Abstract
The spectral reflectance signature of living organisms provides information that closely reflects their physiological status. Because of its high potential for the estimation of geomorphic biological parameters, particularly of gross photosynthesis of plants, two-dimensional spectroscopy, via the use of hyperspectral instruments, has been widely used in remote sensing applications. In genetics research, in contrast, the reflectance phenotype has rarely been the subject of quantitative analysis; its potential for illuminating the pathway leading from the gene to phenotype remains largely unexplored. In this study, we employed hyperspectral imaging techniques to identify Arabidopsis mutants with altered leaf pigment status. The techniques are comprised of two modes; the first is referred to as the ‘targeted mode’ and the second as the ‘non-targeted mode’. The ‘targeted’ mode is aimed at visualizing individual concentrations and compositional parameters of leaf pigments based on reflectance indices (RIs) developed for Chls a and b, carotenoids and anthocyanins. The ‘non-targeted’ mode highlights differences in reflectance spectra of leaf samples relative to reference spectra from the wild-type leaves. Through the latter approach, three mutant lines with weak irregular reflectance phenotypes, that are hardly identifiable by simple observation, were isolated. Analysis of these and other mutants revealed that the RI-based targeted pigment estimation was robust at least against changes in trichome density, but was confounded by genetic defects in chloroplast photorelocation movement. Notwithstanding such a limitation, the techniques presented here provide rapid and high-sensitive means to identify genetic mechanisms that coordinate leaf pigment status with developmental stages and/or environmental stress conditions.
Keywords: Arabidopsis, Hyperspectral imaging, Mutant identification, Pigment, Quantitative phenotyping
Introduction
The color of living organisms is often diagnostic of their physiological status. Combined with morphology, it accounts for much of the visible phenotype. In plants, for example, adverse growth environments, such as low temperatures and macronutrient deficiencies, lead to leaf color changes from green to yellow-green, and sometimes even to red (Diaz et al. 2006, Hasdai et al. 2006). These changes can result from passive necrotic cell death, but most are considered relevant as active adaptive responses aimed at evading photooxidative damage (Niyogi 1999, Havaux and Kloppstech 2001, Steyn et al. 2002, Dall’Osto et al. 2010). While leaf color is governed both by cellular structure and biochemical components, the concentration and composition of pigments, including Chls a and b, carotenoids (Cars) and anthocyanins (Anths) are the primary determinants of leaf color. Accordingly, there has been a long line of research trying to make full use of the color, namely the visible spectroscopic signature, for absolute quantification of these pigments (for reviews, see Porra 2002, Ustin et al. 2009).
Traditionally used wet chemical pigment analysis included leaf extraction with organic solvents and spectrophotometric absorbance measurement of the extracts. In order to eliminate the necessity for chromatographic separation, equations for calculating individual pigment concentrations in crude extracts have been elaborated (Arnon 1949, Lichtenthaler 1987, Gitelson et al. 2001, Porra 2002). However, such ‘gold standard’ methods still require time- and labor-intensive procedures for sample preparation. Moreover, due to the invasive nature of the procedures, leaf samples once used for chemical pigment analysis are no longer able to be used for additional biochemical assays for enzymatic activities and/or quantification of other metabolites, and to temporal analysis of pigment dynamics in response to environmental and/or developmental cues.
In the research area of remote sensing, alternative solutions for leaf pigment analysis with non-invasive optical methods have been proposed (for reviews, see Blackburn 2007, Ustin et al. 2009). Particular efforts have been devoted to the development of robust algorithms for total Chl determination based on spectral reflectance measurements (e.g. Chappelle et al. 1992, Blackburn 1998, Datt 1998, Richardson et al. 2002, Sims and Gamon 2002, Gitelson et al. 2003, le Maire et al. 2004, Gitelson et al. 2006a, and references therein), as they help estimate photosynthetic activity, and accordingly the gross primary production, from the leaf to canopy scale (Chappelle et al. 1992, Gitelson et al. 2006b). Although much less frequent, algorithms for estimating total Car and Anth concentrations from the spectral reflectance signature have also been reported (Chappelle et al. 1992, Blackburn 1998, Datt 1998, Gitelson et al. 2001, Gitelson et al. 2002, Sims and Gamon 2002, Gitelson et al. 2006a). However, except for the occasional exploratory use of commercial Chl meters, optical methods for leaf pigment analysis have yet to become a widespread technique in the research area of plant genetics. At least to our knowledge, there have been no reports utilizing genetic bioresources with altered leaf pigment metabolism to adjust the algorithms for optical pigment estimation. Also, no biological studies applying such optical methods to identify genetic defects in leaf pigment metabolism have been carried out. This may be partly because the superiorities and limitations of the methods are not well recognized across different research areas. In this respect, it seems of great significance to demonstrate how they can contribute to genetic investigations, especially when two-dimensional (2D) imaging spectrophotometry is used.
In the recent post-genome research, quantitative phenotype description is becoming increasingly important, heading toward understanding the whole biological system from genome to phenome (Edwards and Batley 2004, Kuromori et al. 2009). Of the major elements of the visible phenotype, morphological traits have become the subject of computational analysis in a model plant Arabidopsis, and their developmental dynamics and ecotype-specific characteristics have been defined quantitatively (Kaminuma et al. 2004, Mündermann et al. 2005). On the other hand, although physical definition and measuring methods of colors, the spectroscopic traits, appear simpler and more pervasive than those of morphology, to our knowledge, systematic phenomic analysis of leaf color has yet to be undertaken. It is important to note, in this context, that conventional digital imaging procedures usually involve considerable reduction and deterioration of color information. For example, in a standard red–green–blue (RGB) image, each of the three bands has a wide and partially overlapping spectral sensitivity (i.e. low spectral resolution) and therefore the subtle color information is not retained. Moreover, the absence of a standardized white balancing procedure, also referred to as normalization or calibration, makes it impractical to attempt a quantitative comparison of colors between images captured using different equipment and photographic conditions.
In this study, we explored the potential of hyperspectral imaging techniques as a method of quantitative genetics for evaluating leaf color phenotypes in Arabidopsis. Based on quantitative spectral reflectance data, an experimental system for visualizing individual concentrations and compositional parameters of major leaf pigments was constructed. On the other hand, the ratio of spectral reflectance (relative to the reference spectra) was used to highlight subtle color differences between and within leaves. These techniques are not only effective in identifying mutants with an altered leaf pigment status, but also serve as unique building blocks toward enabling chemometric imaging and profiling of wider biochemical components in Arabidopsis leaves.
Results
Targeted determination of individual pigment concentrations: technical background
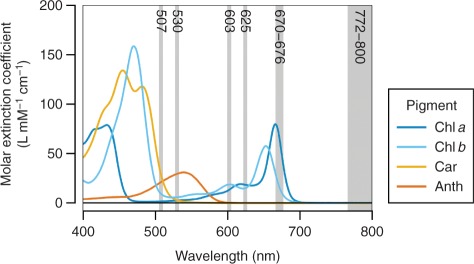
To achieve high-accuracy estimation of leaf physiological status based on spectral reflectance measurement, it is of practical interest to investigate reflectance indices (RIs) that help determine individual pigment concentrations. There are, however, several difficulties that need to be resolved in developing pigment-specific RIs. First, we still do not have much knowledge about in situ spectroscopic properties of individual pigments, as their absorption spectra can be influenced by interaction with solvents and other solutes (Lichtenthaler 1987, Porra 2002). Secondly, there is considerable overlap between the absorption spectra of different pigments (Fig. 1), which demands advanced spectral decomposition techniques to estimate the accurate relative contribution of a pigment of interest to the total reflectance. Thirdly, there is naturally a close interrelationship between the concentrations of different pigments (see Supplementary Fig. S1A); a series of leaf samples with diverse pigment compositions are needed to establish the specificity of the RIs. Fourthly, the range of factors other than pigments that influence the reflectance spectra must be taken into consideration; it is particularly important that the measured spectra be adjusted for differences in backscatter between leaf samples.
Fig. 1.
Absorption spectra of the major leaf pigments. The spectra are plotted in terms of molar extinction coefficient. Standards of Chls and Anth (cyanidin chloride) were prepared in neutral and acid methanol, respectively. Car (β-carotene) was dissolved in acetone, because of its low solubility and therefore the difficulty in preparing a homogeneous solution in methanol from its dried form. The maxima are adjusted to the values reported by Lichtenthaler (1987).
Through continued efforts to overcome these difficulties, several empirical models for relating pigment concentrations to the reflectance spectra have been proposed (for reviews, see Blackburn 2007, Ustin et al. 2009). At the individual leaf scale, when applied to limited numbers of species grown under naturally occurring environmental conditions, some of these have been proven to be reasonable indicators of actual pigment concentrations. On the other hand, there has been little agreement between different studies on the target plant species, the equipment and conditions for spectral reflectance measurements, and the methods employed for spectral signal analysis.
Hyperspectral reflectance imaging
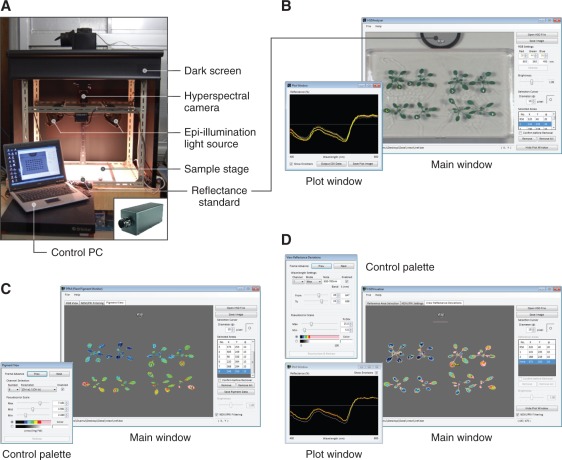
The configuration of the hyperspectral imaging set-up used in this study is basically identical to the one proposed previously (Lenk et al. 2007) (Fig. 2A). This configuration was originally applied to multispectral rather than hyperspectral imaging. Both of these spectral imaging techniques allow extraction of information that cannot be retained in the standard RGB images, due to the higher spectral resolution of the techniques. Multispectral imaging, however, produces discrete, not contiguous, spectral information. Hence, the reflectance images were captured using a line-scanning hyperspectral camera (HSC1700, the earliest commercial model manifactured in 2007; Hokkaido Satellite). This camera is capable of aquiring 8-bit VGA (640×480 pixel) images for 72 contiguous wavebands from 400 to 800 nm (5.6 nm bandwidth). In our set-up, the scanning area was approximately 140×120 mm; the resulting spatial resolution of the images was 116 and 100 dots per inch in the horizontal and vertical dimensions, respectively. As the epi-illumination light source, two double-ended 250 W halogen lamps were used. When capturing Arabidopsis images, a 50% reflectance standard (SRS-050-010; Labsphere) was placed in the same visual field. A hyperspectral image of a sheet of homogenous white paper (No. 526; ADVANTEC) was captured under the same conditions, and was used to calibrate the Arabidopsis images for spatial non-uniformity of illumination.
Fig. 2.
Equipment for hyperspectral reflectance imaging and data analysis. The programs shown in B–D are provided as Supplementary File S1 of this article (see Supplementary Text S1 for legends and methods of operation). (A) Hyperspectral image acquisition system. The system is basically composed of a hyperspectral camera directed downward to the sample stage and epi-illumination halogen lights. The inset shows the VIS/NIR hyperspectral camera HSC1700 (early 2007 model). (B) Screenshot of HSD Analyzer software. The software facilitates calibration and extraction of numerical reflectance data from the areas of interest in hyperspectral images. (C) Screenshot of PPM software. The software allows visualization of individual concentrations and compositional parameters of major pigments in Arabidopsis leaves. (D) Screenshot of HSD Visualizer software. The software displays the deviation of spectral reflectance relative to the reference spectra (usually from the wild type) as a pseudocolor image.
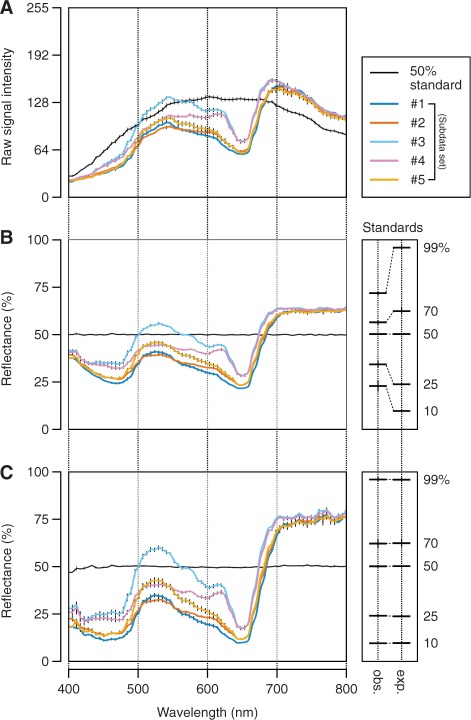
Fig. 3A shows an example of raw reflective signals recorded in the areas of the standard and leaves of Arabidopsis plants from five different subdata sets (Table 1), which are described in detail in the following sections. As is evident from the figure, the irradiation spectrum from the light source was not flat over wavelengths (Fig. 3A, black line). Hence, the reflectance spectrum in each pixel of the images was calibrated by linear regression against the 50% reflectance standard (Fig. 3B). To semi-automate these calibration processes and thus facilitate retrieval of numerical spectral reflectance values from the hyperspectral images, we developed the software HSD Analyzer (Hyperspectral Data Analyzer; Fig. 2B), which is provided as Supplementary File S1 (see Supplementary Text S1 for legends and methods of operation). The subsequent development of RI-based equations for ‘targeted’ pigment estimation (Equations 2–12) and of PPM software (Fig. 1C) depended primarily on the analysis using HSD Analyzer.
Fig. 3.
Retrieval of reflectance data from hyperspectral images. Data in each plot are the mean ± SD (SD is shown by a vertical bar) derived from measurements in five different leaves of Arabidopsis plants from each subdata set or 25 different areas in the 50% reflectance standard. Details of each subdata set (#1–5) are summarized in Table 1. Plants grown under normal conditions (as defined in Table 1) were used. (A) Raw signal output from the hyperspectral imaging sensor. Values are in the range 0–255 (8-bit). Note that the signal detected in the areas of the standard is proportional to the irradiation intensity of the light source. (B) Reflectance spectra of leaves as determined by linear regression against the 50% reflectance standard. The means ± SD of the calculated (observed, abbreviated as obs.) and actual (expected, abbreviated as exp.) reflectance of five different standards (10, 25, 50, 70 and 99%) through the entire 72 wavebands (400–800 nm) are indicated on the right. Expected values are from the calibration certificate of the standards provided by the manufacturers. Except for the 50% standard, the observed and expected reflectance did not agree with each other at this step. (C) Reflectance spectra of leaves after second-order correction for the sensor’s non-linearity. The values of the coefficients in quadratic functions used to transform the reflectance at each waveband in B to that in C are shown in Supplementary Table S1. Here, the observed and expected reflectance of the five standards agree closely with each other as shown on the right.
Table 1.
Combinations of genotypes and growth conditions used to obtain the five subdata sets
| Subdata set | Genotype | Growth mediuma |
Phenotypes | |
|---|---|---|---|---|
| Primary | Secondary | |||
| #1 | WT | A | B | No apparent Anths |
| #2 | WT | A | Cb | Accumulate Anths |
| #3 | ch1-3 | A | B | Chl b-deficient mutant, no apparent Anths |
| #4 | ch1-3 | A | Cb | Chl b-deficient mutant, accumulate Anths |
| #5 | tt4 | A | B | Anth-deficient mutant |
Plants germinated on the primary medium were transplanted on day 8 to the secondary medium. They were grown for another 6–10 d before being subjected to hyperspectral reflectance imaging and chemical pigment analysis. WT, wild type.
aCompositions of growth media indicated by each symbol are as follows: A, 1× MS (0/1× NH4NO3, 1× KH2PO4), 1% (w/v) sucrose; B, 1× MS (0/1/2× NH4NO3, 1×KH2PO4), 0/1/5 μM norflurazon; C, 1× MS (0/1/2× NH4NO3, 10−3× KH2PO4). 1× concentrations for NH4NO3 and KH2PO4 are 20.6 and 1.25 mM, respectively. Nutrient concentrations regarded as ‘normal’ are indicated in bold.
bSome of the plants were wounded in root tissues after being transplanted to the secondary medium.
However, although the calibration made so far, and accordingly the output of HSD Analyzer, was sufficient to detect subtle differences in reflectance phenotypes between leaf samples, the absolute reflectance values are not yet correct, largely due to the sensor’s non-linearity (see Fig. 3B, right side). Thus, more accurate reflectance values were calculated by second-order correction using quadratic functions (y = ax2 + bx + c, where x and y are thought of as reflectance values before and after the correction, respectively) regressed for each waveband from a measurement of five different reflectance standards [50%, Labsphere; 10% (SG3083/24), 25% (SG3089/52), 70% (SG3089/41), and 99% (SG3089/9), SphereOptics] (Fig. 3C). The coefficients in the quadratic functions used and the determination coefficient (r2) of the correction are shown in Supplementary Table S1. Such a second-order correction, however, will not necessarily be required if recent high-performance hyperspectral cameras are used; the optical systems have been improved enough to keep high sensor linearity in their measurable light intensity range.
Besides the sensor’s non-linearity, the low bit-depth of the camera was another difficulty for our system. Due to the uneven spectral distribution of the light source, the maximum and minimum raw signal intensities recorded in the 50% standard were 131.5 ± 2.32 (at 603 nm) and 28.9 ± 1.29 (at 400–406 nm), respectively (n = 25, where each measurement is the averaged intensity from a 20 pixel diameter area) (Fig. 3A). Accordingly, after the non-linearity correction, the reflectance resolution was 0.39–1.0% in the wavelength range above 439 nm, whereas the resolution was no better than 1.2–2.5% in the shorter wavelengths.
Reflectance index model
While most empirical models for optical pigment estimation have been aimed at relating the concentration of a single pigment to the reflectance spectra, Gitelson and his colleagues proposed a flexible conceptual model that is applicable to the estimation of multiple pigment concentrations (Gitelson et al. 2003, Gitelson et al. 2006a). We therefore started with their ‘three-band model’ to develop RIs for Chl a and b, Anth and Car estimation in Arabidopsis leaves. The model is expressed as:
| (1) |
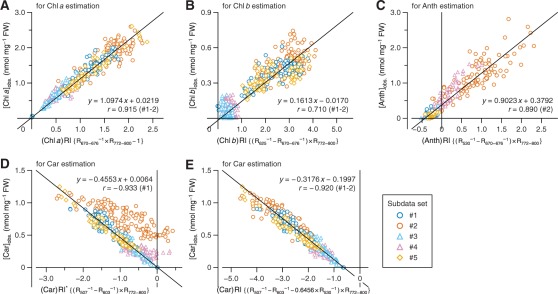
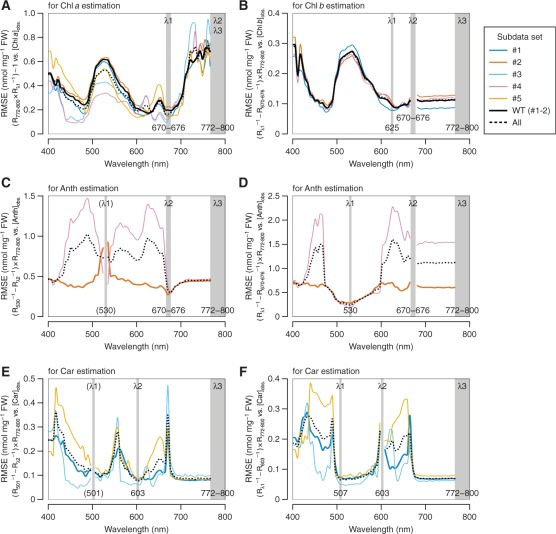
where Cp denotes the target pigment concentration, and asks to find three wavebands (λ1–λ3) for each pigment that maximize the linear correlation between Cp and the RI on the right-hand side. At the first waveband (λ1), the reflectance (Rλ) should be highly sensitive to absorption by the pigment of interest. The second waveband (λ2) is introduced with the purpose of removing the extraneous effects of other pigments on Rλ1. The third waveband (λ3) is selected from the spectral range where the backscattering rather than pigment absorption controls the total reflectance. If the resulting correlation was not satisfactory enough, a custom modification was made to the model. This was the case for the development of the Car-specific RI, designated as (Car)RI, that is robust against the presence of Anths (Equation 10; see Fig. 5E).
Fig. 5.
Relationship between pigment concentration and reflectance index (RI) in the calibration data set. Details of each subdata set (#1–5) are summarized in Table 1. The vertical axis represents the chemically determined (observed, abbreviated as obs.) concentration of the indicated pigment. The horizontal axis represents the value of RI developed for Chl a (A), Chl b (B), Anth (C) or Car (D and E) estimation. (Car)RI* in D is a tentative index, which was improved to (Car)RI in E. The equation of each RI is indicated in the horizontal label. For each plot, a regression line fitted to the wild-type subdata set(s) (#1 and/or #2) is shown, along with the equation and correlation coefficient (r) of the regression line. The statistical details of the analysis are summarized in Table 2.
Data sets
To collect a series of leaf samples with different Chl concentrations, plants were grown on agar media containing varying amounts of nitrogen (N) source (Table 1; subdata sets #1, 3 and 5). This is reasonable because a certain fraction of total leaf N is allocated to Chls (Evans 1989). The Anth concentrations were negligibly low in all plants in these subdata sets, suggesting that the minimum N concentration used here (18.8 mM as potassium nitrate) was sufficient to prevent N deficiency-induced Anth accumulation (Diaz et al. 2006). To cover the lower Chl concentration range, plants treated with norflurazon, which induces chlorosis without accompanying Anth accumulation (Jung 2004), were also included in these subdata sets. A series of leaf samples with different Anth concentrations was prepared by growing the plants on low-phosphate (P) media, or by mechanical wounding in root tissues (Trull et al. 1997, Steyn et al. 2002) (subdata sets #2 and 4). Unlike Chl a and Cars, complete depletion of Chl b and Anths does not lead to plant lethality (Espineda et al. 1999, Abrahams et al. 2002). Hence, the mutants ch1 (subdata sets #3 and 4) and tt4 (subdata set #5), which are deficient in Chl b and Anths, respectively, were used to evaluate if RI-based pigment estimation works equally well regardless of the presence or absence of these pigments. In contrast, it was difficult to disturb the strong linear correlation found between the concentrations of Chl a and Cars (Supplementary Fig. S1A). The ratio of Cars to Chl a, however, was somewhat lower in ch1 plants (subdata sets #3 and 4), especially when grown under Anth-inducing conditions (subdata set #4), than in plants of the other genotypes. We therefore considered that the assembly of the five subdata sets (#1–5) described above represents sufficiently diverse pigment compositions needed to establish specific RIs for the estimation of individual pigment concentrations. Two independent data sets consisting of the same series of subdata sets (#1–5) were prepared for calibration and validation purposes.
Chl a estimation from hyperspectral imagery
According to the precedent in the literature of the ‘three-band model’ (Gitelson et al. 2006a), λ3 was selected from the near-infrared (NIR) region. Here, the spectral range of 772–800 nm was adopted as λ3 for all pigments, because little correlation was found in any subdata sets between the reciprocal reflectance (Rλ−1) in this waveband and the concentration of any individual pigments (Supplementary Fig. S2). It has been hypothesized that Rλ−1 is nearly proportional to the sum of absorption coefficients of all pigments absorbing the light at wavelength λ (Gitelson et al. 2006a). On the other hand, spectral ranges of λ1 and λ2 were derived independently for each pigment. For Chl a, λ2 was set identical to λ3 as in the precedent, while λ1 was selected from the long-wavelength side within the red Chl a absorption band, wherein minimal or no light absorption by pigments other than Chl a takes place (Fig. 1). We also took care in determining λ1 so that the values of RI (right-hand side of Equation 1) steadily exhibit high correlation with observed (chemically determined) Chl a concentrations ([Chl a]obs.) through the five subdata sets. To meet these requirements, a spectral range of 670–676 nm appeared most suitable for λ1, as is supported by the low root mean square error (RMSE) of Chl a estimation (Fig. 4A). Collectively, the RI for the estimation of Chl a concentration, which is designated as (Chl a)RI, was summarized in the following equation:
| (2) |
Fig. 4.
Optimization of wavebands λ1 and λ2 in the model (Rλ1−1 – Rλ2−1)×Rλ3 for the estimation of individual pigment concentrations. The analysis was performed on the calibration data set. Details of each subdata set (#1–5) are summarized in Table 1. The process of model tuning for Chl a (A), Chl b (B) and Anth (C and D) estimation, or Car estimation in the absence of Anth accumulation (E and F) is indicated. The RMSE was calculated for linear regression of the model indicated in the vertical label vs. the observed concentration of the pigment of interest. A lower RMSE indicates more accurate estimation. In C and E, λ1 is indicated in parentheses due to its temporary nature; the optimal waveband was reconsidered in the second step of model tuning in D and F, respectively. WT, wild type.
The correlation between (Chl a)RI and [Chl a]obs. in the whole calibration data set is shown by a scatter plot in Fig. 5A. A linear regression fitted to the wild-type subdata sets (#1 and 2) gives an equation:
| (3) |
where [Chl a]exp. denotes the expected Chl a concentration. The RMSE of the differences between [Chl a]obs. and [Chl a]exp., and related statistical parameters are summarized in Table 2.
Table 2.
Statistics indicating the accuracy of reflectance-based pigment estimation
| Data set | Concentration (nmol mg−1 FW) |
n | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chl a |
Chl b |
Anth |
Car |
|||||||||||||
| Purpose | Genotype | Subset | Range | Mean ± SD | RMSE | Range | Mean ± SD | RMSE | Range | Mean ± SD | RMSE | Range | Mean ± SD | RMSE | (Chl a) RMSEa | |
| Calibration | WT | #1 | 0.019–1.997 | 1.291 ± 0.413 | 0.142 | 0.067–0.608 | 0.365 ± 0.106 | 0.078 | – | – | – | 0.000–0.905 | 0.568 ± 0.200 | 0.071 | 0.071 | 93 |
| WT | #2 | 0.977–2.597 | 1.670 ± 0.412 | 0.218 | 0.245–0.743 | 0.459 ± 0.102 | 0.102 | 0.116–3.098 | 1.037 ± 0.591 | 0.277 | 0.409–1.152 | 0.751 ± 0.189 | 0.098 | 0.099 | 131 | |
| ch1-3 | #3 | 0.260–1.288 | 0.609 ± 0.235 | 0.113 | – | – | – | – | – | – | 0.040–0.522 | 0.236 ± 0.113 | 0.054 | 0.061 | 72 | |
| ch1-3 | #4 | 0.366–1.039 | 0.714 ± 0.153 | 0.133 | – | – | – | 0.006–1.535 | 0.424 ± 0.375 | 0.185 | 0.110–0.354 | 0.235 ± 0.059 | 0.063 | 0.076 | 103 | |
| tt4 | #5 | 0.689–2.606 | 1.250 ± 0.459 | 0.136 | 0.157–0.537 | 0.343 ± 0.089 | 0.097 | – | – | – | 0.272–1.247 | 0.532 ± 0.228 | 0.079 | 0.076 | 66 | |
| WT | #1–2b | 0.019–2.597 | 1.513 ± 0.452 | 0.190 | 0.067–0.743 | 0.420 ± 0.113 | 0.093 | 0.116–3.098 | 1.037 ± 0.591 | 0.277 | 0.000–1.152 | 0.675 ± 0.213 | 0.088 | 0.088 | 224 | |
| WT/chl1-3/tt4 | Allb | 0.019–2.606 | 1.159 ± 0.542 | 0.161 | 0.067–0.743 | 0.402 ± 0.113 | 0.093 | 0.006–3.098 | 0.767 ± 0.591 | 0.241 | 0.000–1.247 | 0.489 ± 0.270 | 0.077 | 0.080 | 465 | |
| Validation | WT | #1 | 0.614–2.084 | 1.330 ± 0.402 | 0.127 | 0.183–0.534 | 0.369 ± 0.102 | 0.065 | – | – | – | 0.268–0.884 | 0.581 ± 0.191 | 0.059 | – | 15 |
| WT | #2 | 1.000–2.181 | 1.600 ± 0.428 | 0.106 | 0.295–0.620 | 0.446 ± 0.102 | 0.072 | 0.301–2.166 | 1.129 ± 0.541 | 0.300 | 0.449–0.985 | 0.725 ± 0.186 | 0.089 | – | 15 | |
| ch1-3 | #3 | 0.286–1.107 | 0.615 ± 0.253 | 0.108 | – | – | – | – | – | – | 0.030–0.445 | 0.235 ± 0.120 | 0.055 | – | 15 | |
| ch1-3 | #4 | 0.379–1.076 | 0.719 ± 0.177 | 0.130 | – | – | – | 0.043–1.268 | 0.414 ± 0.362 | 0.153 | 0.137–0.342 | 0.236 ± 0.057 | 0.072 | – | 15 | |
| tt4 | #5 | 0.678–2.019 | 1.196 ± 0.396 | 0.150 | 0.197–0.462 | 0.338 ± 0.082 | 0.083 | – | – | – | 0.270–0.912 | 0.502 ± 0.185 | 0.077 | – | 15 | |
| WT | #1–2b | 0.614–2.181 | 1.465 ± 0.430 | 0.117 | 0.183–0.620 | 0.408 ± 0.107 | 0.069 | 0.301–2.166 | 1.129 ± 0.541 | 0.300 | 0.268–0.985 | 0.653 ± 0.199 | 0.075 | – | 30 | |
| WT/chl1-3/tt4 | Allb | 0.286–2.181 | 1.092 ± 0.503 | 0.125 | 0.183–0.620 | 0.384 ± 0.104 | 0.074 | 0.043–2.166 | 0.771 ± 0.580 | 0.238 | 0.030–0.985 | 0.456 ± 0.247 | 0.071 | – | 75 | |
| gl1 | −Anth | 0.711–1.549 | 1.141 ± 0.280 | 0.094 | 0.162–0.407 | 0.310 ±0.068 | 0.069 | – | – | – | 0.321–0.732 | 0.520 ± 0.134 | 0.046 | – | 30 | |
| +Anth | 0.916–1.996 | 1.539 ± 0.309 | 0.168 | 0.268–0.590 | 0.448 ± 0.083 | 0.110 | 0.239–1.537 | 0.763 ± 0.354 | 0.167 | 0.389–0.898 | 0.664 ± 0.142 | 0.070 | – | 30 | ||
| Allb | 0.711–1.996 | 1.340 ± 0.355 | 0.136 | 0.162–0.590 | 0.379 ±0.103 | 0.092 | 0.239–1.537 | 0.763 ± 0.354 | 0.167 | 0.321–0.898 | 0.592 ± 0.155 | 0.059 | – | 60 | ||
| iref1 | −Anth | 0.672–1.876 | 1.235 ± 0.347 | 0.195 | 0.171–0.704 | 0.390 ± 0.136 | 0.198 | – | – | – | 0.274–0.782 | 0.528 ± 0.147 | 0.121 | – | 30 | |
| +Anth | 1.283–2.494 | 1.903 ± 0.293 | 0.426 | 0.425–0.740 | 0.564 ± 0.089 | 0.287 | 0.163–1.141 | 0.623 ± 0.212 | 0.132 | 0.514–1.077 | 0.808 ± 0.145 | 0.276 | – | 30 | ||
| Allb | 0.672–2.494 | 1.569 ± 0.463 | 0.331 | 0.171–0.740 | 0.477 ± 0.144 | 0.246 | 0.163–1.141 | 0.623 ± 0.212 | 0.132 | 0.274–1.077 | 0.668 ± 0.202 | 0.213 | – | 60 | ||
| iref3 | −Anth | 0.633–1.717 | 1.259 ± 0.310 | 0.094 | 0.173–0.615 | 0.354 ± 0.104 | 0.074 | – | – | – | 0.279–0.796 | 0.571 ± 0.151 | 0.061 | – | 30 | |
| +Anth | 0.971–2.065 | 1.610 ±0.347 | 0.144 | 0.271–0.649 | 0.432 ±0.104 | 0.072 | 0.123–1.390 | 0.735 ± 0.407 | 0.176 | 0.394–0.926 | 0.715 ± 0.157 | 0.113 | – | 30 | ||
| Allb | 0.633–2.065 | 1.435 ± 0.371 | 0.121 | 0.173–0.649 | 0.393 ± 0.110 | 0.073 | 0.123–1.390 | 0.735 ± 0.407 | 0.176 | 0.279–0.926 | 0.643 ± 0.169 | 0.091 | – | 60 | ||
| iref4-D | −Anth | 0.678–1.646 | 1.238 ± 0.261 | 0.115 | 0.218–0.568 | 0.349 ± 0.083 | 0.096 | – | – | – | 0.260–0.738 | 0.541 ± 0.126 | 0.066 | – | 30 | |
| +Anth | 1.354–2.377 | 1.852 ± 0.294 | 0.191 | 0.331–0.627 | 0.499 ± 0.075 | 0.106 | 0.201–0.888 | 0.457 ± 0.171 | 0.157 | 0.567–1.079 | 0.810 ± 0.141 | 0.122 | – | 30 | ||
| Allb | 0.678–2.377 | 1.545 ± 0.414 | 0.158 | 0.218–0.627 | 0.424 ± 0.109 | 0.101 | 0.201–0.888 | 0.457 ± 0.171 | 0.157 | 0.260–1.079 | 0.675 ± 0.189 | 0.098 | – | 60 | ||
The range, mean and SD of pigment concentrations, and root mean square error (RMSE) of pigment estimation using reflectance-based equations developed for Chl a (Equation 3), Chl b (Equation 5), Anths (Equation 7) and Cars (Equations 11 and 12a) in individual and summed subdata setsb are indicated. WT, wild type.
aRMSE of Car estimation based on the linear relationship between Car and Chl a concentrations using Equation 12.
bSummed subdata sets.
Chl b estimation from hyperspectral imagery
To develop an RI for the estimation of Chl b concentration, which is hereafter designated as (Chl b)RI, we attempted to select λ1 from the long-wavelength side within the red Chl absorption band, wherein minimal or no light absorption by pigments other than Chls a and b takes place (Fig. 1). Because λ1 in (Chl a)RI (Equation 2) was selected from the spectral range where Chl a absorption predominates over the absorption by other pigments, λ2 in the (Chl b)RI was set identical to this waveband, 670–676 nm. Then the optimal spectral range of λ1 was explored from the wavelength range above 600 nm by correlating the observed Chl b concentration ([Chl b]obs.) with the model (Rλ1−1 – R670–676−1)×R772–800. A preferable result, that is not greatly affected by the presence or absence of Anths, was obtained when λ1 was set to 625 nm (Fig. 4B). Collectively, (Chl b)RI was summarized in the following equation:
| (4) |
The correlation between (Chl b)RI and [Chl b]obs. in the whole calibration data set is shown by a scatter plot in Fig. 5B. A linear regression fitted to the wild-type subdata sets (#1 and 2) gives an equation:
| (5) |
where [Chl b]exp. denotes the expected Chl b concentration. The RMSE of the differences between [Chl b]obs. and [Chl b]exp., and related statistical parameters are summarized in Table 2. As is later confirmed in the validation section, the equation is quite successful in separating the Chl b-deficient ch1 mutants (subdata sets #3 and 4) from the wild type (subdata sets #1 and 2) (see Fig. 6B).
Fig. 6.
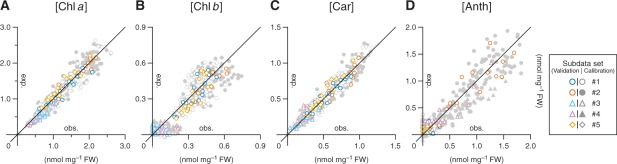
Relationship between observed and expected pigment concentrations in the validation data set. Details of each subdata set (#1–5) are summarized in Table 1. Horizontal and vertical axes, respectively, represent chemically determined (observed, abbreviated as obs.) and optically estimated (expected, abbreviated as exp.) concentrations of Chl a (A), Chl b (B), Car (C) or Anth (D). The background data points in light gray are from the calibration data set and provide a measure of the possible deviation range in the pigment estimation using Equations 3 (Chl a), 5 (Chl b), 7 (Anth) and 11 (Car). The statistical details are summarized in Table 2.
Anthocyanin estimation from hyperspectral imagery
In contrast to Chls a and b, which are single molecular species, Cars and Anths are generic names for the groups of pigments sharing similar basic structures. Hence, there is substantial diversity in the biochemical properties among the members of each group, including absorption spectra, especially with regard to Anths. The composition of Anths in Arabidopsis leaves, however, is far simpler than that in colored petals or deciduous leaves of native plants (Bloor and Abrahams 2002). Moreover, even though spectroscopic properties of Anths can change depending on solvent environment, the major absorption band of Anths in Arabidopsis leaves is probably distributed around 530 nm under physiological conditions. This is evident from the comparison of reflectance spectra of leaf samples and absorption spectra of their extracts derived from Anth-accumulating and -deficient plants (e.g. see Fig. 3C, subdata sets #2 vs. 5), and is apparent in the correlogram between Rλ−1 and observed Anth concentration ([Anth]obs.) (Supplementary Fig. S2D).
To develop an RI for the estimation of Anth concentration, which is hereafter designated as (Anth)RI, we started with an initial setup of λ1 = 530 nm in the ‘three-band model’. The optimal spectral range of λ2 was explored by correlating [Anth]obs. with the model (R530−1 – Rλ2−1)×R772–800. In the Anth-accumulating wild-type subdata set (#2), the highest correlation was achieved when λ2 was set to the spectral range of 670–676 nm (Fig. 4C). Then the optimal spectral range of λ1 was reconsidered by a similar procedure using the model (Rλ1−1 – R670–676−1)×R772–800. A preferable result was obtained when λ1 was set to 530 nm (Fig. 4D), demonstrating that the initial setup of λ1 was no less suited for Anth estimation. Collectively, (Anth)RI was summarized in the following equation:
| (6) |
The correlation between (Anth)RI and [Anth]obs. in each subdata set is shown by a scatter plot in Fig. 5C. A linear regression fitted to subdata set #2 gives an equation:
| (7) |
where [Anth]exp. denotes the expected Anth concentration. The RMSE of the differences between [Anth]obs. and [Anth]exp., and related statistical parameters are summarized in Table 2. As is later confirmed in the validation section, the equation is quite successful in separating the Anth-deficient tt4 mutants (subdata set #5) from the wild type grown under Anth-inducing conditions (subdata set #2) (see Fig. 6D).
Carotenoid estimation from hyperspectral imagery
As shown in Supplementary Fig. S1A, a strong linear correlation was found between Chl a and Car concentrations in the calibration data set. The ratio of Cars to Chl a, however, was appreciably lower in ch1 subdata sets (#3 and 4) than in the others. Hence, once an equation for the estimation of Car concentration was formulated, it was essential to evaluate if it provides as accurate estimate of Cars in ch1 mutants as in plants of the other genotypes.
Absorption spectra of Cars are characterized by unimodal distribution (Fig. 1). There is increasing overlap in the absorption band of Cars with that of Chls a and b at wavelengths below 500 nm. For this reason, we started with an initial set-up of λ1 = 501 nm in the ‘three-band model’ to develop (Car)RI. The optimal spectral range of λ2 was explored by correlating the observed Car concentration ([Car]obs.) with the model (R501−1 – Rλ2−1)×R772–800. Under conditions where Anth accumulation was negligible (subdata sets #1, 3 and 5), a waveband centered at 603 nm appeared most suitable for λ2 (Fig. 4E). The reflectance at this waveband was correlated closely with total Chl concentration, but was independent of the presence or absence of Chl b. This implies that the absorption coefficients of Chls a and b in vivo would equilibrate at this wavelength. Then the optimal spectral range of λ1 was reconsidered by using the model (Rλ1−1 – R603−1)×R772–800. A better result was obtained when λ1 was set to 507 nm, rather than 501 nm (Fig. 4F). Collectively, (Car)RI was summarized tentatively in the following equation (marked with an asterisk to indicate that it is temporary):
| (8) |
At this moment, the relationship between (Car)RI* and [Car]obs. did not fit well to a linear regression when there was substantial Anth accumulation (subdata sets #2 and 4) (Fig. 5D). Hence, to remove the Anth contribution from (Car)RI*, an additional term R530−1, which closely correlates with the Anth concentration, was introduced into Equation 8 with an adjustable constant c as follows:
| (9) |
As shown in Fig. 5E, when a constant of 0.6456 was substituted for c, which was derived by multiregression analysis using the wild-type subdata sets (#1 and 2), the relationship between (Car)RI and [Car]obs. from all subdata sets fell on a single regression line. Here, the equation for the estimation of Car concentration can be generalized as follows:
| (10) |
| (11) |
where [Car]exp. denotes the expected Car concentration. The RMSE of the differences between [Car]obs. and [Car]exp., and related statistical parameters are summarized in Table 2. The slope of Equation 11 is negative (Fig. 5E) because, different from RIs for other pigments, the total reflectance at waveband λ1 (507 nm) is usually higher than at waveband λ2 (603 nm) (Fig. 3C).
As described above, the Car concentration can also be approximated based on its linear correlation with the Chl a concentration using the equation as follows (Supplementary Fig. S1B):
| (12) |
While the RMSE of Car estimation was not greatly different between cases where Equation 11 or 12 was used, the former produced a more accurate estimate in total and especially in ch1 subdata sets (#3 and 4) (Table 2). This provides strong counter-evidence against complete autocorrelation between (Chl a)RI and (Car)RI.
Validation
The equations for the estimation of Chls a and b, Anths and Cars (Equations 3, 5, 7 and 11, respectively) were tested for their predictive performance by using validation data sets composed of plants containing certain amounts of the pigment of interest. While the wild-type (subdata sets #1 and 2), ch1 (subdata sets #3 and 4) and tt4 (subdata set #5) plants were all included in the validation data set for Chl a and Car estimation, ch1 and tt4 mutants were excluded from the validation of Chl b and Anth estimation, respectively. Similar to the case with the calibration data sets, close agreement between the observed and expected concentrations of each pigment was confirmed in the validation data sets by the scatter plot analysis (Fig. 6). Several statistical parameters obtained from the multivariate correlation analysis between the pairs of concentrations in the validation as compared with the calibration data sets are summarized in Table 2. Only small differences in the RMSE of pigment estimation were noted between cases where either data set was used. This indicates that the equations developed above provide reasonable estimates of the concentration of each pigment, at least under the growth conditions used in this study.
Using these equations, the software PPM (Plant Pigment Monitor), which allows 2D monitoring of leaf pigment status, was constructed (Fig. 2C). In addition to the individual concentrations, this also allows visualization of compositional parameters of pigments such as the sum and/or ratio of two or more constituents (e.g. see Fig. 7F). The software is provided as Supplementary File S1 of this article (see Supplementary Text S1 for legends and methods of operation).
Fig. 7.
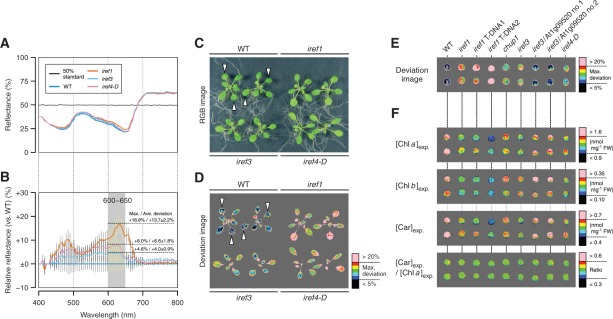
Isolation and characterization of mutants with irregular reflectance (iref) phenotypes. Pseudocolor images in D and E were produced by HSD Visualizer (Fig. 2D), while those in F were produced by PPM (Fig. 2C). The hyperspectral image data used in E and F are provided as Supplementary File S2 of this article. (A) Reflectance spectra of the wild-type (WT) and iref mutant leaves. The reflectance was calculated by linear regression against the 50% reflectance standard (as in Fig. 3B). Data are the mean ± SD (SD is shown by a vertical bar) derived from measurements of five different leaves of Arabidopsis plants from each genotype grown under normal conditions (as defined in Table 1). (B) Ratio reflectance spectra of iref mutants relative to the wild type. Data are derived from the spectra shown in A. As an example, maximum and average deviations of spectral reflectance in a wavelength range of 600–650 nm are shown for each iref mutant. (C) RGB image of iref mutants grown under normal conditions. (D) Visualization of iref phenotypes. The plants shown in C were subjected to a hyperspectral imaging procedure. The deviation of spectral reflectance in iref mutants relative to averaged reference spectra from the wild-type leaves (indicated by open triangles in C and D) is displayed in a color gradient from black (smallest) to pink (largest). Here, the values of maximum deviation in a wavelength range of 600–650 nm were used for visualization. (E) Confirmation of iref1 and iref3 loci through an allelism test and genetic complementation. The image was produced by the same procedure as in D, except detached leaves from the indicated plant lines were used. iref1 T-DNA1, WiscDsLox457-460P9 homozygous line; iref1 T-DNA2, SAIL_574_B09 homozygous line; iref3/At1g09520 Nos. 1 and 2, T2 individuals from transgenic iref3 lines carrying a wild-type genomic fragment of the At1g09520 locus. (F) Visualization of predicted pigment status in iref mutants. Individual concentrations and compositional parameters of pigments indicated on the left were calculated using Equations 3 (Chl a), 5 (Chl b) and 11 (Car). The range of parameters displayed in a color gradient from black (smallest) to pink (largest) is indicated alongside the scale bar on the right.
Non-targeted rapid detection of altered leaf pigment status: isolation of irregular reflectance mutants using hyperspectral imagery
While PPM was developed aiming for ‘targeted’ screening of mutants with defects in leaf pigment metabolism and/or its regulation in response to environmental and/or developmental cues, it required a certain amount of time and effort until the equations implemented in this software had been established. We therefore developed in parallel an additional analytical technique of hyperspectral imagery in order to detect subtle differences in reflectance phenotypes that are possibly associated with altered leaf pigment status. The technique is implemented in the software HSD Visualizer (Fig. 2D), which is also packaged in Supplementary File S1 of this article (see Supplementary Text S1 for legends and methods of operation).
Fig. 7A shows an example of reflectance spectra from the leaves of the wild type and mutants isolated via the use of HSD Visualizer (the details of the screening are described later in this section). Here, the reflectance spectra calibrated simply by linear regression against the 50% standard (as in Fig. 3B) were used; as described above, they were sufficient to evaluate qualitative differences in reflectance phenotypes. Using the spectra from the wild type as the baseline, the ratio of spectral reflectance was calculated for each pixel of the hyperspectral images. The derived ratio spectra in the areas of the mutant leaves are shown in Fig. 7B. To cite an example, in a wavelength range of 600–650 nm, the reflectance deviation in a mutant designated as irregular reflectant 1 (iref1) was +16.8% at the maximum and +13.7 ± 2.2% (n = 5) on average relative to the wild type. The software HSD Visualizer automatically calculates such parameters and display the reflectance deviation as a pseudocolor image (e.g. see Fig. 7D, E).
Using this software, we conducted, so to speak, a ‘non-targeted’ screen for mutants with altered reflectance phenotypes with the view to getting information about in what situations the reflectance-based pigment estimation works as expected or fails. During a small-scale ‘pilot’ screening of ethyl methanesulfonate (EMS)-mutagenized M2 lines of Arabidopsis (1,700 individuals), we encountered a number of mutants with altered reflectance phenotypes that are readily identifiable from their colors and/or associated morphological defects. Here, we focused exclusively on mutants with weak iref phenotypes that are hardly identifiable without using hyperspectral imagery. Combined with an additional screening of T-DNA activation-tagged lines (7,000 individuals), three mutants exhibiting iref phenotypes under normal growth condition (as defined in Table 1) were isolated (Fig. 7C, D).
Genetic localization of irregular reflectance loci
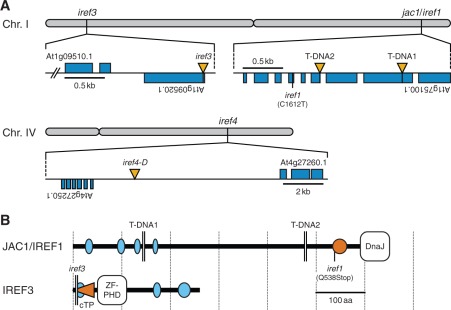
Genetic mapping of a recessive EMS allele, iref1, localized the mutation to a 134 kb region between two single nucleotide polymorphisms, PERL0254266 and PERL0255054 (Perlegen), on chromosome 1. Sequencing of this region revealed that an open reading frame (ORF) At1g75100.1 of the iref1 genome contained a C-to-T nonsense transition in the sixth exon (Fig. 8A), which results in a truncation of the C-terminal 114 amino acids of the gene product (Fig. 8B). To confirm further if this base transition is responsible for the iref phenotype of the mutant, two recessive T-DNA insertion alleles of the same ORF were analyzed. As shown in Fig. 7E, all homozygous T-DNA individuals exhibited the phenotype comparable with that of the iref1 mutant. This provides unequivocal evidence that the At1g75100 locus corresponds to the IREF1 gene.
Fig. 8.
Identification of irregular reflectance (iref) loci. (A) Chromosomal positions of iref mutations. Rectangles (blue) and vertical triangles (yellow) indicate exons and T-DNA insertions, respectively. The iref1 mutation causes a C-to-T transition at nucleotide 1,612 (excluding introns) in an ORF At1g75100.1. T-DNA1 and 2 indicate T-DNA positions in WiscDsLox457-460P9 and SAIL_574_B09 lines, respectively. The iref3 genome contains a T-DNA 20 bp downstream from the translation start site of an ORF At1g09520.1. The iref4-D mutation is linked to a T-DNA located within an intergenic region between ORFs At4g27250.1 and At4g27260.1 (2.1 and 7.1 kb apart, respectively). (B) The structure of JAC1/IREF1 and IREF3 proteins. The diagram is based on the information available from the InterPro (http://www.ebi.ac.uk/interpro/) and SMART (http://smart.embl.de/) database, and a TargetP prediction (http://www.cbs.dtu.dk/services/TargetP/). Ellipses indicate segments of low compositional complexity (light blue) and a coiled-coil region (orange). The horizontal triangle (orange) in IREF3 indicates the putative chloroplast transit peptide (cTP). The iref1 mutation truncates the C-terminal 114 amino acids (aa) of the JAC1/IREF1 protein (651 aa). T-DNA1 and 2 truncate the protein at aa 145 and 478, respectively. The iref3 T-DNA truncates the IREF3 protein (260 aa) at aa six. DnaJ, DnaJ molecular chaperone homology domain; ZF-PHD, zinc finger plant homeodomain-type signature.
In contrast to the iref1 mutant, iref3 and iref4 mutants were isolated from the T-DNA population. Accordingly, inverse PCR amplification of the T-DNA flanking regions followed by sequencing of the PCR products led to identification of the locations of T-DNA tags that co-segregate with the iref phenotypes of the mutants. In the genome of the recessive iref3 mutant, T-DNA was inserted 20 bp downstream from the translation start site of an intron-less ORF At1g09520.1 (Fig. 8A). Because no other T-DNA knockout allele of this ORF was available, the identity of the IREF3 gene was tested by genetic complementation of the iref3 mutant. As shown in Fig. 7E, introduction of a genomic fragment covering the wild-type At1g09520 locus into the mutant resulted in the disappearance of the iref phenotype, demonstrating that this locus corresponds to the IREF3 gene.
On the other hand, iref4 was a dominant mutant, and therefore the corresponding mutation is designated as iref4-D. According to segregation analysis, the iref phenotype of this mutant was linked to the activation T-DNA tag located within an intergenic region between the ORFs At4g27250.1 and At4g27260.1 (Fig. 8A; 2.1 and 7.1 kb apart from the respective ORFs). However, reverse transcription–PCR (RT–PCR) analysis revealed that expression of these flanking genes in leaves was neither activated nor repressed at the transcriptional level. Identification of the bona fide IREF4 gene still awaits further genetic and expression analysis of a wider genomic region flanking the T-DNA tag on chromosome 4.
Evaluation of limit situations for reflectance-based pigment estimation
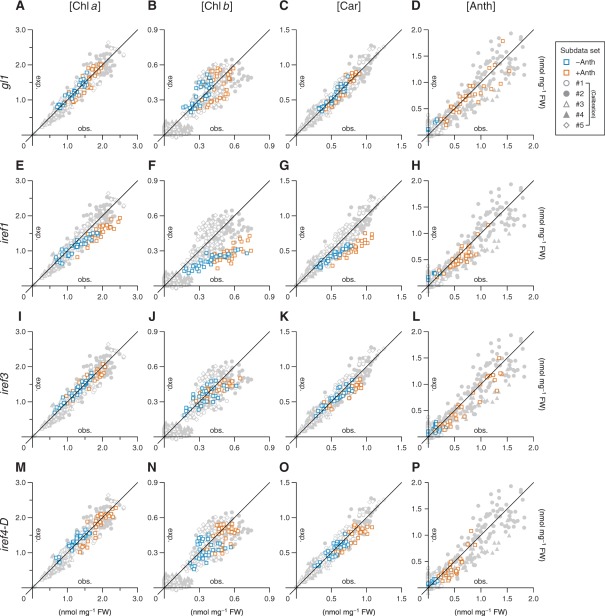
In a previous report, it was warned that changes in leaf surface structure such as trichome density can mislead reflectance-based assessment of leaf chemistry and physiology, including the values of RIs conventionally used to approximate leaf pigment compositions (Levizou et al. 2005). To test if this is true of the ‘targeted system’ developed in this study, the predictive performance of the equations for the estimation of individual pigment concentrations (Equations 3, 5, 7 and 11) was evaluated in a trichome-less gl1 mutant. As shown by scatter plots in Fig. 9A–D, the correlation between the observed and expected concentrations of each pigment under routine growth conditions, i.e. the conditions used to obtain subdata sets #1 and 2 (Table 1), was comparable in the gl1 plants and the wild-type plants. This, in combination with the related statistical parameters (Table 2), corroborates the robustness of our ‘targeted’ system for leaf pigment estimation at least against the decrease in trichome density.
Fig. 9.
Applicability of reflectance-based pigment estimation in mutants with altered leaf surface structure and irregular reflectance (iref) phenotypes. Each plot shows the relationship between observed (horizontal axis, abbreviated as obs.) and expected (vertical axis, abbreviated as exp.) concentrations of pigment indicated at the top in mutant line gl1 (A–D), iref1 (E–H), iref3 (I–L) or iref4-D (M–P). Squares in orange indicate the data obtained from plants grown under Anth-inducing conditions, while those in blue are from plants kept in non-inducing conditions. The background data points in light gray are from the calibration data set and provide a measure of the possible deviation range in the estimation using Equations 3 (Chl a), 5 (Chl b), 7 (Anth) and 11 (Car). Details of each calibration subdata set (#1–5) are summarized in Table 1. The statistical details are summarized in Table 2.
An example of a physiological process that limits reflectance-based pigment estimation became evident from the characterization of the iref1 mutant. Actually, the mutant locus At1g75100 has been formerly identified as being required for chloroplast photorelocation movement. The encoded protein, JAC1, an auxilin-like J-domain protein (Fig. 8B), is involved in the process of low light-induced accumulation of chloroplasts in the outermost cytoplasmic layer of palisade mesophyll cells (Suetsugu et al. 2005). Because all iref1 alleles tested in this study lead to truncation of the characteristic J-domain at the C-terminus (Fig. 8B), as in the cases of jac1 alleles (Suetsugu et al. 2005, Takano et al. 2010), the iref phenotype seen in the mutant is most likely to be the result of loss of function of the JAC1 protein. A similar but somewhat weaker iref phenotype was also seen in a loss-of-function allele (Schmidt von Braun and Schleiff 2008) of another chloroplast photorelocation mutant, chup1 (Fig. 7E). This mutant is also deficient in the low light-induced chloroplast accumulation response (Oikawa et al. 2003). Under routine growth conditions, where chloroplast accumulation is mostly stimulated in the wild-type plants, the reflectance-based method resulted in severe underestimation of the concentrations of photosynthetic pigments (Chls a and b, and Cars) in the iref1 mutant (Fig. 9E–G; Supplementary Fig. S3). These exemplify a limit situation for the reflectance-based pigment estimation, and also suggest that, if better predictive performance is desired for this method, plants should be acclimated to low-light conditions before being subjected to hyperspectral reflectance imaging.
In contrast to the case of the iref1 mutant, the aberrancies in pigment composition in the iref3 and iref4-D mutants as predicted from their hyperspectral images were consistent with those elucidated by chemical pigment analysis. Under the normal growth conditions defined in this study (Table 1), the observed concentrations of some photosynthetic pigments were slightly but significantly lower (P < 0.05) in both iref3 and iref4-D mutants than in the wild-type plants (Supplementary Fig. S3). As for the iref3 mutant, the mutant locus At1g09520 encodes an uncharacterized protein with a plant homeodomain-type zinc finger motif and a putative chloroplast transit peptide (cTP) at the N-terminus (Fig. 8B). Because the T-DNA tag in the iref3 mutant is located within the cTP coding region (Fig. 8B), the iref phenotype of the mutant is most likely to be the result of loss of function of the IREF3 protein. As shown in Fig. 7F, the subtle decrease in the photosynthetic pigment concentrations in these mutants relative to those in the wild type (Supplementary Fig. S3) could be detected readily and separately for each pigment using PPM. Thus, although there are some limiting situations for the reflectance-based ‘targeted’ pigment estimation, it provides rapid and highly sensitive means to identify altered leaf pigment status resulting from physiological responses to environmental and/or developmental factors, or from genetic mutations affecting homeostatic mechanisms of the metabolism of leaf pigments.
Discussion
In this study, we showed that hyperspectral imaging techniques provide far more information than the eyes can see with regard to leaf pigment status, largely due to its quantitative capability. One of the concerns about the applicability of reflectance-based pigment estimation, however, resides in the equations for Anth and Car estimation (Equations 7 and 11), as these pigments are in nature composed of heterogeneous chemical compounds with somewhat different spectroscopic properties, and the composition of these compounds can change in response to environmental and/or developmental cues. As for Cars, it has been shown that defects in metabolic steps leading to an increase in their structural diversity, such as lutein and β,β-xanthophyll biosynthetic pathways, do not result in large alterations in the total Car concentration. This is because the decrease or increase in certain Car compounds is usually compensated by corresponding changes in the amount of other compounds (Pogson et al. 1998, Kim and DellaPenna 2006). The spectral resolution of the hyperspectral camera used in this study was not high enough to reach 5 nm or below throughout the measuring wavelength range (400–800 nm). While the absorption coefficients of different Car compounds at wavebands used in the Car estimation (Equation 11) differ, at least to some extent, from each other, it may be that such a ‘wide’ bandwidth rather ensured the robustness of total Car estimation against compositional changes in the Car compounds. Even so, hyperspectral cameras are still the product of emerging technology; higher spectral resolution and bit depth are much needed to facilitate model tuning of RIs and to accomplish more accurate estimation of individual pigment concentrations.
The applicability of the equation for Anth estimation (Equation 7) could be even more restricted to plants grown under temperate, non-extreme conditions. At present, the equation is devoid of the terms that specifically remove the contribution of Chl b and Cars to the total reflectance. Hence, the observed accuracy of Anth estimation could be the result of low (though non-negligible) absorption coefficients of pigments other than Anths at 530 nm (Fig. 1). Moreover, it is well known that changes in the solvent environment surrounding the Anth molecules, such as vacuolar acidification, greatly affect their coloration properties (Castañeda-Ovando et al. 2009), possibly including their absorption coefficient at 530 nm. Considering such uncertainties in the reflectance-based Anth estimation, adoption of Anth-less genetic backgrounds, such as tt4, would be a feasible solution if the research target is not Anths themselves and is considered as being independent of Anth metabolism.
Despite a certain chemical diversity in leaf pigments, especially in Cars and Anths as described above, biomolecules, so-called pigments, account for only a minor fraction of the entire metabolites in plant cells. In contrast, almost all metabolites, being organic compounds, have their own absorption spectra in the infrared range, which are correlated with their intrinsic molecular vibration spectra. The NIR range extended to about 2,500 nm is particularly advantageous for non-invasive in situ analysis of primary metabolites directly associated with plant growth, such as soluble and polymeric sugars, and organic nitrogen compounds. Actually, NIR spectroscopy has been widely applied in food industries for quality management of agricultural products by using handheld or inline NIR analyzers (Huang et al. 2008). In addition to the adoption of wider wavelength ranges, acquisition of different types of hyperspectral images, such as transmission and fluorescence images, and even polarized reflectance images, would be achievable in a laboratory environment. Such series of ‘multimode’ hyperspectral images would allow the retention of a large amount of information with regard to the physiological status of plants used in any published work; these can be revisited by other researchers to verify theoretical perspectives of their own field of interest.
Presently, further breakthrough is required in the methodology of spectral analysis in order for it to be able to draw out comprehensive information about the metabolic status of plants from their hyperspectral signature in both quantitative and qualitative terms. Even so, quantitative description of leaf color phenotypes based on reflectance and/or other spectra in the visible (VIS) range, and extension of the wavelength range from the ‘colored’ VIS to ‘colorless’ NIR range will provide a technical interface allowing for correlation analysis between leaf metabotypes and phenotypes. Similar to morphological traits, spectroscopic traits, including colors, can account for a large fraction of plant phenotypes, and are highly sensitive to environmental and/or developmental factors. Hence, a comprehensive ‘spectrome’ catalog describing the relationship between spectroscopic traits of plant tissues and the growth conditions they experienced will help to improve our understanding of how genetic variation leads to an effect on phenotypic traits at the individual level. Thus, the prevalence of such a ‘spectromics’ viewpoint facilitates one of the most essential issues of post-genome research to be resolved, i.e. data integration from genome to phenome.
Materials and Methods
Plant materials and growth conditions
All lines of Arabidopsis thaliana described here were descended from the Columbia wild type. The homozygous T-DNA insertion lines, SM_3_15530 (The John Innes Centre) and SALK_039478C (The Salk Institute), respectively, were used as Anth-deficient tt4 (Abrahams et al. 2002) and trichome-less gl1 (Wang et al. 2004) mutants in the Columbia background. The ch1-3 line from the Arabidopsis Biological Resource Center was used as the Chl b-deficient mutant (Espineda et al. 1999). The T-DNA lines WiscDsLox457-460P9 (The University of Wisconsin) and SAIL_574_B09 (Syngenta) were obtained as alleles of the jac1/iref1 mutant, and SALK_129128 as an allele of the chup1 mutant. A genomic fragment corresponding to nucleotides 52,346–54,691 of the bacterial artificial chromosome (BAC) clone F14J9 (GenBank accession No. AC003970) was subcloned into the binary vector pKGW (Karimi et al. 2002) using Gateway™ (Invitrogen) technology, and applied for complementation of the iref3 mutant via Agrobacterium-mediated transformation (Clough and Bent 1998). Kanamycin-resistant T2 individuals from two independent transgenic iref3 lines were subjected to further analysis. Plants were routinely grown on Murashige and Skoog (MS)-based solid media containing 0.5% (w/v) gellan gum (Kanto Chemical) at 22°C under continuous fluorescent light (80 μmol m−2 s−1). To obtain leaf samples with various pigment compositions, plants initially grown on P-replete media with or without ammonium nitrate (20.6 mM) were transferred to secondary media containing varying concentrations of N and P or 1–5 μM norflurazon (see Table 1 for details).
Pigment analysis
The concentrations of individual leaf pigments were determined from the same leaf samples that were used for hyperspectral reflectance imaging. Leaf tissues (approximately 3 mg FW) were frozen and crushed in liquid nitrogen into fine powder, and then extracted with 600 μl of 100% methanol. After centrifugation, one 200 μl aliquot of the supernatant was acidified with 1 μl of half-saturated (approximately 5.66 M) hydrogen chloride for the determination of total Anth concentration, while another aliquot was used for determination of the concentrations of Chls a and b, and total Cars. An 80 μl sample of each aliquot was dispensed into a 96-well half-area microplate (MICROLON 200; Greiner Bio-One) that ensures a 0.5 cm light path length with this sample volume. Absorbance measurement was performed using a microplate reader equipped with data analysis software (Infinite M200A and Magellan; Tecan). For calculating the concentration of each pigment, equations reported by Lichtenthaler (1987) for Chls a and b, and total Cars in 100% methanol, and the equation reported by Gitelson et al. (2001) for total Anths in acid methanol were used. The concentration was expressed on a fresh weight basis. The reagents used to analyze the absorption spectra of these pigments (Fig. 1) were purchased from Sigma [Chl a, C5753, purity 88% (w/w); Chl b, C5878, purity 99.6% (w/w); cyanidin chloride, 79457, purity 95% (w/w)] and Nacalai Tesque [β-carotene, 07312-81, purity 98% (w/w)].
Hyperspectral reflectance imaging
The configuration of the hyperspectral imaging set-up is outlined in the Results (Fig. 2A). To add further detail, a blue color conversion filter (LBB-4; Kenko) was mounted in front of the camera lens so as to prevent saturation of the detector at longer wavelengths, resulting from the irradiation with ‘reddish’ halogen lights. For applications requiring extreme throughput, such as correlation analysis between RIs and chemically determined pigment concentrations, first rosette leaves were detached at the petiole and aligned on a solid medium containing 0.5% (w/v) gellan gum and 3 mM CaCl2 (for gelatinization). This ‘invasive’ procedure allowed 50 or more leaves per 136×96 mm square plate (AW2000, EIKEN CHEMICAL) to be analyzed at a time. Otherwise, plants grown on solid media were used in their intact forms. For 2-week-old plants, a density of at most 12 individuals per plate was desirable to prevent overlap of adjacent plants.
Software development and data analysis
The self-made software shown in Fig. 2B–D, which is provided as Supplementary File S1 of this article (see Supplementary Text S1 for legends and methods of operation), was developed in managed C++ code using Visual Studio 2005 Standard Edition (Microsoft). Spectral reflectance data were extracted from hyperspectral images using HSD Analyzer (Fig. 2B). They were exported as comma-separated-value (CSV) format files prior to further statistical analysis using Microsoft Excel software. Note that the exported data are not yet corrected for the sensor’s non-linearity (equivalent to the data shown in Fig. 3B). The method to obtain more accurate reflectance values by introducing a second-order correction step is described in the Results (Fig. 3C).
Analysis of mutants with irregular reflectance phenotypes
Mutants with iref phenotypes were isolated through hyperspectral image analysis using HSD Visualizer (Fig. 2D) from 1,700 EMS-mutagenized M2 lines and 7,000 T-DNA activation-tagged lines developed by RIKEN (Nakazawa et al. 2003). The chromosomal position of the EMS-induced iref1 mutation was localized by a map-based cloning strategy using cleaved amplified polymorphic sequence and simple sequence length polymorphism markers. The T-DNA mutations, iref3 and iref4-D, were identified by restriction digestion and self-circularization of the mutant genomic DNA, followed by inverse PCR amplification of the T-DNA flanking sequences, and DNA sequencing of the PCR products.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Sumitomo Foundation [Grant for Basic Science Research Projects (No. 080117 to O.M.)]; Bio-oriented Technology Research Advancement Institution (BRAIN) [Promotion of Basic Research Activities for Innovative Biosciences (to K.I.)]; the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT) [Grants-in-Aid for Scientific Research on Innovative Areas (No. 21114002 to K.I.)]; the Asahi Glass Foundation [Task-oriented Research Grants (to O.M.)].
Supplementary Material
Acknowledgments
We thank Mark Allen Lanoue and David A. Smith (Themis Vision Systems) for technical advice with software development and helpful discussions.
Glossary
Abbreviations
- Anth
anthocyanin
- Car
carotenoid
- CSV
comma-separated-value
- 2D
two-dimensional
- cTP
chloroplast transit peptide
- EMS
ethyl methanesulfonate
- HSD
hyperspectral data
- iref
irregular reflectance
- MS
Murashige and Skoog
- N
nitrogen
- NIR
near-infrared
- ORF
open reading frame
- P
phosphate
- PPM
Plant Pigment Monitor
- RGB
red–green–blue
- RI
reflectance index
- RMSE
root mean square error
- RT–PCR
reverse transcription–PCR
- VIS
visible.
References
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR. Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol. 2002;130:561–576. doi: 10.1104/pp.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn GA. Quantifying chlorophylls and carotenoids from leaf to canopy scales: an evaluation of some hyperspectral approaches. Remote Sens. Environ. 1998;66:273–285. [Google Scholar]
- Blackburn GA. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007;58:855–867. doi: 10.1093/jxb/erl123. [DOI] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S. The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry. 2002;59:343–346. doi: 10.1016/s0031-9422(01)00460-5. [DOI] [PubMed] [Google Scholar]
- Castañeda-Ovando A, Pacheco-Hernández ML, Páez-Hernández ME, Rodríguez JA, Galán-Vidal CA. Chemical studies of anthocyanins: a review. Food Chem. 2009;113:859–871. [Google Scholar]
- Chappelle EW, Kim MS, McMurtrey JE. Ratio analysis of reflectance spectra (RARS): an algorithm for the remote estimation of the concentrations of chlorophyll a, chlorophyll b and carotenoids in soybean leaves. Remote Sens. Environ. 1992;39:239–247. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Cazzaniga S, Havaux M, Bassi R. Enhanced photoprotection by protein-bound vs free xanthophyll pools: a comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant. 2010;3:576–593. doi: 10.1093/mp/ssp117. [DOI] [PubMed] [Google Scholar]
- Datt B. Remote sensing of chlorophyll a, chlorophyll b, chlorophyll a + b, and total carotenoid content in Eucalyptus leaves. Remote Sens. Environ. 1998;66:111–121. [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, et al. Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:74–83. doi: 10.1093/pcp/pci225. [DOI] [PubMed] [Google Scholar]
- Edwards D, Batley J. Plant bioinformatics: from genome to phenome. Trends Biotechnol. 2004;22:232–237. doi: 10.1016/j.tibtech.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Espineda CE, Linford AS, Devine D, Brusslan JA. The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1999;96:10507–10511. doi: 10.1073/pnas.96.18.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia. 1989;78:9–19. doi: 10.1007/BF00377192. [DOI] [PubMed] [Google Scholar]
- Gitelson AA, Merzlyak MN, Chivkunova OB. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001;74:38–45. doi: 10.1562/0031-8655(2001)074<0038:opaneo>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gitelson AA, Zur Y, Chivkunova OB, Merzlyak MN. Assessing carotenoid content in plant leaves with reflectance spectroscopy. Photochem. Photobiol. 2002;75:272–281. doi: 10.1562/0031-8655(2002)075<0272:accipl>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gitelson AA, Gritz Y, Merzlyak MN. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003;160:271–282. doi: 10.1078/0176-1617-00887. [DOI] [PubMed] [Google Scholar]
- Gitelson AA, Keydan GP, Merzlyak MN. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006a;33:L11402. [Google Scholar]
- Gitelson AA, Vina A, Verma SB, Rundquist DC, Arkebauer TJ, Keydan G, et al. Relationship between gross primary production and chlorophyll content in crops: implications for the synoptic monitoring of vegetation productivity. J. Geophys. Res. 2006b;111:D08S11. [Google Scholar]
- Hasdai M, Weiss B, Levi A, Samach A, Porat R. Differential responses of Arabidopsis ecotypes to cold, chilling and freezing temperatures. Ann. Appl. Bot. 2006;148:113–120. [Google Scholar]
- Havaux M, Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta. 2001;213:953–966. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- Huang H, Yu H, Xu H, Ying Y. Near infrared spectroscopy for on/in-line monitoring of quality in foods and beverages: a review. J. Food Eng. 2008;87:303–313. [Google Scholar]
- Jung S. Effect of chlorophyll reduction in Arabidopsis thaliana by methyl jasmonate or norflurazon on antioxidant systems. Plant Physiol. Biochem. 2004;42:225–231. doi: 10.1016/j.plaphy.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Kaminuma E, Heida N, Tsumoto Y, Yamamoto N, Goto N, Okamoto N, et al. Automatic quantification of morphological traits via three-dimensional measurement of Arabidopsis. Plant J. 2004;38:358–365. doi: 10.1111/j.1365-313X.2004.02042.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl Acad. Sci. USA. 2006;103:3474–3479. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol. 2009;50:1215–1231. doi: 10.1093/pcp/pcp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire G, François C, Dufrêne E. Towards universal broad leaf chlorophyll indices using PROSPECT simulated database and hyperspectral reflectance measurements. Remote Sens. Environ. 2004;89:1–28. [Google Scholar]
- Lenk S, Chaerle L, Pfündel EE, Langsdorf G, Hagenbeek D, Lichtenthaler HK, et al. Multispectral fluorescence and reflectance imaging at the leaf level and its possible applications. J. Exp. Bot. 2007;58:807–814. doi: 10.1093/jxb/erl207. [DOI] [PubMed] [Google Scholar]
- Levizou E, Drilias P, Psaras GK, Manetas Y. Nondestructive assessment of leaf chemistry and physiology through spectral reflectance measurements may be misleading when changes in trichome density co-occur. New Phytol. 2005;165:463–472. doi: 10.1111/j.1469-8137.2004.01250.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Mündermann L, Erasmus Y, Lane B, Coen E, Prusinkiewicz P. Quantitative modeling of Arabidopsis development. Plant Physiol. 2005;139:960–968. doi: 10.1104/pp.105.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, et al. Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 2003;34:741–750. doi: 10.1046/j.1365-313x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, et al. CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Niyogi KK, Björkman O, DellaPenna D. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl Acad. Sci. USA. 1998;95:13324–13329. doi: 10.1073/pnas.95.22.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002;73:149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
- Richardson AD, Duigan SP, Berlyn GP. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 2002;153:185–194. [Google Scholar]
- Schmidt von Braun S, Schleiff E. The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta. 2008;227:1151–1159. doi: 10.1007/s00425-007-0688-7. [DOI] [PubMed] [Google Scholar]
- Sims DA, Gamon JA. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002;81:337–354. [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Kagawa T, Wada M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005;139:151–162. doi: 10.1104/pp.105.067371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Suetsugu N, Wada M, Kohda D. Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1372–1376. doi: 10.1093/pcp/pcq089. [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J. The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant Cell Environ. 1997;20:85–92. [Google Scholar]
- Ustin SL, Gitelson AA, Jacquemoud S, Schaepman ME, Asner GP, Gamon JA, et al. Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009;113:S67–S77. [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, et al. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell. 2004;16:2323–2334. doi: 10.1105/tpc.104.024844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.