [Google Scholar]
- 16.Guttman A, Chen F-TA, Evangelista RE, Cooke N. High-resolution capillary gel electrophoresis of reducing oligosaccharides labeled with 1-aminopyrene-3,6,8-trisulfonate. Analytical Biochemistry. 1996;233:234–242. doi: 10.1006/abio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 17.Rudd PM, Colominas C, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF, Dwek RA. A high-performance liquid chromatography based strategy for rapid, sensitive sequencing of N-linked oligosaccharide modifications to proteins in sodium dodecyl sulphate polyacrylamide electrophoresis gel bands. Proteomics. 2001;1(2):285–294. doi: 10.1002/1615-9861(200102)1:2<285::AID-PROT285>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Guttman A, Ulfelder KW. Exoglycosidase matrix-mediated sequencing of a complex glycan pool by capillary electrophoresis. J Chromatogr A. 1997;781(1–2):547–554. doi: 10.1016/s0021-9673(97)00724-3. [DOI] [PubMed] [Google Scholar]
- 19.Mittermayr S, Bones J, Doherty M, Guttman A, Rudd PM. Multiplexed analytical glycomics: rapid and confident IgG N-glycan structural elucidation. J Proteome Res. 2011;10(8):3820–3829. doi: 10.1021/pr200371s. [DOI] [PubMed] [Google Scholar]
- 20.von Pawel-Rammingen U, Johansson BP, Bjorck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002;21(7):1607–1615. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo Z, Guttman A, Rejtar T, Karger BL. Improved sample preparation method for glycan analysis of glycoproteins by CE-LIF and CE-MS. Electrophoresis. 2010;31(8):1389–1395. doi: 10.1002/elps.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royle L, Radcliffe CM, Dwek RA, Rudd PM. Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods Mol Biol. 2006;347:125–143. doi: 10.1385/1-59745-167-3:125. [DOI] [PubMed] [Google Scholar]
- 23.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics. 2009;9(15):3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 24.Szabo Z, Guttman A, Bones J, Karger BL. Rapid high-resolution characterization of functionally important monoclonal antibody N-glycans by capillary electrophoresis. Analytical chemistry. 2011;83(13):5329–5336. doi: 10.1021/ac2007587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24(9):1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 26.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325(5):979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 27.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 28.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 29.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]