Abstract
LAGLIDADG homing endonucleases (LHEs) are a family of highly specific DNA endonucleases capable of recognizing target sequences ∼20 bp in length, thus drawing intense interest for their potential academic, biotechnological and clinical applications. Methods for rational design of LHEs to cleave desired target sites are presently limited by a small number of high-quality native LHEs to serve as scaffolds for protein engineering—many are unsatisfactory for gene targeting applications. One strategy to address such limitations is to identify close homologs of existing LHEs possessing superior biophysical or catalytic properties. To test this concept, we searched public sequence databases to identify putative LHE open reading frames homologous to the LHE I-AniI and used a DNA binding and cleavage assay using yeast surface display to rapidly survey a subset of the predicted proteins. These proteins exhibited a range of capacities for surface expression and also displayed locally altered binding and cleavage specificities with a range of in vivo cleavage activities. Of these enzymes, I-HjeMI demonstrated the greatest activity in vivo and was readily crystallizable, allowing a comparative structural analysis. Taken together, our results suggest that even highly homologous LHEs offer a readily accessible resource of related scaffolds that display diverse biochemical properties for biotechnological applications.
INTRODUCTION
LAGLIDADG homing endonuclease (LHE) genes are mobile genetic elements that code for rare cleaving DNA enzymes, which in turn are responsible for catalyzing their mobility, known as homing. The homing process relies on the generation of DNA double strand breaks in an allele lacking the LHE gene insertion, which stimulates homologous recombination (HR) using the LHE-containing allele as the template (1,2). As an LHE’s physiological recognition sequence is ∼20 bp in length, it appears on average only once every ∼1012 bases. Even after accounting for an LHE's promiscuity at individual DNA bp positions, the overall specificity of these enzymes appears to be at least approximately one in 109. Consequently, LHEs have drawn attention as rare cleaving nucleases for use in diverse site-specific genome engineering applications, particularly for organisms with large genomes (3–5).
An important limitation to widespread application of LHEs in genome engineering is the requirement to modify a starting native LHE (‘scaffold’) to create variants of that scaffold that cleave at specific desired target sites. Although computational design methods and selection protocols for this purpose are now quite advanced (6–10), it remains challenging to consistently produce variants with high levels of in vivo activity. One constraint on present approaches for engineering LHE's is their narrow application to a small set of previously reported, well characterized, native LHE scaffolds: I-SceI, I-CreI, I-DmoI, I-AniI and I-OnuI (11–15). We hypothesized that because members of this small group were not originally identified based on specific biotechnologically useful properties, that homologous proteins might represent a source of closely related scaffolds that possess a desirable range of such properties. To address this question, we searched public sequence databases to identify open reading frames (ORFs) encoding proteins homologous to the LHE I-AniI and surveyed the properties of a subset of these proteins. Individual proteins were assessed using an assay that relies upon yeast surface display and that reports upon protein folding, expression, DNA binding and cleavage (15,16). Each of these properties can then be assayed by flow cytometric analysis in high throughput, detecting binding or cleavage of fluorescently labeled oligonucleotides. A separate in vivo genome engineering reporter assay was then used to measure targeted gene modification activity in transfected human cells (16–18). These analyses revealed that I-AniI's close homologs exhibit a broad spectrum of in vitro and in vivo activities. The best-performing enzyme in this group, I-HjeMI, was readily expressed, purified and crystallized, facilitating a comparative structural analysis of the two enzyme scaffolds. These results delineate a robust approach for identifying related LHE scaffolds and illustrate the value of this approach for identifying scaffolds with optimal biotechnological properties.
MATERIALS AND METHODS
Yeast surface display expression constructs and flow cytometric expression analysis
The ability of an LHE to bind and cleave a broad panel of DNA target sequences can be readily assayed using enzyme constructs that are displayed on the surface of yeast, as described in Jarjour et al. (16). Yeast surface display of I-AniI homologs on EBY100 Saccharomyces cerevisiae was achieved using the standard vector backbones and methods described previously (17). Putative LHE ORF sequences were selected, corresponding to full-length I-AniI beginning three to four amino acids before the first LAGLIDADG helix. Corresponding DNA sequences were synthesized and cloned into the pETCON2 vector (map available on addgene.org) between N-terminal hemagglutinin (HA) tag and C-terminal Myc tag coding sequences using NheI and XbaI; clones were verified by sequencing. Accession numbers for the protein sequences of I-AchMI, I-HjeMI, I-PnoMI, I-TasMIP, I-TinMIP and I-VinIP are AAX34413, BK008014, ABU49435, BK008015, BK008016 and AAB95258, respectively. Strains harboring these vectors were grown in media containing 2% raffinose + 0.1% glucose at 30°C for 1 day before induction in 2% galactose for 2–3 h at 30°C and 18–26 h at 20°C. To measure expression levels, 106 cells were washed in yeast staining buffer (YSB): 180 mM KCl, 10 mM NaCl, 0.2% bovine serum albumin (BSA), 0.1% galactose and 10 mM Hepes, pH 7.5. Cells were then stained with a 1:100 dilution of ICL Labs’ αMyc-FITC antibody and a 1:250 dilution of biotinylated αHA (Covance) antibody in YSB for 30 min at 4°C. Cells were washed and counterstained with streptavidin–PE (BD Biosciences) in YSB for 15 min at 25°C, washed again and run on a BD LSRII™ cytometer (BD Biosciences). The output was analyzed using FloJo software (Tree Star) for the percentage FITC-positive cells when compared with an unstained population.
Immunoprecipitation and western blot of surface-released protein
Approximately 250 million expressing yeast cells (induced as above) were harvested, washed twice in 1 × phosphate-buffered saline (PBS, Thermo Scientific) and incubated for 1 h at 30°C in 1 ml 2 mM dithiothreitol in PBS with protease inhibitor (complete mini EDTA free, Roche) to liberate the LHEs; this is accomplished by reducing the disulfide bond anchoring the Aga2P-LHE fusion to the surface expressed Aga1P protein (Supplementary Figure S5). The release reaction was quenched with 10 mM iodoacetamide for 10 min at 25°C to allow subsequent immunoprecipitation. The LHE-containing supernatant was incubated with 1:100 monoclonal rabbit αHA antibody (C29F4, Cell Signaling) for 1 h at 4°C and precipitated with protein A-conjugated Sepharose (GE Healthcare) by incubation overnight at 4°C. Samples were treated with PNGaseF (New England BioLabs) according to the manufacturer's protocol to remove glycosyl residues and allow proper migration on a gel. Samples were prepared by boiling in 1× Laemmli buffer (Bio-Rad).
Denaturing polyacrylamide gel electrophoresis and western blot to a polyvinylidene fluoride membrane were performed using standard protocols. The blot was stained with a 1:1000 dilution of rabbit αHA antibody (Cell Signaling), washed and counter-stained with a 1:5000 dilution of donkey αRabbit, horseradish peroxidase antibody (GE Healthcare) for imaging with the ECL system using Kodak Biomax light film.
Flow cytometric cleavage assay, end-holding and specificity profiling
The catalytic activity of each LHE was measured by tethering Alexa647-fluorescent target dsOligo to the surface expressed LHE and measuring the decrease in fluorescence associated with dsOligo cleavage. Biotinylated fluorescent dsOligo is tethered to the HA epitope via an antibody-streptavidin bridge. Approximately 5 × 105 cells were first stained with 1:250 dilution biotinylated αHA (Covance) and 1:100 fluorescin isothiocyanate (FITC)-conjugated αMyc (ICL Labs) for 30 min at 4°C in the YSB. Preconjugated streptavidin–PE:Biotin-dsOligo-A467 was then bound to the yeast via the HA–biotin–streptavidin–PE interaction. This secondary stain was performed in the same buffer plus 400 mM KCl to allow biotin–streptavidin conjugation while disallowing the LHE to bind the dsOligo directly. Cells were washed in the cleavage solution: 10 mM NaCl, 113 mM K-Glutamate, 0.05% BSA and 10 mM HEPES and pH 8.2. Cells were resuspended in the cleavage buffer and split into two wells each. Each pair of wells were centrifuged and resuspended in cleavage buffer plus 2 mM either MgCl2 (cleavage permissive) or CaCl2 (cleavage restrictive); fluorescence loss due to magnesium-dependent cleavage of the dsOligo can subsequently be measured in these otherwise identical sample pairs. After a 20-min cleavage incubation at 37°C, cells were pelleted and resuspended in cold secondary stain buffer plus 4 mM EDTA to aid release of cleaved substrate and mitigate any end-holding effects on dsOligo-fluorophore release. In subsequent experiments, end-holding was determined by an increased loss in fluorescence when the fluorophore was conjugated to the plus half of the DNA substrate compared with when it was conjugated to the minus half during the flow cleavage assay; the final high-salt wash was not performed.
Sample fluorescence was measured on a BD LSRII™ cytometer, and the resulting data were analyzed using Flowjo. Each sample was normalized for enzyme concentration by applying an identical narrow FITC gate. Cells were then controlled for initial substrate concentration by adjusting a narrow PE gate for each non-cleaving Ca++ sample until the median A647 fluorescence intensity was matched for all samples. Relative cleavage efficiencies were derived for this normalized population by dividing the median DNA-A647 fluorescence value of the Mg++ sample (reduced fluorescence due to cleavage) by the corresponding median fluorescence value of the Ca++ matched pair (no cleavage). Higher Ca++/Mg++ ratios indicate more cleavage.
Specificity profiles were produced by determining cleavage of each of the 60 possible target sequences wherein each base at each of the 20 positions was substituted with each of the alternate three bases, as in Jarjour et al.’s (17) original description of this assay. In these experiments, all Ca++/Mg++ ratios were normalized to the Ca++/Mg++ ratio of the native target site.
Assessment of in vivo gene modification activity
Each LHE's target site was ligated into the truncated green fluorescent protein (GFP) of the traffic light reporter (18) using annealed, phosphorylated dsOligo (Supplementary Figure S1a). Lentivirus containing this construct was used to transduce HEK 293T cells at limiting dilution to obtain a population of cells with single copy chromosomal integration events. Cells were sorted against GFP and mCherry fluorescence to ensure that the reported started in the ‘off’ state. Endonuclease expression/GFP repair template vectors were generated by cloning each LHE from the yeast surface display vectors into the Lentiviral backbone containing the GFP repair fragment (Supplementary Figure S1b). ORFs were ligated in frame with a self-cleaving T2A peptide sequence, followed by a blue fluorescent protein, mTagBFP, to allow expression levels to be measured. On Day 0, 1 × 105 HEK cells of each reporter cell line were plated. On Day 1, each reporter cell line was transfected with 400 ng of LHE expression/repair plasmid with polyethylenimine at a wt/wt ratio of 4:1 in a pH 7, 150 mM NaCl, 5 mM HEPES buffer. Cell medium was replaced on day 2, and cells were allowed to accumulate conversion events until Day 4, when they were analyzed by flow cytometry on a BD LSRII™. Using FloJo software, each expressing population was defined by mTagBFP fluorescence. GFP+ and mCherry+ statistics, representing HR and mutagenic non-homologous end-joining events, respectively, were tabulated for these populations. mTagBFP positivity was determined in comparison with non-transfected cells for each cell line; GFP and mCherry in comparison with non-expressing populations in the transfected cells. To ensure that the non-expressing population was truly not expressing the construct, a small number of the highest mTagBFP-low cells were excluded from the non-expressing population.
Protein expression and purification
The IHjeMI reading frame was ligated into a commercially available pET15b expression plasmid (Novagen, Inc) that incorporates an N-terminal 6-histidine affinity purification tag and subsequent thrombin cleavage site prior to the endonuclease reading frame. One-point mutation was incorporated into the I-HjeMI coding sequence (corresponding to L232K), based on the knowledge that a similar mutation at that position increases the solubility of the homologous I-AniI (19). The I-AniI construct used for parallel expression experiments under the same conditions was as described previously (20). Both I-HjeMI and I-AniI constructs were expressed in Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene Inc.), using a method described previously for automatic induction of protein expression (21).
Harvested cells were collected by centrifugation, resuspended in 500 mM NaCl, 50 mM Tris–HCl, pH 8.0, 5% glycerol with 0.2 mM phenylmethylsulfonyl fluoride and benzonase and lysed by sonication. After a second centrifugation step, the clarified cell lysate was filtered (45 µ pore size), purified using a single Heparin affinity purification chromatography step (HiTrap Heparin HP, GE Healthcare Life Sciences) and eluted with an increasing gradient of 0.5–1.0 M NaCl (Supplementary Figure S2). The resulting protein was exchanged into thrombin cleavage buffer and the N-terminal His-tag was proteolytically removed. The homing endonuclease protein was then purified from the thrombin cleavage products by incubating the sample with nickel-NTA agarose resin (to bind the cleaved histidine tag and linked fusion polypeptide), followed by size exclusion chromatography.
Crystallographic analysis
The DNA oligonucleotides used for cocrystallization (5′-GCG CTG AGG AGG TTT CTC TGT TAA GCG A-3′ and 5′-CGC TTA ACA GAG AAA CCT CCT CAG CGC T-3′) were synthesized by Eurofins MWG Operon Inc (desalted; 50 nmol scale syntheses). The oligonucleotides were dissolved in 10 mM Tris–EDTA buffer pH 7.8, to a final concentration of the resulting DNA duplex of 1 mM, and the complementary DNA strands were annealed by incubation at 95°C for 5 min and cooling to 25°C, over a 2-h period. Purified I-HjeMI protein described above was mixed with 1.2-fold molar excess of the DNA substrate for a final concentration of 4.5 mg/ml protein, in the presence of 1 mM CaCl2, 400 mM NaCl and 50 mM Tris–HCl. The protein–DNA drops were mixed in a 1:1 volume ratio with a reservoir solution containing 0.2 M ammonium sulfate, 0.1 M bis–Tris pH 5.5 and 25% polyethelylene glycol 3350. Crystals grew within 1 week and were frozen by transfer for 1–2 min to crystallization reservoir solution supplemented with 30% sucrose (w/v), followed by direct submersion into liquid nitrogen. The space group of the crystals corresponded to P21212; a = 181.54; b = 73.58; c = 82.0 Å. The crystals diffracted up to ∼2.5 Å resolution at the ALS beamline 5.0.2 (Lawrence Berkeley National Laboratory). Data sets were processed using the HKL2000 software package (22). The structure of the I-HjeMI/DNA complex was solved by molecular replacement using the protein data bank (PDB) coordinates of the WT I-AniI/DNA complex (PDB: 2QOJ), and was modeled using COOT (Crystallography Object-Oriented Toolkit) (23) and refined using REFMAC/CCP4i (24). Due to significant disorder displayed by one of the two independent copies of the protein–DNA complex in the crystal asymmetric unit (which resulted in poor refinement statistics across the upper resolution shells), the final modeling and refinement was carried out to 3 Å resolution. While the values for Rwork and Rfree were still elevated at this resolution (0.28/0.36), the quality of all other refinement metrics and the fit of the well-ordered complex to experimental electron density were excellent.
RESULTS
Identification of I-AniI homologs and their target sites
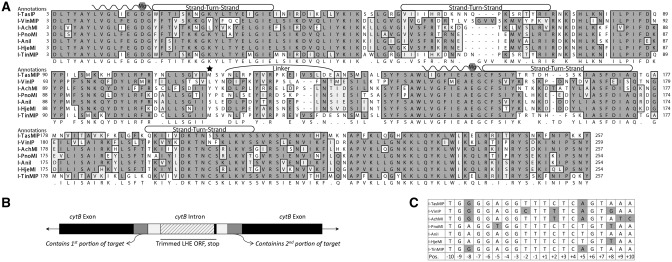
To identify LHE-coding sequences homologous to I-AniI, the National Center for Biotechnology Information ‘tblastn’ function was used to identify multiple putative LHEs of varying similarity in public sequence databases. Six homologs, each identified in different fungal mitochondrial genomes, and whose alignments are shown in Figure 1A, were selected based on the conservation of catalytic Mg++-coordinating residues within the LAGLIDADG motif to increase chances of finding an active LHE. The introns containing these putative LHEs also had flanking sequences differing from I-AniI, suggesting slightly altered cleavage specificities (25,26) (Figure 1B). These homologs were named according to the conventions put forth by Roberts et al. (27); notably, a ‘P’ suffix denotes a homolog of unverified enzymatic functionality. The additional suffix M was added to avoid redundancy in nomenclature relative to previously identified restriction or homing endonucleases, and to also denote that the host genome that harbored the LHE gene was mitochondrial. Figure 1C shows the putative target sequences of the six homologs as determined by the comparison of the sequence flanking the LHE insertion, where the I-AniI target sequence is found.
Figure 1.
Predicted homologs and targets. (A) The alignment of the I-AniI homologs with the residues shaded by chemical similarity. LAGLIDADG motifs are marked by waved lines. Conserved Mg++-coordinating residues and DNA-contact rich strand-turn-strand regions are also annotated. The homologous serine 111, a residue important for increased catalytic activity in I-AniI, is starred. The map was generated by MacVector using Gonnet-weighted pairwise and multiple sequence alignments with residue-specific and hydrophilic penalties. Residue numbering was matched to I-AniI, based on the first LAGLIDADG motif. (B) Schematic depicting the original host gene (black) with intron insertion (white) from which the LHE ORF sequences were taken and the exon/intron junctions used to predict target sequences (gray). (C) Predicted targets for each homolog, derived by comparing flanking intron/exon regions for each intronic LHE with those from I-AniI; differences therefrom are shaded.
Assessment of nuclease functionality
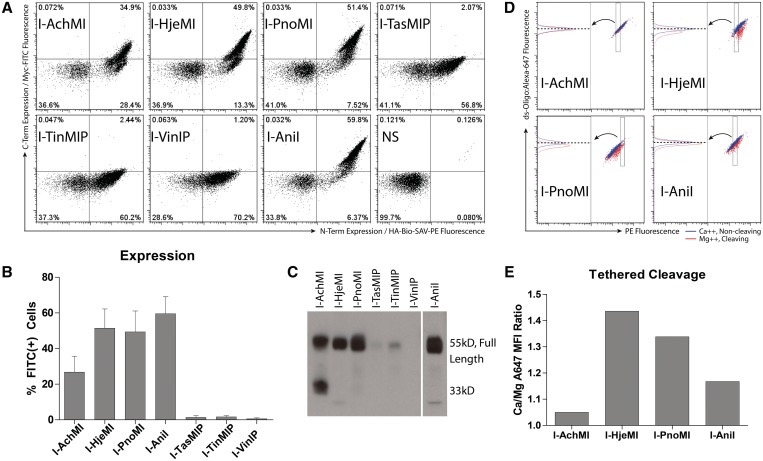
Yeast surface display represents a convenient method for characterizing putative LHEs in high throughput, as it provides facile access to quantitative information on protein folding and stability, DNA binding and cleavage, without the need for large scale enzymatic purification (17,28). In this approach, the enzyme is fused to an inducible surface displayed protein, Aga2p, which is anchored to the yeast's exterior by two disulfide bonds (29). Transit through the ER quality control and secretory pathways helps ensure that only LHEs which are stably folded at the induction temperature (20–30°C) are expressed on the yeast surface; dysfunctional variants which do not fold correctly are retained in direct proportion to their thermal stability (30). To compare the properties of six of the closest I-AniI homologs identified in Figure 1, yeast codon optimized ORFs were synthesized (Supplementary Table S1) and subcloned to the pCTCON2 vector. Relative expression levels were assessed using staining for N-terminal hemaggluTinMIn and C-terminal myc epitope tags (28,30) (Figure 2A). Three of the six homologs, I-AchMI, I-HjeMI and I-PnoMI, expressed full-length proteins on the yeast surface; the latter two very well, as determined by the level of C-terminal epitope tag expression (Figure 2B). I-TasMIP, I-TinMIP and I-VinIP surface expressed poorly, presumably because they were insufficiently stable at the 30°C induction temperature. Consistent with this interpretation, poor surface expression correlated with the accumulation of heterogeneously truncated proteins containing only the N-terminal tag, a pattern confirmed by western blot of the surface released protein (Figure 2C) and congruent with previous observations of surface-expressed proteins of low thermostability (31–34). Notably, the level of surface expression correlated with the level of amino acid sequence homology to I-AniI (Supplementary Figure S3).
Figure 2.
LHE characterization by flow cytometry. (A) Expression of full-length protein, determined by flow cytometry in a representative experiment. Staining against a C-terminal epitope tag and associated measured fluorescence intensity (Y-axis) and N-terminal epitope tag (X-axis) allows qualitative and quantitative assessment of surface expression. I-TasMIP, I-TinMIP and I-VinIP show minimal full-length protein (quadrant I and dual-positive), indicating reduced thermostability and/or poor folding. (B) Percentage of expressing (dual-positive) cells, as in panel (A), is summarized for five replicates (three for I-TasMIP, I-TinMIP and I-VinIP) with standard deviation plotted. (C) A western blot using antibodies against the N-terminal epitope tag allowed visualization of full length and truncated protein. Much of I-AchMI was expressed as ∼33 kDa protein fragment. Only minimal full-length protein and primarily heterogeneously truncated I-TasMIP, I-TinMIP and I-VinIP products were expressed, while I-Hje, I-PnoMI and I-AniI were primarily full length and in great abundance. (D) Demonstration of the gating strategy used to normalize substrate for the flow cleavage assay. These displayed populations are already normalized for enzyme concentration by a uniform, narrow FITC (C-terminal epitope) gate (data not shown). Equivalent amounts of tethered dsOligo across samples was selected by finding a streptavidin–PE level (rectangle) for each sample for which all DNA-A647 median fluorescence intensities (dashed horizontal line) were equal in the Ca++ sample (blue population). This gate was held constant for the matched pair Mg++ sample (red population), allowing quantification of magnesium-dependent loss of the DNA-conjugated fluorophore. The left half of the plot shows the population in the rectangular PE gate from the right plot (follow arrow). (E) Dividing the median Alexa647 fluorescence intensity of the calcium-containing sample (blue) by that of the magnesium-containing sample (red) yields a ratio proportional to the amount of enzymatic activity for a given LHE.
The three homologs with detectable surface expression, I-AchMI, I-PnoMI and I-HjeMI, were further assayed for binding and cleavage properties using fluorescently labeled oligonucleotide containing the predicted target site. Each bound their predicted native target with similar affinity to I-AniI (Supplementary Figure S4). Cleavage analysis was assessed using a previously described tethered oligonucleotide assay (17,20), depicted in Supplementary Figure S5, wherein enzyme and substrate levels were normalized (Figure 2D). I-HjeMI and I-PnoMI demonstrated catalytic activity against their putative DNA target sequences at levels comparable with, or slightly greater than, that of I-AniI against its target; I-AchMI showed a reduced level of activity (Figure 2E). Each enzyme's ability to specifically cleave its substrate was also evaluated with a solution-type assay following release of yeast surface expressed protein to validate the flow cytometry (Supplementary Figure S6).
DNA recognition specificity profiles
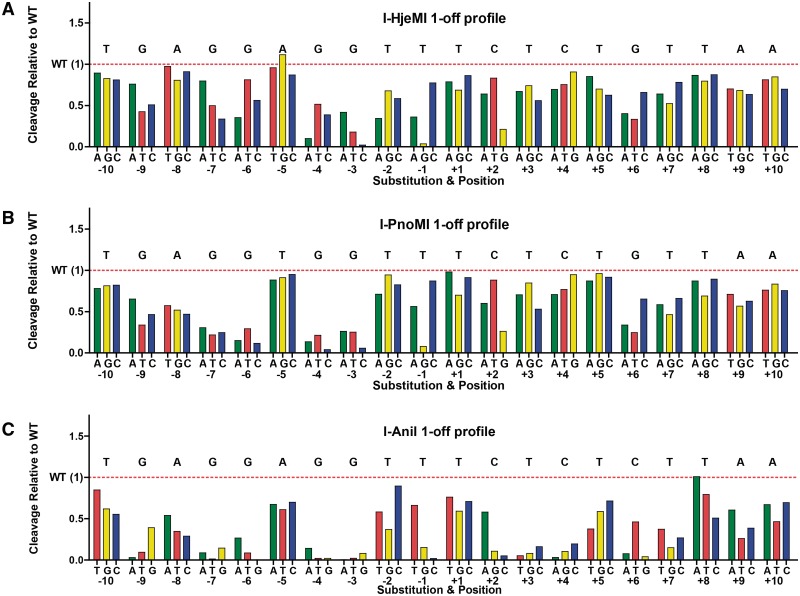
The above data indicate that an appreciable fraction of raw LHE ORFs identified in public databases by sequence similarity possess potentially useful enzymatic activities. To compare the biochemical properties of these enzymes in more detail, a ‘one-off’ cleavage specificity profile was determined for WT I-AniI and each of the two highly active enzymes, I-HjeMI and I-PnoMI, using the yeast tethered DNA cleavage assay (Figure 3). In this assay, a panel of DNA substrates, each harboring a single base pair mismatch relative to the LHE's physiological target, are assessed for relative cleavability by the expressed enzyme. This assessment revealed that, as expected, I-HjeMI and I-PnoMI exhibit overall I-AniI-like profiles with localized variances in positions where their predicted targets sites differ from that of I-AniI. For example, I-HjeMI exhibited elevated specificity at position −2 compared with the other two enzymes, but reduced specificity at −8, and to a lesser extent, −7 and −6, while I-PnoMI preferred a ‘T’ at −5, one of the two differences in its cognate target from I-AniI. Some small idiosyncratic differences were also observed, such as I-HjeMI preferring a ‘G’ at the −5 position, despite the fact that its predicted native target site has an ‘A’.
Figure 3.
Specificity profiles for (A) I-HjeMI, (B) I-PnoMI and (C) I-AniI. The impact of each possible single-base pair substitution is shown relative to wild-type cleavage efficiency (red dashed line, wild-type base noted above). Values at or close to zero denote minimal tolerance of the mismatch and therefore minimal cleavage, while values above one indicate a target is cleaved more efficiently than the predicted target.
We also assessed the potential for ‘end holding’, a property in which one DNA half-site is bound (and retained after cleavage) with particularly high affinity by the LHE when compared with the opposing half-site. This behavior is particularly notable for I-AniI and has been exploited for computational design purposes (14). Similar to I-AniI, both I-HjeMI and I-PnoMI were found to 'end-hold' the minus (or left) half of their DNA substrates (Supplementary Figure S7). This asymmetric pattern suggests that these homologs use a similar nucleotide discrimination mechanism to I-AniI (14), consistent with the high conservation of amino acid identity in the protein/DNA interface among the three enzymes in the beta sheets regions of the strand-turn-strand domains (20) (Figure 1A).
In vivo LHE activity
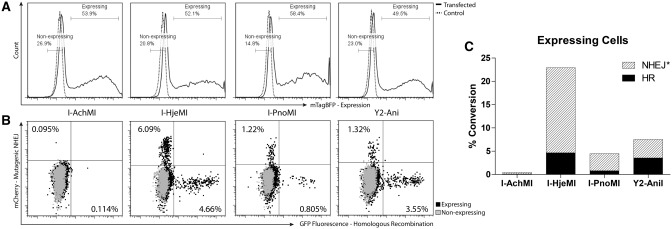
The three enzymes that exhibited detectable surface expression and cleavage activity (I-AchMI, I-PnoMI and I-HjeMI) were also assayed for their potential for endogenous DNA targeting and genome engineering using a reporter system in a human cell line. For this purpose, a recently described reporter system (18) was used to determine the relative ability of each LHE to induce mutagenic non-homologous end-joining (NHEJ) or HR, key genome engineering events. The reporter system is comprised of two parts: a chromosome-embedded reporter and an endonuclease expression and repair template vector (Supplementary Figure S1). If a break is generated in the reporter, it can be repaired by HR using the template GFP sequence to restore a functional GFP protein and the cell will be green. If the break is repaired by mutagenic NHEJ with a frameshift to the +3 reading frame, the GFP will be read-through and the mCherry will now be properly translated in-frame, producing a red cell. Nuclease expression and donor delivery is tracked by a blue fluorescent protein linked in translation via a T2A self-cleaving sequence.
To implement the assay, polyclonal cell lines were generated which harbored integrated single copies of reporters possessing each respective enzymes’ target site. Next, each of these cell lines were transfected with equal amounts of a donor template plasmid which also drives expression of the respective homing endonuclease. This resulted in similar distributions and sums of nuclease expression and repair template copy number, as assessed by the expression (fluorescence) of a monomeric blue fluorescent protein, mTagBFP (Figure 4A). I-AchMI exhibited little to no in vivo activity, consistent with its poor performance in the yeast tethered flow cleavage assay—this may reflect either an actual reduced catalytic efficiency, or that an impaired protein folding and/or thermal stability limits accumulation of active enzyme in cells cultured at 37°C. For these reasons, I-AchMI should be considered a compromised engineering scaffold for in vivo applications.
Figure 4.
LHE functionality in vivo. (A) Nuclease-expression histogram. Number of cells (Y-axis) of a given mTagBFP fluorescence (X-axis) are shown to be uniform for all transfected cells (solid line) and are compared with an untransfected control (dashed line). Gates used for comparison of expressing and non-expressing populations in panel (B) are shown. (B) Mutagenic NHEJ and HR repair events are shown for each nuclease-expressing population (black) compared with the non-expressing (gray). NHEJ events are mCherry(+) (Y-axis) and HR events are GFP(+) (X-axis). (C) As each mCherry(+) cell represents approximately one-third of the actual mutagenic NHEJ events (18) (Supplementary Figure S1d), a corrected value is plotted for NHEJ events, calculated by multiplying the number of mCherry(+) cells by three. Cells with converted loci, by event type, are shown as a percentage of the total expressing population.
In contrast, I-PnoMI and especially I-HjeMI, demonstrated repair of the GFP reporter by HR at frequencies much higher than native I-AniI and comparable with the previously reported increased activity variant Y2-Ani (35) (Figure 4B and C). Furthermore, remarkably high levels of mutagenic NHEJ were observed for I-HjeMI in the traffic light reporter assay: levels ∼3-fold higher than those stimulated by Y2-Ani and I-PnoMI. Thus, biotechnologically relevant activities appear to vary substantially among this group of closely related proteins.
I-HjeMI crystal structure
Based on I-HjeMI's enhanced in vitro and in vivo functional properties, we were curious whether it might possess structural differences from I-AniI that could be identified and correlated with its performance characteristics. Thus, we expressed I-HjeMI in bacteria, purified it to homogeneity and placed it into crystallization trials using a spectrum of standard conditions. In striking contrast to I-AniI, which we have found to be prone to chronic aggregation that required multiple solubilizing mutations to ameliorate, I-HjeMI was easily produced in large quantities and remained soluble, even at a 20 mg/ml concentration of the purified protein. The structure of the resulting complex of I-HjeI bound to its DNA target was determined at 3.0 Å resolution.
Two separate copies of the I-HjeMI/DNA complex are found within the asymmetric unit of the crystal form as described in ‘Materials and Methods' section. One of the complexes (corresponding to protein ‘chain A’ and DNA ‘chains B and C’ in the resulting model) was extremely well ordered, and displayed crystallographic packing contacts in which the overhanging A and T bases of the DNA duplex formed a continuous, Watson–Crick matched pseudocontinuous helix. The density for the entire complex (with the exception of several disordered residues in the linker that connects the two protein domains) was very clear, and the resulting model displayed excellent Ramachandran distribution and equally outstanding correlation to the density maps (Supplementary Figure S8). However, the second copy of the complex (corresponding to protein chain B in the resulting model) was very poorly ordered and displayed obvious clash at the ends of neighboring crystallographically related DNA molecules that resulted in a disruption of the base pairing at both ends of the duplex. As a result, the overall fit of the model to the second copy of the complex was of much lower quality. The poor quality of density across the second copy of the complex and the equally challenging model fit to that density preventing the overall refinement R-factors from being reduced to their usual acceptable values (Rwork and Rfree correspond to 0.28 and 0.36, respectively, see Table 1). However, the high sequence identity (85%) of I-HjeMI to the I-AniI homing endonuclease [which has previously been solved and refined in multiple independent space groups to high resolution (19,20,35)] and the excellent quality of electron density for the well-ordered complex of I-HjeMI to its DNA target nevertheless allowed us to generate an unambiguous comparison of the structures of the two homologous homing endonucleases (Figure 5). The results below are based on the analysis of only the well-ordered complex of I-HjeMI.
Table 1.
Data collection and refinement statistics
| Data collection | |
| ALS beamline | BL5.0.1 |
| Wave length (Å) | 1.00000 |
| Space group | P21212 |
| Unit cell dimension (Å) | a = 181.6, b = 73.6, c = 82.0, |
| Asymmetric unit content | Two complexes |
| Total reflections | 85 960 |
| Unique reflections | 22 491 |
| Resolution (Å)a | 50.00–3.00 (3.11–3.00) |
| Completeness (%)a | 98.4 (94.5) |
| Redundancya | 3.9 (3.6) |
| Rmergea,b | 0.046 (0.088) |
| Average I/sIa | 22.8 (13.8) |
| Refinement | |
| Rwork (%)c | 0.28 |
| Rfree (%)c | 0.36 |
| Protein residues | 504 |
| Nucleotides | 112 |
| Water molecules | 175 |
| Metal ions | 5 Ca++ |
| RMSD bond length (Å) | 0.0103 |
| RSMD bond angle (°) | 1.704 |
| Ramachandran distribution (%) | 90% preferred, 7% allowed, 3.0% outliers |
| Ramachandran distribution (Copy A) | 93% preferred, 6% allowed, 1% outliers |
| Average B-factors (A2) | 24.5 |
aHighest resolution shell values in parenthesis.
bRmerge = Σ|Ihi − <Ih>|/ΣIh, where Ihi is the ith measurement of reflection h, and <Ih> is the average measured intensity of reflection h.
cRwork/Rfree = Σh|Fh(o) - Fh(c)|/Σh|Fh(o)|, where Rfree was calculated with 5% of the data excluded from refinement.
Figure 5.
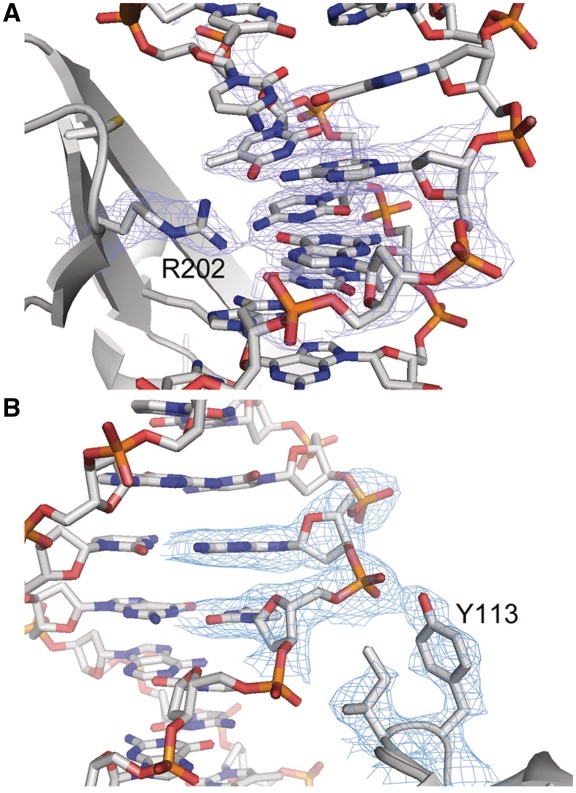
I-HjeMI model and electron density map. There is high-quality density (blue mesh) in the well-ordered complex around (A) R202 (K200 in Ani) and (B) Y113 (Y111 in Ani).
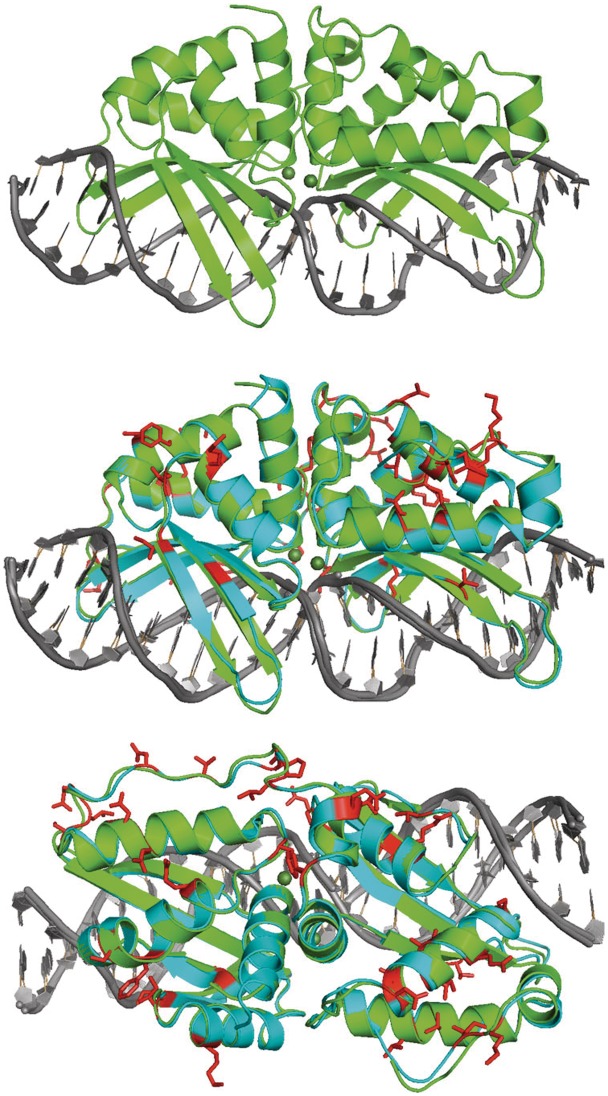
As expected, I-HjeMI displays a very similar overall structure to the structure of I-AniI (Figure 6), except for the few final residues of their C-termini and a short region of extended peptide sequence (spanning residues 123–129 in I-HjeMI) that links the N-terminal and the C-terminal domains of the two enzymes. The regions of folded secondary structure across the two enzymes and in particular the two central α-helices that contain the ‘LAGLIDADG’ sequence motifs, are closely superimposable [root mean square deviation (RMSD) less than 1 Å between all α-carbons] while the overall RMSD for all α-carbons across the superimposed proteins is ∼1.6 Å. The overall bend angles of the DNA and the geometric values of individual base pairs (i.e. propeller twist, roll, etc.) in the I-HjeMI and I-AniI complexes were also very similar.
Figure 6.
Structure of I-HjeMI. The solved structure of I-HjeMI (green) is shown bound to its target DNA (gray). This structure has been aligned to that of I-AniI (cyan) with differences highlighted red.
Of the 37 amino acid substitutions that distinguish I-HjeMI from I-AniI, 11 are located in the N-terminal folded domain (residues 1–110), 18 are located in the C-terminal domain (residues 126–254) and 8 are located in the linker that connects the two (residues 111–125). Of those substitutions, none are located in the LAGLIDADG helices and very few are buried in the hydrophobic core (the exception being I212, I213, L215 and L235 in the core of the I-AniI C-terminal domain, which are instead V212, V213, I216 and I235 in I-HjeMI). The remainder of amino acid differences involves residue positions that are partially or fully surface accessible. Four substitutions appear to involve residues that are involved in DNA contacts: I55, S111, R172 and K200 in I-AniI are instead K55, Y111, K172 and R200 in I-HjeMI. Of these substitutions, two (I55K and S111Y) result in additional nonspecific contacts to the DNA backbone, one (R172K) appears to have little effect on the structural mechanism of DNA recognition, and one (K200R) involves a side chain that appears to make contacts to nucleotide bases in the DNA target site (the arginine in I-HjeMI is located within H-bond distance to −4 A and −5G on one strand of the DNA target). The substitution of a tyrosine for serine at residue 111 (S111Y, which results in a nonspecific interaction to the DNA backbone outside of the 22 base pair target site) corresponds to a mutation that was previously introduced into I-AniI during a selection experiment for improved cleavage of its own wild-type target site (35).
DISCUSSION
This study demonstrates that a survey of evolutionary sequence space around a specific LHE is able to rapidly identify variants with a range of desirable properties for gene targeting applications. These enzymes displayed varying levels of thermostability, solubility, crystalizability, cleavage activity and capacity to induce different rates of site specific genetic alterations when expressed in a human cell line. These results highlight the considerable utility of surveying evolutionarily information as a supplement to rational protein engineering of novel LHE variants with specific properties.
The use of yeast surface display allowed multiple properties of each LHE ORF to be rapidly assessed, including thermal stability, binding and cleavage activity. Importantly, enzymes whose surface expressed well and exhibited significant activity in the tethered cleavage assay tended to perform very well in vivo, the notable exception being the originally described family member, I-AniI. Additionally, by combining an initial yeast surface display assessment with an in vivo reporter assay, we were able to identify two new enzymes that exhibit in vivo performance on par or better than Y2-Ani—an engineered variant specifically identified to have improved cleavage properties (35). As engineering attempts to modify an LHE's target specificity are often associated with reductions of catalytic efficiency toward the new target site, the availability of scaffolds with improved in vivo performance may provide both optimized starting points for engineering, as well as information on protein modifications that can be made to improve the performance of engineered variants. Furthermore, I-HjeMI's high solubility and crystalizability allowed rapid structural analysis, which can be a powerful tool when compiling many changes to a scaffold (6).
We chose our original search parameters to include both highly related homologs and those exhibiting locally altered substrate specificities, with the goal that sequence information from related scaffolds with differing specificities would help to inform engineering of the scaffolds to cleave new target sites. As exemplified in a contemporaneous work by Szeto et al. (36), small specificity-determining pockets which have been evolutionarily selected can be elucidated by comparing homologs at places of divergent sequence specificity, and these changes can be grafted onto related enzymes. In addition to revealing locally altered substrate specificities and activities, evolutionary sequence information may also help to inform us about the plasticity of enzymes at certain positions. This idea is supported by the improved specificity (versus I-AniI) that we observed at position −2 for I-HjeMI, the improved specificity of I-PnoMI at −8 relative to I-HjeMI and the reverse relationship at the position +5. Therefore, these locations may be identified as particularly amenable to modification, thus facilitating protein engineering of the scaffold group.
An interesting question raised by our results that warrants further investigation is whether there are distinguishing features of those proteins that did not surface express well or perform well in vivo, or those that performed exceptionally well. Strikingly, I-HjeMI and I-PnoMI natively harbor a tyrosine at the position analogous to 111 in I-AniI, one of two changes in Y2-AniI identified by directed evolution to significantly enhance the activity of the original I-AniI enzyme (28). As polar surface residues have been found to play critical roles in protein folding and stability (37–40), we generated homology models of the six I-AniI homologs analyzed here and used them to predict surface exposed residues. Consistent with the importance of these residues in promoting stable folding, I-HjeMI, I-PnoMI and I-AniI-I were predicted to possess fewer overall solvent-exposed hydrophobic residues outside of the protein–DNA interface (19,19,21) than any of I-VinIP, I-TasMIP and I-TinMIP (24,23,25). Incorporation of such analysis may allow to rapidly pare down a list of homologous LHE's identified in public sequence databases to those most likely to possess biotechnologically relevant or other positive attributes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–8 and Supplementary Methods.
FUNDING
National Institutes of Health (NIH) (RL1CA133832 to A.M.S. and RL1CA133833 to B.L.S.). Funding for open access charges: NIH (grant RL1CA133832 to A.M.S.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 2.Dujon B, Colleaux L, Jacquier A, Michel F, Monteilhet C. Mitochondrial introns as mobile genetic elements: the role of intron-encoded proteins. Basic Life Sci. 1986;40:5–27. doi: 10.1007/978-1-4684-5251-8_2. [DOI] [PubMed] [Google Scholar]
- 3.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashworth J, Taylor GK, Havranek JJ, Quadri SA, Stoddard BL, Baker D. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ. Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grizot S, Epinat J-C, Thomas S, Duclert A, Rolland S, Pâques F, Duchateau P. Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds. Nucleic Acids Res. 2010;38:2006–2018. doi: 10.1093/nar/gkp1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin A, Buckle M, Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO J. 1993;12:2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurica MS, Monnat RJ, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 13.Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 14.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi R, Lambert AR, Mak AN-S, Jacoby K, Dickson RJ, Gloor GB, Scharenberg AM, Edgell DR, Stoddard BL. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl Acad. Sci. USA. 2011;108:13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volna P, Jarjour J, Baxter S, Roffler SR, Monnat RJ, Stoddard BL, Scharenberg AM. Flow cytometric analysis of DNA binding and cleavage by cell surface-displayed homing endonucleases. Nucleic Acids Res. 2007;35:2748–2758. doi: 10.1093/nar/gkm182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarjour J, West-Foyle H, Certo MT, Hubert CG, Doyle L, Getz MM, Stoddard BL, Scharenberg AM. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ, Scharenberg AM. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, editor. Macromolecular Crystallography Part A. Vol. 276. USA: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 25.Lucas P, Otis C, Mercier JP, Turmel M, Lemieux C. Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res. 2001;29:960–969. doi: 10.1093/nar/29.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethuraman J, Majer A, Friedrich NC, Edgell DR, Hausner G. Genes within genes: multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol. Biol. Evol. 2009;26:2299–2315. doi: 10.1093/molbev/msp145. [DOI] [PubMed] [Google Scholar]
- 27.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev Sk, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Meth. Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 29.Watzele M, Klis F, Tanner W. Purification and characterization of the inducible a agglutinin of Saccharomyces cerevisiae. EMBO J. 1988;7:1483–1488. doi: 10.1002/j.1460-2075.1988.tb02966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 31.Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb. Chem. High Throughput Screen. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Boder ET. High-throughput engineering and analysis of peptide binding to class II MHC. Proc. Natl Acad. Sci. USA. 2010;107:13258–13263. doi: 10.1073/pnas.1006344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen F, Esteban O, Zhao H. Rapid identification of CD4+ T-cell epitopes using yeast displaying pathogen-derived peptide library. J. Immunol. Methods. 2008;336:37–44. doi: 10.1016/j.jim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szeto MD, Boissel SJS, Baker D, Thyme SB. Mining endonuclease cleavage determinants in genomic sequence data. J. Biol. Chem. 2011;286:32617–32627. doi: 10.1074/jbc.M111.259572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, Makhatadze GI. Rational stabilization of enzymes by computational redesign of surface charge-charge interactions. Proc. Natl Acad. Sci. USA. 2009;106:2601–2606. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor TJ, Vaisman II. Discrimination of thermophilic and mesophilic proteins. BMC Struct. Biol. 2010;10(Suppl 1):S5. doi: 10.1186/1472-6807-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jochens H, Aerts D, Bornscheuer UT. Thermostabilization of an esterase by alignment-guided focussed directed evolution. Protein Eng. Des. Sel. 2010;23:903–909. doi: 10.1093/protein/gzq071. [DOI] [PubMed] [Google Scholar]
- 40.Siddiqui KS, Poljak A, De Francisci D, Guerriero G, Pilak O, Burg D, Raftery MJ, Parkin DM, Trewhella J, Cavicchioli R. A chemically modified alpha-amylase with a molten-globule state has entropically driven enhanced thermal stability. Protein Eng. Des. Sel. 2010;23:769–780. doi: 10.1093/protein/gzq051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.