Abstract
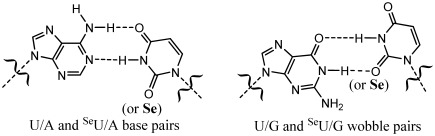
Specificity of nucleobase pairing provides essential foundation for genetic information storage, replication, transcription and translation in all living organisms. However, the wobble base pairs, where U in RNA (or T in DNA) pairs with G instead of A, might compromise the high specificity of the base pairing. The U/G wobble pairing is ubiquitous in RNA, especially in non-coding RNA. In order to increase U/A pairing specificity, we have hypothesized to discriminate against U/G wobble pair by tailoring the steric and electronic effects at the 2-exo position of uridine and replacing the 2-exo oxygen with a selenium atom. We report here the first synthesis of the 2-Se-U-RNAs as well as the 2-Se-uridine (SeU) phosphoramidite. Our biophysical and structural studies of the SeU-RNAs indicate that this single atom replacement can indeed create a novel U/A base pair with higher specificity than the natural one. We reveal that the SeU/A pair maintains a structure virtually identical to the native U/A base pair, while discriminating against U/G wobble pair. This oxygen replacement with selenium offers a unique chemical strategy to enhance the base pairing specificity at the atomic level.
INTRODUCTION
DNA and RNA are crucial genetic information carriers (1,2). The base pairs of DNAs (T/A and C/G) and RNAs (U/A and C/G) need to be highly specific and accurate for the purpose of the precise genetic information storage, replication, transcription and translation. However, the wobble base pairs, where U in RNA (or T in DNA) pairs with G instead of A, may compromise the high specificity of the base pairing. In RNA, especially non-coding RNA, U/G wobble pair (Figure 1) is ubiquitous (3) and sometimes it has the similar stability as the Watson–Crick U/A pair (4,5). U/G wobble pair offers unique structural and thermodynamic features (3–5). On the one hand, the U/G pairing increases structure and function diversities of RNA (6). But on the other hand, it may jeopardize the pairing specificity and can cause potential mutations in RNA transcription and protein translation. Codon–anticodon mismatch or misreading is observed with an error frequency at 10−5 or higher, which may affect the accuracy of synthesized proteins (7–9). For instance, the first position of the codon–anticodon interaction with wobble mismatch (U/G) was discovered in Escherichia coli (error frequency = 0.1%) with 100-fold higher than the normal error level (9). In this mis-incorporation of serine (codon: AGC) (9), glycine codon (GGC) in mRNA is recognized by Ser-charged tRNA (anticodon: GCU) instead of Gly-charged tRNA (anticodon: GCC). Similarly, the second position of the codon–anticodon interaction with wobble mismatch (U/G) was also observed, where Lys (codon: AAA) is mis-incorporated instead of normal incorporation of Arg (codon: AGA), with much higher error frequency (5–12%) (10). To avoid the negative impact of the wobble pairing on the level of protein synthesis, the genetic codes with degeneracy are used to deal with the consequence of the wobble pairing. Thus, wobble pairing is often observed at the third codon position through the codon degeneracy to limit errors. However, the codons forming the Watson–Crick pairs with tRNA anticodons are still preferred (11,12). Study shows that the third codon position with a Watson–Crick base pair can reduce the frequency of amino acid mis-incorporation by nearly 10-fold, and it is much more accurate than that with a wobble pair for the same amino acid (13). Nevertheless, the 3-nt genetic codes that accommodate the wobble pairing are used as the most ideal countermeasure at the level of protein synthesis in living organisms (14). Clearly, on the basis of the chemical principle, this degeneracy strategy properly guarantees the translation accuracy at the protein level by tolerating wobble pairs and silent mutations at the RNA and DNA levels.
Figure 1.
Native and Se-modified U/A pairs and U/G wobble pairs.
Since the 2-exo-oxygen of uridine plays a significant role in U/G wobble pair, we hypothesized that tailoring the steric and electronic effects at this site may discriminate against the wobble pair, enabling the modified U/A base pair with higher specificity. Interestingly, selenium has been discovered in natural tRNAs in the 2-Se-uridine form, i.e. 5-methylaminomethyl-2-selenouridine (mnm5se2U), in the wobble position on the anticodon loop (15,16). The function of such selenium modification is not completely clear yet, though it was proposed that such Se derivatization on tRNAs probably improves the accuracy and efficiency of protein translation (17). Similarly, the corresponding sulfur modification has been observed on natural tRNAs (18). Sulfur was chemically introduced to the 2-position of uridine (19,20). The S-modified U/G pair is slightly less stable than the native U/G pair (5), while the SU/A is more stable over the native U/A pair. Thus, we hypothesized that the 2-oxygen replacement with selenium (SeU, Figure 1) can destabilize and discriminate against the U/G wobble pair, because the atomic size of selenium (1.16 Å) is larger than that of sulfur (1.02 Å) and oxygen (0.73 Å). Moreover, selenium has the least ability to form a hydrogen bond among O, S and Se, which weakens the hydrogen bond originally formed by the 2-oxygen of the wobble pair. Thus, it is expected that this 2-Se-replacement can largely destabilize U/G pair by generating a steric hindrance against the pair and significantly weakening the hydrogen bond. Furthermore, it is expected that the 2-Se-substitution does not significantly affect the hydrogen bonds within the U/A pair, since the 2-oxygen is not directly involved in the U/A base pairing. Therefore, we decided to incorporate selenium into the 2-position of uridine in RNA, in order to atom-specifically increase the U/A pair specificity and disrupt the U/G wobble.
MATERIALS AND METHODS
Synthesis of 2-Se-uridine phosphoramidite
1-(5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-methylthiouridine 7
Five grams of dry compound 6 (its synthesis: Scheme S1 in Supplementary Data) was dissolved in dry N, N-dimethylformamide (DMF), followed by addition of iodomethane (5.5 ml, 89 mmol). 1,8-Diazabicyclo[5.4.0]undec-7-ene (2 ml, 13.3 mmol) was then added to the reaction mixture at 0°C. The reaction was monitored by thin layer chromatography (TLC) plate (12% methanol in dichloromethane, Rf = 0.4) and completed in 4 h. Ethyl acetate (50 ml) was poured into the mixture and DMF was removed by washing the organic layer with saturated sodium chloride solution. The organic phase was dried over anhydrous magnesium sulfate and evaporated under reduced pressure. The residue was purified by flash column chromatography (10% methanol in dichloromethane) and pure compound 7 was obtained in 95% yield. 1H NMR (CDCl3) δ: 7.87 (d, J = 7.7 Hz, 1H, H-6), 7.44–7.20 (m, 9H, Ar), 6.85 (m, 4H, Ar), 6.11 (br, 1H, OH), 5.88 (d, J = 6.0 Hz, 1H, H-1′), 5.54 (d, J = 7.7 Hz, 1H, H-5), 4.63 (m, 1H, H-4′), 4.44 (m, 1H, H-3′), 4.24 (d, J = 2.3 Hz, 1H, H-2′), 3.75 (d, J = 3.1 Hz, 6H, OCH3), 3.42 (m, 2H, H-5′), 3.40–3.30 (br, 1H, OH), 2.55 (s, 3H, SCH3); 13C NMR (CDCl3) δ: 169.19 (C-4), 164.36 (C-2), 158.88 (Ar), 144.49 (Ar), 140.13 (C-6), 135.37 (Ar), 135.22 (Ar), 130.41 (Ar), 130.28 (Ar), 128.32 (Ar), 128.28 (Ar), 127.29 (Ar), 113.54 (Ar), 108.92 (C-5), 91.95 (C-1′), 87.35 (C-Ar3), 84.82 (C-4′), 75.24 (C-2′), 71.63 (C-3′), 63.40 (C-5′), 55.40 (OCH3), 15.39 (SCH3); High resolution mass spectra (HRMS) electrospray ionization-time of fight (ESI-TOF) [M + H+] = 577.2003 (calc. 577.2008), Chemical formula: C31H33N2O7S.
Scheme 1.
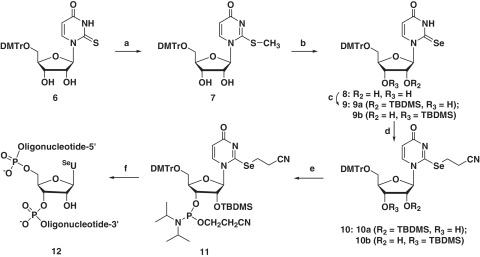
Synthesis of SeU-phosphoramidite 11 and RNAs (12). Reagents and conditions: (a) CH3I, DBU, DMF; (b) Se, NaBH4, EtOH; (c) TBDMS-Cl, imidazole, DMF; (d) ICH2CH2CN, (i-Pr)2NEt, CH2Cl2; (e) (i-Pr2N)2P(Cl)OCH2CH2CN, (i-Pr)2NEt, CH2Cl2; (f) solid-phase synthesis.
1-(5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-selenouridine 8
A solution of NaSeH was generated by addition of absolute ethanol (50 ml) to selenium (6.2 g, 78 mmol) and sodium borohydride (NaBH4, 4.43 g, 0.117 mol) at 0°C. The reaction was completed in 2 h and a clear solution was formed. The ethanolic solution was added to compound 7 (4.5 g, 7.80 mmol) and the mixture was stirred for 8 h under argon. The reaction mixture was then concentrated under reduced pressure and ethyl acetate (50 ml) was added to the residue. The organic layer was washed with water several times (5 × 30 ml), and then dried over anhydrous magnesium sulfate. Purification was performed by flash column chromatography (4% methanol in dichloromethane) and the light yellow compound (8) was obtained (85% yield). 1H NMR (CDCl3) δ: 10.95 (s, 1H, NH), 8.24 (d, J = 8.2 Hz, 1H, H-6), 7.44–7.19 (m, 9H, Ar), 6.84 (m, 4H, Ar), 6.48 (s, 1H, H-1′), 5.66 (d, J = 8.1 Hz, 1H, H-5), 4.48 (m, 2H, H-4′,H-3′), 4.22 (m, 1H, H-2′), 3.89 (s, 1H, OH), 3.79 (s, 6H, OCH3), 3.58 (dd, J = 23.6, 9.2 Hz, 2H, H-5′), 2.97 (br, 1H, OH); 13C NMR (CDCl3) δ: 175.74 (C-2), 159.21 (C-4), 158.98 (Ar), 158.94 (Ar), 144.45 (Ar), 140.82 (C-6), 135.38 (Ar), 135.18 (Ar), 130.35 (Ar), 130.27 (Ar), 128.30 (Ar), 128.28 (Ar), 127.45 (Ar), 113.58 (Ar), 108.37 (C-5), 96.86 (C-1′), 87.38 (C-Ar3), 84.41 (C-4′), 76.33 (C-2′), 69.19 (C-3′), 61.20 (C-5′), 55.48 (OCH3). HRMS (ESI-TOF) [M-H+]− = 609.1136 (calc. 609.1140), Chemical formula: C30H29N2O7Se; UV (MeOH): λmax = 311 nm (in methanol).
1-(2′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-selenouridine 9a and 1-(3′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-selenouridine 9b
5′-DMTr-2-selenouridine 8 (0.5 g, 0.82 mmol) was dissolved in dry DMF, then tert-butyldimethylsilyl chloride (TBDMSCl, 0.15 g, 0.98 mmol) and imidazole (0.11 g, 1.64 mmol) were added into the solution under nitrogen gas. The reaction was monitored by TLC plate (15% ethyl acetate in dichloromethane, Rf = 0.8). The mixture was stirred overnight at room temperature and then directly poured into ethyl acetate (20 ml) and washed with water (2 × 20 ml). The organic layer was dried by anhydrous magnesium sulfate and evaporated under reduced pressure. Two compounds, 9a and 9b, were obtained. The two regional isomers (ratio 1:1) were purified together by flash column chromatography (10% ethyl acetate in dichloromethane) and were not further separated. Since, it was both challenging and unnecessary to separate each isomer, we decided to move to the next step of synthesis without separation of these two isomers. HR-MS (ESI-TOF, 9a and 9b) [M-H+]− = 723.1990 (calc. 723.2005). Chemical formula: C36H43N2O7SeSi.
1-(2′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-cyanoethylselanyluridine 10a and 1-(3′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-cyanoethylselanyluridine 10b
The mixture (0.52 g, 0.72 mmol) of 9a and 9b was dissolved in dried dichloromethane at 0°C. Iodopropionitrile (0.78 g, 4.31 mmol) was added to the solution, followed by addition of diisopropylethylamine (0.37 ml, 2.15 mmol). The reaction was monitored by TLC plates (30% ethyl acetate in dichloromethane). After 4-h reaction, the solvent was removed under reduced pressure and the residue was partitioned between ethyl acetate (20 ml) and water (20 ml). The organic phase was dried over anhydrous magnesium sulfate and evaporated into dryness. Two crude products were obtained: 1-(2′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-cyanoethylselanyluridine 10a (Rf = 0.35) and 1-(3′-O-tert-butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-cyanoethylselanyluridine 10b (Rf = 0.30). These two compounds can be separated by flash column chromatography (15% ethyl acetate in dichloromethane). 10a was obtained in 0.228 g (41% yield) and 10b was obtained in 0.235 g (42% yield). 10a: 1H NMR (CDCl3) δ: 7.96 (d, J = 7.7 Hz, 1H, H-6), 7.53–7.11 (m, 9H, Ar), 6.85 (m, 4H, Ar), 5.71 (d, J = 7.7 Hz, 1H, H-5), 5.60 (d, J = 6.5 Hz, 1H, H-1′), 4.61–4.49 (m, 1H, H-4′), 4.31 (m, 2H, H-3′, H-2′), 3.80 (s, 6H, OCH3), 3.54–3.34 (m, 4H,H-5′, SeCH2CH2CN), 3.01 (m, 2H, SeCH2CH2CN), 2.91 (s, 1H, OH), 0.94 (s, 9H, SiCMe3), 0.09 (d, 6H, SiMe2); 13C NMR (CDCl3) δ: 167.70 (C-4), 159.04 (C-2), 158.51 (Ar), 144.17 (Ar), 139.19 (C-6), 134.91 (Ar), 134.74 (Ar), 130.26 (Ar), 130.17 (Ar), 128.30 (Ar), 128.11 (Ar), 127.58 (Ar), 118.78 (CN), 113.58 (Ar), 110.61 (C-5), 93.13 (C-1′), 87.82 (C-Ar3), 85.39 (C-4′), 77.27 (C-2′), 72.40 (C-3′), 63.82 (C-5′), 55.44 (OCH3), 25.84 (SiCMe3), 24.06 (SeCH2CH2CN), 18.89 (SeCH2CH2CN), 18.14 (SiCMe3), −4.56 (SiCH3), −4.91 (SiCH3). HRMS (ESI-TOF) [M+H+]+ = 778.2464 (calc. 778.2427). Chemical formula: C39H48N3O7SeSi. 10b: 1H NMR (CDCl3) δ: 8.09 (d, J = 7.7 Hz, 1H, H-6), 7.32 (m, 9H, Ar), 6.88 (m, 4H, Ar), 5.75 (d, J = 7.7 Hz, 1H, H-5), 5.65 (d, J = 3.7 Hz, 1H, H-1′), 4.46 (m, 1H, H-2′), 4.21 (dd, J = 9.3, 5.1 Hz, 1H, H-4′), 4.18–4.08 (m, 1H, H-3′), 3.83 (s, 6H, OCH3), 3.70 (m, 1H, H-5′), 3.55 (m, 1H, H-5′), 3.41 (m, 2H, SeCH2CH2CN), 3.22 (d, J = 5.5 Hz, 1H, OH), 3.08 (m, 2H, SeCH2CH2CN), 0.90 (s, 9H, SiCMe3), 0.11 (d, 6H, SiMe2). 13C NMR (CDCl3) δ: 167.90 (C-4), 159.05 (C-2), 157.82 (Ar), 143.99 (Ar), 138.96 (C-6), 135.02 (Ar), 134.89 (Ar), 130.35 (Ar), 130.34 (Ar), 128.37 (Ar), 128.29 (Ar), 127.58 (Ar), 118.95 (CN), 113.56 (Ar), 113.53 (Ar), 110.49 (C-5), 93.84 (C-1′), 87.57 (C-Ar3), 84.72 (C-4′), 76.17 (C-2′), 71.05 (C-3′), 61.71 (C-5′), 55.48 (OCH3), 25.82 (SiCMe3), 24.12 (SeCH2CH2CN), 18.92 (SeCH2CH2CN), 18.17 (SiCMe3), −4.59 (SiCH3), −4.60 (SiCH3). HRMS (ESI-TOF) [M + H+]+ = 778.2401 (calc. 778.2427). Chemical formula: C39H48N3O7SeSi.
1-[2′-O-tert-butyldimethylsilyl-3′-O-(2-cyanoethyl-N,N-diisopropylamino) phosphoramidite-5′-O-(4,4′-dimethoxytrityl-β-d-ribofuranosyl)]-2-cyanoethylselanyluridine 11
Diisopropylethylamine (15.5 mg, 0.12 mmol) and 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (26 mg, 0.11 mmol) were added to a solution of 10a (100 mg, 0.10 mmol) in dry dichloromethane (5 ml) at room temperature under nitrogen gas. The mixture was monitored by TLC (15% ethyl acetate in dichloromethane). When the reaction was completed in 4 h, rapid Al2O3 column chromatography (dichloromethane as the eluent) was performed to remove the organic salts. The solvent was then evaporated under reduced pressure and the residue was dissolved in 0.5 ml dichloromethane and precipitated in dry hexane under vigorous stirring. The precipitate was collected by filtration, dried under reduced pressure and directly used for solid-phase synthesis. 1H NMR (CDCl3) δ: 7.95 (d, J = 7.7 Hz, 1H, H-6), 7.30 (m, 9H, Ar), 6.84 (m,4H, Ar), 5.82 (d, J = 7.6 Hz, 1H, H-1′), 5.63 (d, J = 7.7 Hz, 1H, H-5), 4.68 – 4.43 (m, 1H, H-4′), 4.43 – 4.29 (m, 1H,), 4.24 (s, 1H), 3.98 (dd, J = 15.6, 8.4 Hz, 2H), 3.80 (s, 6H, OCH3), 3.63 (dd, J = 19.8, 7.3 Hz, 4 H), 3.42 (dd, J = 19.3, 8.7 Hz, 5H), 3.01 (s, 2H), 2.70 (d, J = 5.8 Hz, 2H), 1.20 (d, J = 6.7 Hz, 18H), 1.08 (d, J = 6.5 Hz, 6H), 0.92 (s, 13H), 0.14 – 0.02 (m, 9H); 13C NMR (CDCl3) δ: 167.83 (C-4), 159.04 (C-2), 158.71 (Ar), 144.16 (Ar), 139.02 (Ar), 134.96 (Ar), 134.74 (Ar), 130.21 (Ar), 130.14 (Ar), 128.38 (Ar), 128.04 (Ar), 127.58 (Ar), 118.86 (SeCH2CH2CN), 117.75 (OCH2CH2CN), 113.67 (Ar), 110.69 (C-5), 92.50 (C-1′), 87.92 (C-Ar3), 85.43 (C-4′), 77.15 (C-2′), 72.81 (C-3′), 63.72 (C-5′), 59.39 (OCH2CH2CN), 55.48 (OCH3), 43.22-43.10 (NCMe2), 29.90 (OCH2CH2CN), 26.12-25.97 (NCMe2), 24.85 (SiCMe3), 24.02 (SeCH2CH2CN), 18.94 (SeCH2CH2CN), 18.37 (SiCMe3), −4.35 (SiCH3), −4.56 (SiCH3); 31P NMR (CDCl3) δ: 148.81, 152.30. HRMS (ESI-TOF) [M + H+]+ = 978.3528 (calc. 978.3505). Chemical formula: C48H65N5O8PSeSi.
Solid-phase synthesis of the 2-Se-functionalized RNAs
ABI3400 DNA/RNA Synthesizer was used for all the RNA oligonucleotides synthesis (1.0 µmol scale). All the non-modified nucleoside phosphoramidite reagents used were ultra-mild (Glen Research). RNA oligonucleotides were synthesized in DMTr-on form, cleaved from the beads and deprotected by the treatment of 0.05 M K2CO3 methanol solution for 10 h at room temperature. After evaporating the solution to dryness, the 2′-TBDMS deprotection was performed in TBAF (0.5 ml, 1 M) for 14 h at room temperature. Then the RNAs were treated with 1 M Tris–HCl buffer (0.5 ml, pH 7.5) for 5 min, followed by concentrating to 0.5 ml and desalting using G-25 Sephadex column. The 5′-DMTr deprotection was then performed using Glen-Pek RNA column, followed by desalting using Sep-Pak Vas column.
HPLC analysis and purification
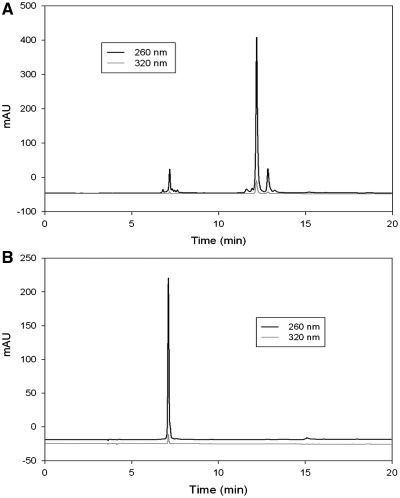
The RNA oligonucleotides were analyzed and purified by reversed-phase high performance liquid chromatography (RP-HPLC), flow rate 6 ml/min [buffer A: 20 mM triethylammonium acetate (TEAAc, pH 7.1) in water; buffer B: 20 mM TEAAc (pH 7.1) in 50% acetonitrile]. The HPLC analysis was performed with a linear gradient from buffer A to 100% buffer B in 20 min. Native RNAs were purchased from Integrated DNA Technologies. The concentrations of the native, Se-modified RNAs were adjusted to 1.0 mM in water. The Se-RNA samples were characterized by matrix assisted laser desorption/ionization-time of fight mass spectrometry (MALDI-TOF MS) (Table 1) and HPLC (Figure 2).
Table 1.
MALDI-TOF MS of 2-Se-U RNAs
| Entry | Oligonucleotide molecular formula | Measured (calc.) m/z |
|---|---|---|
| 1 | 5′-rGUAUASeUAC-3′ | [M + H]+ = 2558.7 (2558.5) |
| C76H94N29O53P7Se | ||
| 2 | 5′-rAUCACCSeUCCUUA-3′ | [M+H]+ = 3740.3 (3740.2) |
| C111H141N38O82P11Se | ||
| 3 | 5′-rAAUGCSeUGCACUG-3′ | [M + H]+ = 3859.4 (3859.3) |
| C114H142N45O81P11Se |
Figure 2.
HPLC analysis of 2-Se-U modified RNA 12-mer (5′-rAUCACCSeUCCUUA-3′). (A) The crude DMTr-on Se-RNA, retention time was 12.2 min. (B) The pure DMTr-off Se-RNA with same gradient and buffer, retention time was 7.1 min. Samples were eluted with a linear gradient from buffer A (20 mM triethylammonium acetate, pH 7.1) to 70% buffer B (50% acetonitrile, 20 mM triethylammonium acetate, pH 7.1) in 10 min, to 100% buffer B in 12 min and continuous 100% buffer B to 20 min.
pH titration curve of 2-selenouridine
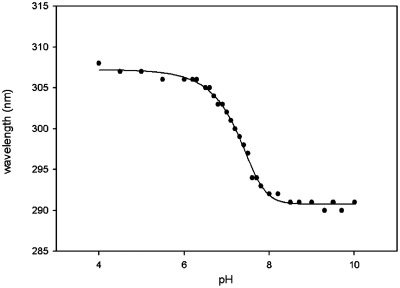
2-Selenouridine was prepared through detritylation of 1-(5′-O-4,4′-dimethoxytrityl-β-d-ribofuranosyl)-2-selenouridine (8) by acid treatment. The 2-selenouridine solutions were adjusted to desired pH values in the buffer of 50 mM Na2HPO4 at room temperature. The UV–Vis spectra were recorded every 0.1 pH unit between pH 6–8 and every 0.2–0.5 pH unit between pH 4–6 and pH 8–10. The pH of each solution was measured before and after its UV–Vis spectrum collection and the error was within ± 0.02 pH unit. The titration data was plotted and shown in Figure 3.
Figure 3.
Plot of wavelength (nm) versus pH for 2-selenouridine nucleoside. The fitted titration curve yields the pKa value (7.29 ± 0.02).
Thermodenaturization of duplex RNAs
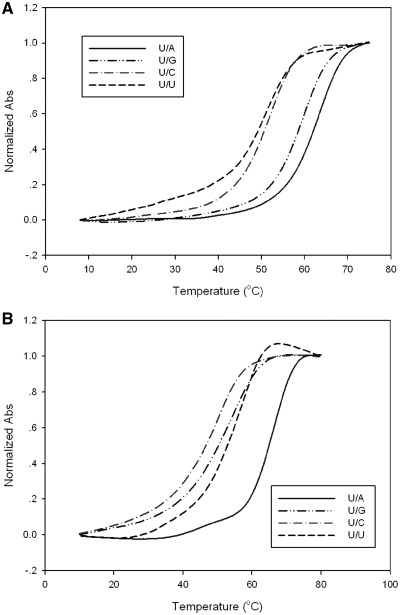
The UV-melting temperature studies were carried out by Cary 300 UV–Vis Spectrophotometer and a temperature control module. The samples (2 μM RNA duplexes) were dissolved in buffer of 150 mM NaCl, 2 mM MgCl2 and 10 mM Na2HPO4–NaH2PO4 (pH 6.8). The samples were heated to 80°C and cooled to room temperature slowly and then kept in 4°C overnight. The heating rate of melting experiment was 0.5°C per min. The melting temperature data and curves of the matched and mismatched duplexes are presented in Table 2 and Figure 4.
Table 2.
Melting temperatures (Tm) of the native, S- and Se-modified RNA duplexes
| Entry | Sequences | Base pairs | Tm (°C) |
|---|---|---|---|
| 1 | I: 5′-rAUCACCUCCUUA-3′ | ||
| 2 | I + 3′-rUAGUGGAGGAAU-5′ | U/A | 62.8 |
| 3 | I + 3′-rUAGUGGGGGAAU-5′ | U/G | 62.5 |
| 4 | I + 3′-rUAGUGGCGGAAU-5′ | U/C | 50.6 |
| 5 | I + 3′-rUAGUGGUGGAAU-5′ | U/U | 48.8 |
| 6 | II: 5′-rAUCACCSeUCCUUA-3′ | ||
| 7 | II + 3′-rUAGUGGAGGAAU-5′ | SeU/A | 65.8 |
| 8 | II + 3′-rUAGUGGGGGAAU-5′ | SeU/G | 58.5 |
| 9 | II + 3′-rUAGUGGCGGAAU-5′ | SeU/C | 50.3 |
| 10 | II + 3′-rUAGUGGUGGAAU-5′ | SeU/U | 57.3 |
| 11 | III: 5′-rAAUGCUGCACUG-3′ | ||
| 12 | III + 3′-rUUACGACGUGAC-5′ | U/A | 64.1 |
| 13 | III + 3′-rUUACGGCGUGAC-5′ | U/G | 59.4 |
| 14 | III + 3′-rUUACGCCGUGAC-5′ | U/C | 52.0 |
| 15 | III + 3′-rUUACGUCGUGAC-5′ | U/U | 51.5 |
| 16 | IV: 5′-rAAUGCSeUGCACUG-3′ | ||
| 17 | IV + 3′-rUUACGACGUGAC-5′ | SeU/A | 66.5 |
| 18 | IV + 3′-rUUACGGCGUGAC-5′ | SeU/G | 55.5 |
| 19 | IV + 3′-rUUACGCCGUGAC-5′ | SeU/C | 51.9 |
| 20 | IV + 3′-rUUACGUCGUGAC-5′ | SeU/U | 58.0 |
Bold and underlined sequences indicate the pairing and mis-pairing sites.
Figure 4.
Normalized UV-melting curves of RNA duplexes. (A) Native RNA (5′-rAAUGCUGCACUG-3′) paired with matched and mismatched strands. (B) Se-RNA (5′-rAAUGCSeUGCACUG-3′) with matched and mismatched strands.
Crystallization
The purified RNA oligonucleotide (5′-GUAUA-SeU-AC-3′, 1 mM) was heated to 80°C for 2 min, and cooled down slowly to room temperature. The Nucleic Acid Mini Screen Kit (Hampton Research) was applied to screen the crystallization conditions at different temperatures (10, 20 and 25°C) using the hanging-drop method by vapor diffusion.
Data collection
Perfluoropolyether was used as a cryoprotectant during the crystal mounting, and data collection was taken under the liquid nitrogen stream at −174°C. The Se-RNA crystal data were collected at beam line X12B and X12C in NSLS of Brookhaven National Laboratory. A number of crystals were screened to identify the one with strong anomalous scattering at the K-edge absorption of selenium. The distance of the detector to the crystals was set to 150 mm. The wavelength of 0.9795 Å was chosen for selenium SAD phasing. The crystals were exposed for 10 or 15 s/image with 1° oscillation, and a total of 180 images were taken for each data set. All the data were processed using HKL2000 and DENZO/SCALEPACK (21).
Structure determination and refinement
The structure of the Se-RNA [5′-GUAUA-SeU-AC-3′]2 was solved by molecular replacement with both CNS (22) and Phaser (23). The refinement protocol includes simulated annealing, positional refinement, restrained B-factor refinement, and bulk solvent correction. The stereo-chemical topology and geometrical restrain parameters of DNA/RNA (24) have been applied. The topologies and parameters for modified uridine with 2-selenium (US) were constructed and applied. After several cycles of refinement, a number of highly ordered waters were added. Finally, the occupancies of selenium were adjusted. Cross-validation (25) with a 5–10% test set was monitored during the refinement. The σA-weighted maps (26) of the (2 m|Fo|−D|Fc|) and the difference (m|Fo|−D|Fc|) density maps were computed and used throughout the model building (Table 3).
Table 3.
Data collection and refinement statistics of SeU-RNA
| Structure (PDB ID) | GUAUA-SeU-AC (3S49) |
|---|---|
| Data collection | |
| Space group | R32 |
| Cell dimensions: a, b, c (Å), α, β, γ (°C) | 47.095, 47.095, 424.655, 90, 90, 120 |
| 90, 90, 120 | |
| Resolution range (Å) (last shell) | 50.0–2.28 (2.37–2.28) |
| Unique reflections | 9870 (959) |
| Completeness (%) | 99.8 (99.4) |
| Rmerge (%) | 7.7 (23.5) |
| I/σ(I) | 21.0 (3.9) |
| Redundancy | 18.8 (10.7) |
| Refinement | |
| Resolution range (Å) | 30.0–2.3 |
| Rwork(%) | 21.4 |
| Rfree (%) | 26.9 |
| Number of reflections | 8206 |
| Number of atoms | |
| Nucleic acid (double) | 1162 |
| Heavy atoms and ion | 7 Se |
| Water | 71 |
| RMS deviations | |
| Bond length (Å) | 0.007 |
| Bond angle | 1.169 |
RESULTS AND DISCUSSION
Synthesis of 2-Se-uridine phosphoramidite
Though selenium was incorporated into uridine four decades ago (27,28), RNA containing 2-Se-uridine (SeU) has not been synthesized because of the synthetic challenge. Recently, our laboratory has successfully developed a novel strategy to incorporate the selenium functionality to the 2-position of thymidine in DNA (29). This successful strategy has encouraged us to introduce the selenium functionality to the 2-position of uridine in RNA. Herein, we report the first synthesis of the 2-selenouridine derivatives and RNAs. The synthesis (Scheme S1 in Supplementary Data) started from the glycosidation (30) of the acylated ribofuranose (1) with silylated 2-thiouracil (3), followed by benzoyl deprotection and trityl protection of the 5′-hyroxyl group to offer 6 (31). After methylation of 6 to activate the 2-thio-functionality (29), NaSeH was used to displace the 2-S-functionality and offer the 2-Se-uridine 8 in 85% yield. Following the protections of the 2′-hydroxyl group and the 2-Se-functionality with ICH2CH2CN, the Se-phosphoramidite 11 was synthesized by phosphitylation of 10 a (29,32,33). The SeU-phosphoramidite was finally incorporated into RNAs by solid-phase synthesis. The synthesized SeU-RNAs (12) were deprotected, purified and confirmed by HPLC and MS (Table 1 and Figure 2). The characterization of the Se-nucleosides and Se-nucleotides is presented in Supplementary Figures S1–S23.
Characterization of the 2-Se-functionalized RNA
After cleavage from solid support and deprotection, the crude DMTr-on RNAs were purified by RP-HPLC and lyophilized to dryness. As shown in Figure 2A, the coupling yield is ∼90%. The 5′-DMTr deprotection of the oligonucleotides was then performed using Glen-Pek RNA column. The HPLC analysis of the DMTr-off RNA was shown in Figure 2B. All the pure seleno-RNA oligonucleotides were characterized by MALDI-TOF MS (Table 1).
Thermodenaturization study of 2-Se-uridine RNAs containing match and mismatch base pairs
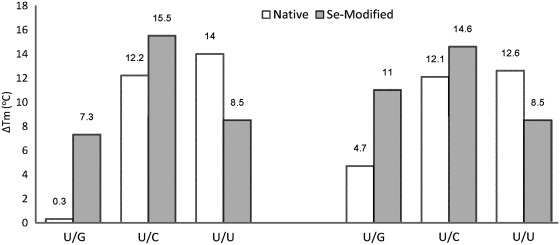
UV-melting temperatures (Tm) of the native and Se-modified duplexes with match and mismatch sequences are shown in Table 2, Figures 4 and 5. Tm of the Se-RNA duplex containing the SeU/A Watson–Crick pair was 3.0°C higher for one duplex (or 2.4°C higher for the other duplex) than those of the corresponding duplexes containing native U/A pair (Table 2). Comparing with native U/G, the SeU/G pair is ∼4°C less stable than the native U/G pair, suggesting that SeU discourages the SeU/G pair formation. While the SeU/C mis-pair is slightly less stable than the native U/C mis-pair, the SeU/U mis-pair is more stable than the native U/U mis-pair. The higher stability may be attributed to the higher acidity of the imino group (3-NH) of SeU [pKa = 7.29 ± 0.02, Figure 3, compared to that of the native uridine (pKa = 9.18 ± 0.02) (34)], which may promote U/U interaction via hydrogen bond. In addition, considering a selenium atom is 0.43 Å larger in atomic radius than an oxygen atom, the 2-Se atom may strengthen the stacking interaction between SeU and its 3′-nucleobase (Supplementary Figure S24).When directly comparing the Watson–Crick base pairs (U/A and SeU/A) with their own corresponding mis-pairs, it is clear that SeU/A pair has the balanced discrimination against all mis-pairs, with the Tm differences of SeU/G (7.3°C for one duplex or 11°C for the other in Table 2, and Figure 5), SeU/C (15.5°C or 14.6°C), and SeU/U (8.5°C for both sequences). While maintaining fine discrimination against U/C pair (Tm difference: 12.2 or 12.1°C) and U/U pair (Tm difference: 14 or 12.6°C), the native U/A pair poorly discriminates against U/G wobble pair (the Tm differences: 0.3 or 4.7°C in Figure 5 and Table 2). Therefore, in general, SeU/A has higher base pair fidelity than the native U/A pair.
Figure 5.
Differencesa of melting temperatures (Tm) of the native and Se-modified U/A pairs and their corresponding mis-pairs. Native (white bar) refers to the Tm difference between the native U/A pair and the other mis-pairs (U/G, U/C and U/U); Se-Modified (gray bar) refers to the Tm difference between the SeU/A pair and the other modified mis-pairs (SeU/G, SeU/C and SeU/U).
Since the Tm differences between the native U/A pair and U/G wobble pair were relatively small (0.3 and 4.7°C in Figure 5). The small Tm differences indicate possible changes between U/A and U/G pairs without a significant decrease in duplex stability. This is consistent with the ubiquitous presence of U/G wobble pair in RNAs, which diversifies the structure and function of RNAs, especially non-coding RNAs. Such small thermostability difference between native U/A pair and U/G wobble pair has been previously observed in the literature (4,5). On the contrary, the Tm differences between the SeU/A and SeU/G pairs were significant, such as 7.3°C (versus 0.3°C in the native) and 11°C (versus 4.7°C in the native) in Figure 5. The single selenium atom replacement directly decreases the thermal stability of the U/G wobble pair by 4.0 and 3.9°C in RNA duplexes (Table 2). This experimental observation reveals that the U/G wobble pair can be greatly discriminated by incorporating a selenium atom to the 2-position of uridine. The strong discrimination against U/G pair is mainly attributed to the selenium disruption of the hydrogen bond formed by the 2-oxygen (Figure 1) and to the steric effect of the bulky selenium atom at the 2-position. Our results indicate that the 2-Se-modification on uridine significantly increases the high specificity of the U/A base pair.
Crystallization and data collection of Se-RNA
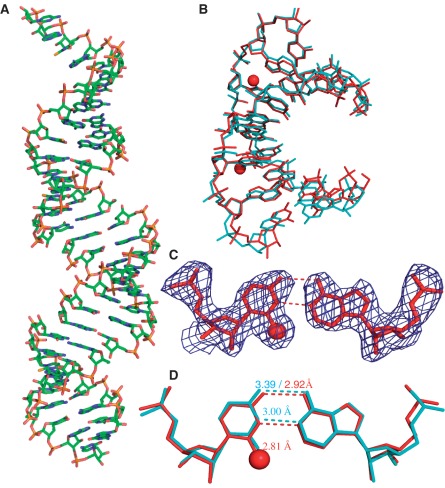
Consistently, our crystal structure study of the SeU-RNA [5′-rGUAUA-SeU-AC-3′] supports the biophysical results of SeU/A pairing. Similar to the native, the Se-RNA crystal is also in rhombohedral space group R32. The Se-RNA structure, determined at 2.3 Å resolution, is virtually identical to the native one (35) (at 2.2 Å resolution, Figure 6). Interestingly, the Se-RNA crystals grew much faster than the native ones. In six days, the Se-RNA formed diffraction-quality crystals in decent sizes (approximately 0.05 × 0.05 mm), while the corresponding native did not crystallize in 3–4 weeks under the same conditions. Moreover, the Se-RNA crystals could form in broader buffer conditions (12 out of 24 conditions in Hampton buffers) than the corresponding native (2 out of 24 conditions). This observation of faster crystal growth of the Se-RNA is consistent with the Se-facilitated duplex stability. As shown in Figure 6A, there are seven self-complementary RNA molecules in a unit cell, and the overall shape of the duplexes is almost linear (∼8° inclination to the screw axes). Although this assembling pattern results in the discontinued backbones and grooves, the duplexes stack on top of each other in a head-to-tail fashion, and a peudo-fiber is formed. The data collection and structure refinement statistics are summarized in Table 3.
Figure 6.
Global and local structures of the SeU-containing RNA r[5′-GUAUA(SeU)AC-3′]2 with a resolution of 2.3 Å. (A) The overall structure of duplex. (B) The superimpose comparison of one SeU-RNA duplex (red; PDB ID: 3S49) with its native counterpart [5′-r(GUAUAUA)-dC-3′]2 (cyan; PDB ID: 246D) with a RMSD value 0.55. The two red balls represent the selenium atoms. (C) The experimental electron density of SeU6/A11 base pair with σ = 1.0. (D) The superimpose comparison of the local base pair SeU6/A11 (red) and the native U6/A11 (cyan). The numbers indicate the distance between the corresponding atoms.
Since 2-exo-oxygen of uridine is not involved in the hydrogen bond interactions of U/A pairing, it's expected that the U/A pair will accommodate the larger selenium atom at this position (Figures 1 and 6C). The Se-modification also leads to the acidity increase of the 3-imino group (NH) in the 2-Se-uridine, which strengthens the hydrogen bond between N3 of U6 and N1 of A11. Indeed, after the selenium modification, the U/A hydrogen bond length between N3 of U6 and N1 of A11 is shortened from the native distance (3 Å) to the Se-modified distance (2.81 Å). Moreover, after the Se-modification (Figure 6D), the U/A hydrogen bond length between O4 of U6 and N6 of A11 decreases by 0.47 Å from the native distance (3.39 Å) to the Se-modified distance (2.92 Å). The shortened H-bond lengths indicate stronger H-bonds, which may explain the increase of duplex stability after the Se-modification. On the contrary, the distance between Se2 of U6 and C2 of A11 in the Se-modified duplex is slightly increased. This distance increase is likely due to a steric effect. This steric clash at the position 2 of the Se-uridine can be a driving force to increase SeU/A pair specificity. Consistent with our biophysical study, our structure study has indicated that the selenium bulkiness at the uridine 2-postion discourages the U/G wobble pairing. Moreover, due to the electronic effect of a selenium atom, the inability of a Se atom to form a stable hydrogen bond is another main factor responsible for the discrimination against U/G wobble pair.
CONCLUSIONS
In summary, we have first synthesized the SeU-RNAs as well as the SeU-phosphoramidite. Our biophysical and structural studies on the SeU-RNAs indicate that the native and Se-modified structures are virtually identical. Moreover, the 2-Se-modification can largely discriminate against the U/G wobble pair without significant impact on U/A pair, thereby providing a unique chemical strategy to further enhance base pair fidelity. Furthermore, the 2-Se-modification will provide a useful tool in X-ray crystal structure studies of RNAs and their protein complexes. The atom-specific mutagenesis with selenium opens a new research avenue for investigating base-pair recognition, fidelity and RNA modification. This novel base pair (SeU/A) with higher specificity likely enables better preservation of genetic information at the RNA level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–24 and Supplementary Methods.
FUNDING
USA National Foundation (NSF CHE-0750235 and MCB-0824837), USA National Institute of Health (NIH GM095086) and Georgia Cancer Coalition Distinguished Cancer Clinicians and Scientists award. Funding for open access charge: National Science Foundation (CHE-0750235).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Alex Soares, the mail-in program and PXrr in Brookhaven National Laboratory (BNL). The National Synchrotron Light Source (NSLS) at BNL is funded by NIH's National Center for Research Resources and the Department of Energy's (DOE's) Office of Biological and Environmental Research.
REFERENCES
- 1.Watson JD, Crick FHC. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD. The involvement of RNA in the synthesis of proteins. Science. 1963;140:17–26. doi: 10.1126/science.140.3562.17. [DOI] [PubMed] [Google Scholar]
- 3.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freier SM, Kierzek R, Caruthers MH, Neilson T, Turner DH. Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry. 1986;25:3209–3213. doi: 10.1021/bi00359a019. [DOI] [PubMed] [Google Scholar]
- 5.Testa SM, Disney MD, Turner DH, Kierzek R. Thermodynamics of RNA-RNA duplexes with 2-or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry. 1999;38:16655–16662. doi: 10.1021/bi991187d. [DOI] [PubMed] [Google Scholar]
- 6.Varani G, McClain WH. The GU wobble base pair - a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Reports. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. EMBO J. 1983;2:1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 1998;282:13–24. doi: 10.1006/jmbi.1998.1976. [DOI] [PubMed] [Google Scholar]
- 9.Toth MJ, Murgola EJ, Schimmel P. Evidence for a unique first position codon-anticodon mismatch in vivo. J. Mol. Biol. 1988;201:451–454. doi: 10.1016/0022-2836(88)90152-0. [DOI] [PubMed] [Google Scholar]
- 10.Seetharam R, Heeren RA, Wong EY, Braford SR, Klein BK, Aykent S, Kotts CE, Mathis KJ, Bishop BF, Jennings MJ, et al. Mistranslation in IGF-1 during over-expression of the protein in Escherichia coli using a synthetic gene containing low frequency codons. Biochem. Biophys. Res. Commun. 1988;155:518–523. doi: 10.1016/s0006-291x(88)81117-3. [DOI] [PubMed] [Google Scholar]
- 11.Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. J. Mol. Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 12.Bernardi G. Codon usage and genome composition. J. Mol. Evol. 1985;22:363–365. doi: 10.1007/BF02115693. [DOI] [PubMed] [Google Scholar]
- 13.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Mol. Biol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crick FHC. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 15.Wittwer AJ, Tsai L, Ching WM, Stadtman TC. Identification and synthesis of a naturally occurring selenonucleoside in bacterial tRNAs: 5-[(methylamino)methyl]-2-selenouridine. Biochemistry. 1984;23:4650–4655. doi: 10.1021/bi00315a021. [DOI] [PubMed] [Google Scholar]
- 16.Ching WM, Alznerdeweerd B. A selenium-containing nucleoside at the 1st position of the anticodon in seleno-transfer RNA Glu from clostridium-sticklandii. Proc. Natl Acad. Sci. USA. 1985;82:347–350. doi: 10.1073/pnas.82.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim VI. Analysis of action of wobble nucleoside modifications on codon-anticodon pairing within the ribosome. J. Mol. Biol. 1994;240:8–19. doi: 10.1006/jmbi.1994.1413. [DOI] [PubMed] [Google Scholar]
- 18.Agris PF. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie. 1991;73:1345–1349. doi: 10.1016/0300-9084(91)90163-u. [DOI] [PubMed] [Google Scholar]
- 19.Dunkel M, Cook PD, Acevedo OL. Synthesis of novel C-2 substituted pyrimidine nucleoside analogs. J. Heterocyclic. Chem. 1993;30:1421–1430. [Google Scholar]
- 20.Seio K, Sasami T, Tawarada R, Sekine M. Synthesis of 2-O-methyl-RNAs incorporating a 3-deazaguanine, and UV melting and computational studies on its hybridization properties. Nucleic Acids Res. 2006;34:4324. doi: 10.1093/nar/gkl088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method. Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson G, Vojtechovsky J, Clowney L, Brunger A, Berman H. New parameters for the refinement of nucleic acid-containing structures. Acta. Crystallogr. D: Biol. Crystallogr. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 26.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta. Cryst. A. 1986;42:140–149. [Google Scholar]
- 27.Wise D, Townsend L. Synthesis of the selenopyrimidine nucleosides 2 seleno and 4 selenouridine. J. Heterocyclic. Chem. 1972;9:1461–1462. [Google Scholar]
- 28.Shiue CY, Chu SH. A facile synthesis of 1-beta-D-arabinofuranosyl-2-seleno- and -4-selenouracil and related compounds. J. Org. Chem. 1975;40:2971–2974. doi: 10.1021/jo00908a032. [DOI] [PubMed] [Google Scholar]
- 29.Hassan A, Sheng J, Zhang W, Huang Z. High fidelity of base pairing by 2-selenothymidine in DNA. J. Am. Chem. Soc. 2010;132:2120–2121. doi: 10.1021/ja909330m. [DOI] [PubMed] [Google Scholar]
- 30.Vorbrüggen H, Strehlke P. Nucleosidsynthesen, VII. Eine einfache Synthese von 2 Thiopyrimidin nucleosiden. Chem. Ber. 1973;106:3039–3061. [Google Scholar]
- 31.Kumar RK, Davis DR. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997;25:1272–1280. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salon J, Jiang JS, Sheng J, Gerlits OO, Huang Z. Derivatization of DNAs with selenium at 6-position of guanine for function and crystal structure studies. Nucleic Acids Res. 2008;36:7009–7018. doi: 10.1093/nar/gkn843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salon J, Sheng J, Jiang JS, Chen GX, Caton-Williams J, Huang Z. Oxygen replacement with selenium at the thymidine 4-position for the Se base pairing and crystal structure studies. J. Am. Chem. Soc. 2007;129:4862–4863. doi: 10.1021/ja0680919. [DOI] [PubMed] [Google Scholar]
- 34.Knobloch B, Da Costa CP, Linert W, Sigel H. Stability constants of metal ion complexes formed with N3-deprotonated uridine in aqueous solution. Inorg. Chem. Commun. 2003;6:90–93. [Google Scholar]
- 35.Wahl M, Ban C, Sekharudu C, Ramakrishnan B, Sundaralingam M. Structure of the purine-pyrimidine alternating RNA double helix, r (GUAUAUA) d (C), with a 3′-terminal deoxy residue. Acta Crystallogr. D: Biol. Crystallogr. 1996;52:655–667. doi: 10.1107/S0907444996000248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.