Abstract
FliZ, a global regulatory protein under the control of the flagellar master regulator FlhDC, was shown to antagonize σS-dependent gene expression in Escherichia coli. Thereby it plays a pivotal role in the decision between alternative life-styles, i.e. FlhDC-controlled flagellum-based motility or σS-dependent curli fimbriae-mediated adhesion and biofilm formation. Here, we show that FliZ is an abundant DNA-binding protein that inhibits gene expression mediated by σS by recognizing operator sequences that resemble the −10 region of σS-dependent promoters. FliZ does so with a structural element that is similar to region 3.0 of σS. Within this element, R108 in FliZ corresponds to K173 in σS, which contacts a conserved cytosine at the −13 promoter position that is specific for σS-dependent promoters. R108 as well as C(−13) are also crucial for DNA binding by FliZ. However, while a number of FliZ binding sites correspond to known σS-dependent promoters, promoter activity is not a prerequisite for FliZ binding and repressor function. Thus, we demonstrate that FliZ also feedback-controls flagellar gene expression by binding to a site in the flhDC control region that shows similarity only to a −10 element of a σS-dependent promoter, but does not function as a promoter.
INTRODUCTION
A fundamental biological principle seems to be that rapid proliferation of cells and high levels of stress resistance are mutually exclusive (1–4). In the generally nutrient-limiting natural environments of bacteria, this is reflected in switching between alternative life-styles. On the one hand, this is the ‘foraging’ state of post-exponentially growing motile cells, in which nutrient scavenging is optimized. However, when nutrients become even scarcer, bacteria enter into the stationary phase life-style, which is characterized by a maintenance metabolism, multiple stress resistance and adhesion to other cells or surfaces. Overall, this life-style resembles conditions in a biofilm [for a review, see (2)].
At the molecular level, these life-style transitions are a reflection of σ subunit competition for limiting amounts of RNA polymerase (RNAP) core enzyme (5–8). In Escherichia coli, genes for high affinity nutrient scavenging and motility are usually activated by RNAP containing the vegetative σ subunit σ70 (RpoD) and/or the flagellar σ factor σ28 (FliA), whereas generating multiple stress resistance and adhesion via curli fimbriae is the domain of RNAP containing the general stress σ factor σS (RpoS) (1,2,4,9,10). Internal and external signals can affect σ factor competition through regulation of σ levels or their availability for binding to RNAP core enzyme. In addition, accessory agonists [such as Crl for σS; (11)] and antagonists, i.e. anti-σ factors (12), are involved.
The σ factor σS (RpoS) begins to accumulate already during the post-exponential growth phase and reaches highest levels in stationary phase cells (2). About 10% of the genes in E. coli are under direct or indirect positive control of σS (13). Many of these genes are involved in generating multiple stress resistance and in adapting energy metabolism to slow or no growth (2). Moreover, σS is the master regulator of the regulatory network that controls the expression of two major biofilm components, i.e. adhesive curli fimbriae and cellulose. Curli fimbriae play a central role in switching from the planktonic and motile life-style to the adhesive state during the transition from post-exponential growth to early stationary phase.
Two modes of regulation contribute to the inverse coordination of flagellum-based motility and curli-associated adhesion in E. coli (Supplementary Figure S1). One operates with the bacterial second-messenger c-di-GMP, which is synthesized by diguanylate cyclases (DGCs) containing GGDEF domains and is degraded by phosphodiesterases (PDEs) comprising EAL domains (14–17). During entry into stationary phase, σS stimulates the expression of several DGCs, whereas the PDE YhjH is down-regulated (10,18,19). This results in a c-di-GMP-mediated reduction of flagellar activity (10,20) and stimulates the expression of the curli gene activator CsgD (10,18).
The other mode of antagonistic regulation of motility and adhesion involves FliZ protein, which is under control of the flagellar gene hierarchy. FliZ antagonizes σS-activity during post-exponential growth, when flagellar gene expression and motility peak. As a result, FliZ gives priority to motility and the planktonic lifestyle over σS-dependent gene expression (including curli fimbriae expression) (10). The mechanism underlying this relatively general effect of FliZ on the σS regulon had not been clarified. Here, we show that FliZ is an abundant DNA-binding protein that antagonizes expression of many σS-dependent genes by recognizing DNA sequences that ressemble the extended −10 promoter regions of these genes which conform to the consensus sequence TCTATACTTAA (with the core −10 hexamer in italics) (2). Moreover, FliZ binds to DNA using a structural element that shows similarity to the DNA-binding element in σS, which allows σS (within the RNAP holoenzyme complex) to recognize the extended −10 promoter region. However, while many FliZ binding sites correspond to σS-dependent promoters, promoter activity is not a prerequisite for FliZ binding and repressor function. As an example, we show that in the control region of flhDC, FliZ binds to a site that is similar to a −10 element of a σS-dependent promoter but that is not used as such, resulting in feedback-control of flagellar gene expression.
MATERIALS AND METHODS
Standard molecular biology and biochemical techniques
For the purification of FliZ protein, SDS-PAGE, immunoblot analysis, electrophoretic mobility shift assays (EMSA), DNaseI footprint analysis and the determination of β-galactosidase activity in cellular extracts, standard procedures were applied. All materials and technical details are described in the Supplementary data.
Microbiological techniques
Growth of bacteria was monitored by measuring optical density at 578 nm (OD578). Bacterial motility was tested on swim plates containing 0.5% bacto-tryptone, 0.5% NaCl and 0.3% agar. 100 µg/ml ampicillin was added when strains carrying plasmids were tested. Three microliters of overnight cultures (adjusted to an OD578 of 4.0) were inoculated into the swim plates, and cells were allowed to grow and swim for 4–5 h at 28°C.
RESULTS
FliZ is a global repressor with a DNA sequence specificity overlapping that of σS–containing RNA polymerase
An extended C-terminal part of FliZ is similar to the DNA-binding N-terminal domain of various phage integrases and recombinases (Supplementary Figure S2A) (21,22) and recent publications indicate that FliZ acts as a DNA-binding regulator in other bacterial species (23). This suggested that the ability of FliZ to antagonize σS-dependent gene expression (10) might be due to binding to σS-controlled promoters rather than to σS itself. This possibility would also raise a secondary question, namely how FliZ may be able to discriminate between σS-dependent and vegetative promoters, since these two classes of promoters have very similar sequences.
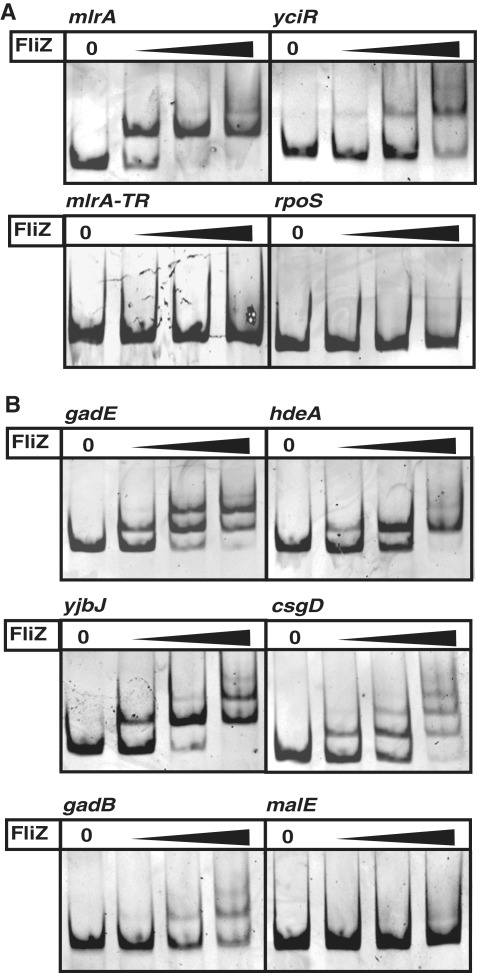
In order to test whether FliZ is able to bind to DNA in vitro, the E. coli protein was purified and subject to EMSA with DNA fragments containing the promoters of mlrA and yciR. The mlrA gene encodes a MerR-like regulator essential for transcriptional activation of the central curli regulator CsgD (Supplementary Figure S1) which shows premature induction in post-exponential phase in a fliZ mutant (10). Similarly, the yciR gene, which specifies a cyclic di-GMP-specific PDE involved in the regulation of csgD expression (18), was previously shown to be repressed by FliZ (10). FliZ bound to both promoter regions (Figure 1A). By contrast, FliZ did not bind to a control fragment comprising part of the translated region of the mlrA gene, nor to a region including the σ70-dependent promoter of the rpoS gene, which encodes σS (Figure 1A).
Figure 1.
FliZ binding to promoter DNA. Electrophoretic mobility shift assays with FliZ (20, 40, 80 nM) are shown for (A) DNA-fragments (6 nM) comprising the promoter regions of the σS-dependent genes mlrA and yciR and control fragments containing part of the translated region of the mlrA gene (mlrA-TR) and the σ70-dependent rpoS promoter, and (B) DNA-fragments containing promoters from other FliZ-controlled genes previously identified (10).
A large number of FliZ-regulated genes were identified by whole genome transcription profiling, demonstrating a more general effect of FliZ on σS-dependent gene expression (10). Many of these genes are activated by σS and repressed by FliZ (including the curli genes and other σS-controlled genes, i.e. gadBC, gadE, hdeAB and yjbJ). Yet, there are also some negatively FliZ-affected genes that are not under σS control (e.g. the gatYZABCD operon), as well as several positively FliZ-affected genes that are either regulated by σS (chaB, ynhG) or independent of σS (e.g. malE, malK) (10). Gel retardation experiments were performed with promoter DNA from selected genes of these groups. FliZ clearly bound to DNA containing promoters of genes, which, like mlrA and yciR, are expressed from σS-dependent promoters and showed negative regulation by FliZ in the microarray experiments (Figure 1B). Promoter regions with other patterns of FliZ- and σS-dependence were either not bound at all or in some cases weakly bound when higher concentrations of FliZ were used (Figure 1B and Supplementary Figure S3). The latter may be due to weak binding sites or sequence-unspecific binding. This also seems to apply to the indirectly σS-activated gadB or to the secondary binding sites observed for gadE, yjbJ and csgD which result in multiple band shifts with higher FliZ concentrations (Figure 1B). Overall, FliZ seems to negatively regulate σS-dependent genes by directly binding to σS-controlled promoter regions, while other modes of regulation by FliZ are likely to be indirect.
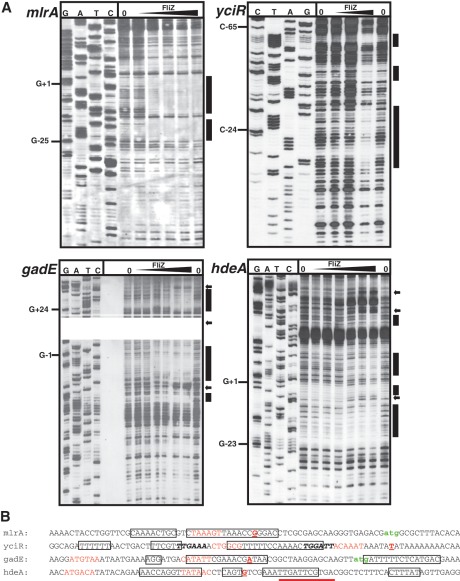
In order to precisely determine the FliZ binding sites, DNaseI-footprint assays were performed with the promoter regions of mlrA, yciR, gadE and hdeA (Figure 2). In the mlrA, gadE and hdeA promoters, the FliZ-protected regions overlapped with the −10 elements and included additional sites further upstream and/or downstream. In the yciR promoter sequence as proposed by Cairrao et al. (24), FliZ bound to the spacer region, the −35 element and further upstream. Interestingly, this FliZ binding site in the yciR promoter contains a sequence that resembles the σS-dependent consensus −10 promoter element (located 6 bp upstream of the proposed −10 region; Figure 2B). Possibly, this sequence might constitute the actual −10 element of the yciR promoter and the 5′-mRNA end identified for yciR by Cairrao et al. (2001) might represent a 5′-end generated by RNA processing. Alternatively, this FliZ binding site only resembles a σS-dependent promoter. Overall, this determination of binding sites for FliZ in σS-dependent promoter regions and/or σS promoter-like sequences suggests that FliZ may have overlapping binding specificity with σS-containing RNAP.
Figure 2.
FliZ binding sites in σS-dependent promoters. (A) Non-radioactive DNase I footprint analysis was performed with FliZ and DIG-labeled DNA fragments containing the promoter regions of mlrA as well as yciR, gadE and hdeA genes. FliZ-binding sites are indicated by bars and were mapped to the promoter sequences and marked by boxes (B). Positions of enhanced DNase cleavage are marked by arrows. Transcriptional start sites have been determined before: mlrA (Supplementary Figure S4), yciR (24), gadE (60), hdeA (61). −10 and −35 elements are colored in red, transcriptional start sites are printed as bold, red, underlined letters; translational start sites are labeled in green; potential alternative −35 and −10 regions in the yciR promoter (see text) are indicated by bold, italic letters; a sequence with partial similarity to a σS-dependent −10 region on the opposite strand of the hdeA promoter region is underlined.
The C(−13) element, which provides σS selectivity to a promoter, also contributes to FliZ binding
With FliZ specifically recognizing sequence elements that correspond to or resemble σS-controlled promoters, the question arose how FliZ is able to distinguish between the very similar σ70- and σS-dependent promoters. The analogous question, how RNAP holoenzymes containing either σ70 or σS themselves discriminate between these promoters, has puzzled scientists for years. Only recently was it understood that modular combinations of certain small promoter features can generate preferred recognition and/or activation of a promoter by EσS (25). The most important promoter element contributing to σS selectivity is a cytosine at position −13, which is not only highly conserved in σS-dependent promoters (13,26,27), but is also directly contacted by a lysine (K173) in σS (26). A thymine at the neighboring position (−14) also contributes to σS selectivity, although to a lesser extent than C(−13) (26).
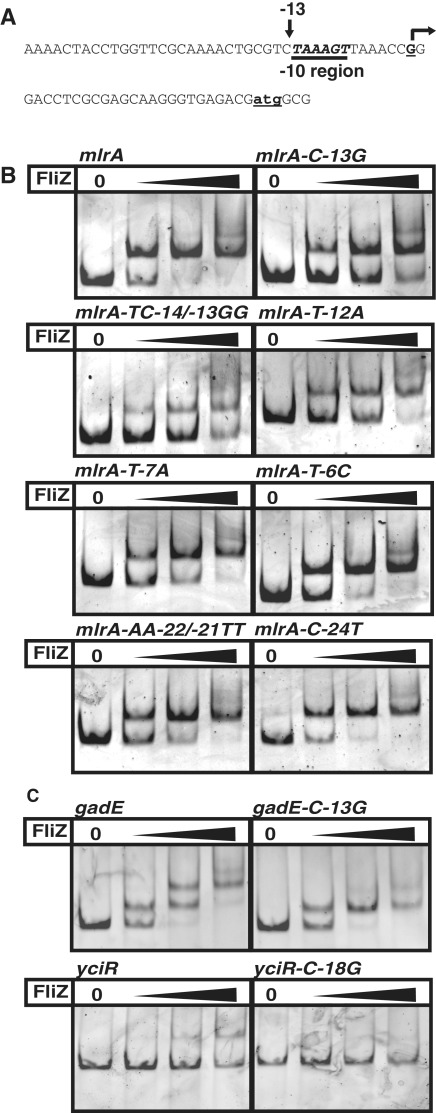
C(−13) and T(−14) are both present in the mlrA promoter (Figure 3A and Supplementary Figure S4). Mutation of the C(−13) strongly reduced FliZ-binding and an additional mutation of the neighboring T(−14) further impaired binding (Figure 3B, with data quantified in Supplementary Figure S5A), indicating that the TC(−14/−13) element also plays a role in FliZ binding. T(−12) and T(−7), which confine the core −10 hexamer, also contribute to FliZ binding, since mlrA promoter mutations at these positions reduced FliZ binding as well (Figure 3B and Supplementary Figure S5A). By contrast, mutations changing T(−6) or several positions up to −24 did not affect FliZ binding (Figure 3B and Supplementary Figure S5A). In conclusion, the specific FliZ binding site in the mlrA promoter region includes the extended −10 region, consisting of the −10 hexamer and the adjacent TC(−14/−13) element.
Figure 3.
Promoter region nucleotides involved in FliZ binding. Several sites in the mlrA promoter sequence (A, B) as well as C(−13) in the gadE promoter sequence and the suggested alternative yciR promoter (or promoter-like sequence) (C) were replaced as indicated and the effects of these mutations on FliZ-binding were tested by EMSA as described in Figure 1.
The gadE promoter also shows a cytosine 13 bp upstream of the transcriptional start site and mutation of this base reduced FliZ-binding (Figure 3C and Suplementary Figure S5B). While the previously reported yciR promoter does not contain this sequence feature (24), there is a second promoter-like sequence located 6 bp upstream (see above) which contains a corresponding cytosine (shown in italics in Figure 2B). Mutation of this cytosine resulted in reduced FliZ-binding (Figure 3C and Supplementary Figure S5B). Together, these results indicate that FliZ and σS bind DNA with overlapping sequence specificity, with a cytosine being a key nucleotide for interaction with both factors, which in an active σS-dependent promoter sequence corresponds to C(−13) in the extended −10 region.
The DNA-binding element in FliZ is similar to the α-helix 3.0 in σS that is crucial for promoter binding in the extended −10 region
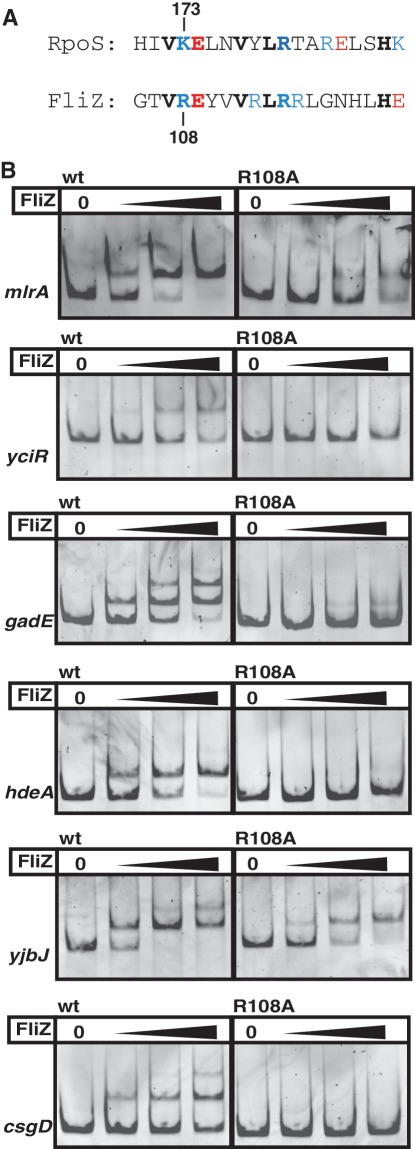
The C-terminal part of FliZ contains a region of 76 amino acids (of a total of 183 amino acids of FliZ) with similarity to the DNA-binding domain of the phage integrase family (Supplementary Figure S2). This domain binds to inverted DNA repeats, which are the core of longer sites targeted by these integrases and recombinases (22). Therefore it seemed likely that the corresponding part of FliZ is involved in DNA binding. Because of the overlapping DNA binding specificity of FliZ and σS, this C-terminal region of FliZ was analyzed for potential similarities with σS that might account for a similar DNA binding mechanism. σS-containing RNAP directly and specifically contacts C(−13) with a lysine (K173) located in the first α-helix of domain 3 of σS (helix 3.0) (26). The C-terminal region of FliZ indeed contains a predicted α-helix that is similar to helix 3.0 of σS (Figure 4A). In this putative α-helix, FliZ also features a positively charged amino acid (R108) at the position corresponding to K173 in σS. R108/K173 as well as a series of identical amino acids (V107/V172, E109/E174, V111/V177, L113/L179 and R114/R180) are all lined up on one side of this α-helix, which in σS is surface-exposed since K173 binds C(−13) in the promoter DNA (26). Overall, these two α-helices in FliZ and σS show an identity of 31.6% and a similarity of 42.1% (for comparison: the respective α-helices in FliZ and XerD, which is the phage integrase family member that shows strongest similarity to FliZ (Supplementary Figure S2A), display 26.3% identity and 36.8% similarity, while the identity and similarity between the respective α-helices of RpoS and the closely related vegetative σ factor RpoD are 21.1% and 63.2%, respectively).
Figure 4.
FliZ and σS use a similar element for DNA binding. (A) Alignment of a putative α-helix in the C-terminal region of FliZ with the extended −10 recognition helix 3.0 of σS (RpoS). Positively charged residues are shown in blue, negatively charged residues in red. (B) FliZ wild-type (wt) and FliZ-R108A binding to FliZ-target promoter DNAs was compared by EMSA (20, 40, 80 nM FliZ).
This resemblance to the extended −10 recognition element in σS prompted us to analyze a potential role of R108 in the interaction of FliZ with σS-dependent promoter regions. The mutant protein FliZ-R108A was indeed strongly reduced in its ability to bind to a series of target promoter fragments (Figure 4B, with data quantified in Supplementary Figure S6); this is due specifically to the loss of R108, since overall stability of the FliZ-R108A protein is similar to that of wild-type FliZ, as indicated by similar digestion patterns and kinetics in limited proteolysis experiments (Supplementary Figure S7).
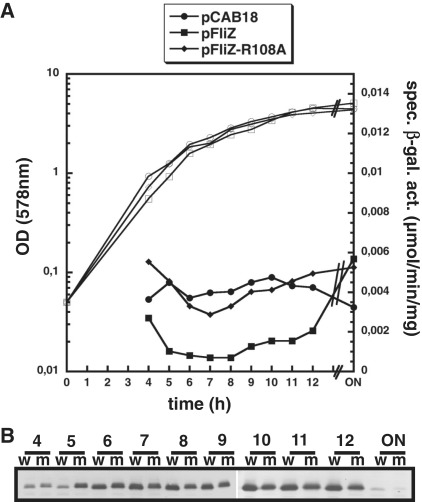
In order to test the effect of the R108A mutation in vivo, we used strains expressing either wild-type FliZ or FliZ-R108A from a low copy plasmid. Since this resulted in higher FliZ levels than in the wild-type, the difference in target gene repression was best seen with a target with moderate affinity for FliZ such as yciR. While plasmid-encoded wild-type FliZ strongly repressed yciR expression, yciR repression was relieved with FliZ-R108A (Figure 5A). As Western-blot analysis demonstrated that both FliZ variants were present at similar levels (Figure 5B), we conclude that FliZ-R108A is unable to efficiently bind and repress yciR in vivo.
Figure 5.
The R108A mutation eliminates FliZ repression of the σS-dependent gene yciR. (A) Expression of a single-copy yciR::lacZ fusion in ΔfliZ mutant (m) strains producing equal levels of either wt FliZ or FliZ-R108A from low copy plasmids or carrying the vector alone (pCAB18) was determined in cells growing in LB medium at 28°C (ON, over night). (B) in parallel, cellular levels of FliZ and FliZ-R108A were determined by immunoblot analysis (w, wt FliZ; m, FliZ-R108A). For technical reasons, two separate blots, comprising samples taken at 4–9 h and 10 h to ON were combined in (B).
Taken together, our in vivo and in vitro data indicate that (i) FliZ antagonizes expression of a series of σS-dependent genes by directly binding within their promoter regions; (ii) the FliZ binding site on the DNA is similar (though not necessarily identical) to the extended −10 promoter region and in particular includes a cytosine, which in an active σS-dependent promoter corresponds to C(−13) and (iii) FliZ uses a structural element (containing R108) for DNA binding that is similar to the extended −10 recognition helix in σS, i.e. region 3.0, which contains the key residue K173 that interacts with C(−13).
Regulation of flhDC by FliZ involves binding to a sequence that is similar to but is not used as a σS-dependent promoter
Overlapping though not completely identical DNA binding specificity of FliZ and σS-containing RNAP may also allow cases where FliZ binds to sequences that only resemble σS-dependent promoters but that are not used as such. As a consequence, not each and every regulatory effect of FliZ would necessarily be linked to σS-dependent expression. Several groups have observed FliZ effects on motility in various species (23,28–31), but motility genes in E. coli and Salmonella are not under the direct control of σS. Thus, in order to analyze the role of FliZ in the control of a process that is not directly σS-regulated, we tested the molecular function of FliZ in motility regulation in E. coli.
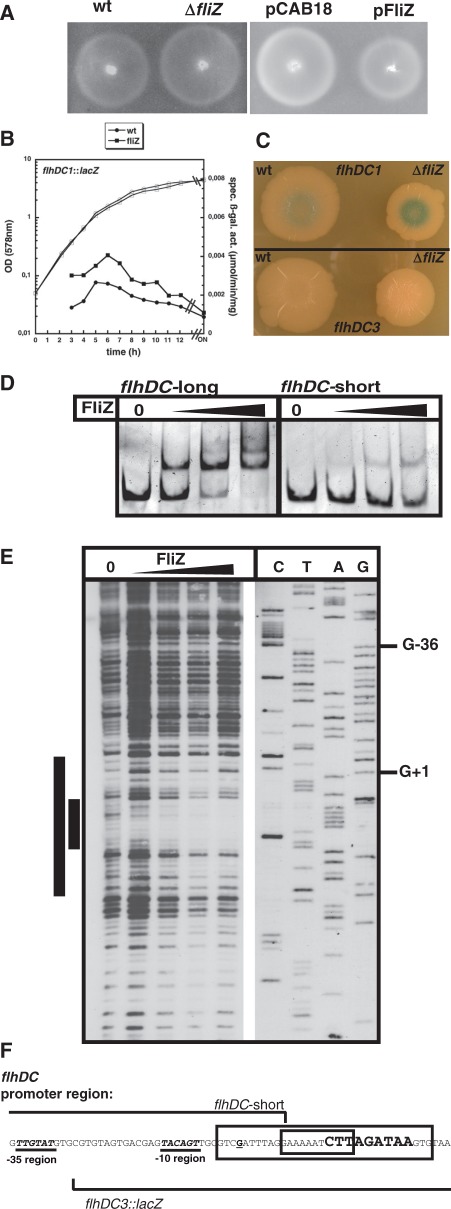
A ΔfliZ mutant showed a subtle but reproducible increase in swimming motility as compared to the wild-type, while FliZ expression from the low-copy plasmid reduced motility (Figure 6A), indicating that in E. coli, FliZ acts as a weak negative regulator of motility. Flagellar assembly is based on the hierarchical expression of three classes of flagellar genes (32) (Supplementary Figure S1). A single-copy chromosomal lacZ fusion to the promoter of the class I flhDC operon, which encodes the flagellar master regulator complex FlhD4C2, showed increased expression in the ΔfliZ mutant (Figure 6B). Similar effects were also observed for lacZ fusions to the class II flgAMN promoter and to the class III gene yhjH (Supplementary Figure S8A). However, FliZ did not bind to classes II and III promoter fragments tested in vitro (Supplementary Figure S8B), indicating that the FliZ effect on the expression of the master regulator FlhDC (class I) is relayed to the classes II and III genes.
Figure 6.
FliZ directly affects motility by repressing flhDC expression. (A) Motility of strain W3110 (wt) and its derivatives carrying ΔfliZ, or the low copy plasmids pCAB18 (vector control) or pFliZ was tested at 28°C. (B) Expression of a single-copy transcriptional lacZ fusion to the flhDC promoter (flhDC1::lacZ) was determined in fliZ+ and fliZ cells growing in LB medium at 28°C. (C) Expression in wild-type (wt) and ΔfliZ cells of the same fusion (flhDC1::lacZ) and of a fusion (flhDC3::lacZ) that does not include the full vegetative flhDC promoter but carries the ‘−10 σS-promoter-like element’ (F); cells were grown on LB/agar plates without salt at 28°C for 7 days. (D) Binding of FliZ to DNA fragments with (flhDC-long) or without (flhDC-short) the ‘−10 σS-promoter-like element’ downstream of the flhDC transcriptional start site was compared by EMSA (80, 160, 320 nM FliZ). (E) The FliZ-binding site in the flhDC upstream regulatory region was determined by non-radioactive DNaseI footprint analysis and the binding site was mapped to the promoter sequence (F). A core binding site and potential upstream and downstream extensions are indicated by smaller and larger bars (E) and boxes (F). The transcriptional start site (62) is printed as a bold, underlined letter and the ‘−10 σS-promoter-like element’ downstream of the flhDC promoter is printed in bold, larger letters. The start of the region present in flhDC3::lacZ and the end of the flhDC-short DNA fragment used in (D) are indicated.
FliZ directly bound to a fragment containing the promoter region of the flhDC operon (Figure 6D). This promoter is known to be σ70-activated. Yet, closer inspection of the flhDC promoter region revealed the presence of a short sequence just downstream of the transcriptional start site that actually resembles the extended −10 element of a σS-dependent promoter (Figure 6F). Consistently, FliZ binding was impaired with a DNA segment that contained the σ70-controlled promoter but lacked this σS-promoter-like element (‘flhDC-short’ in Figures 6D and F). Moreover, DNaseI footprint experiments revealed that FliZ binds downstream of the flhDC promoter overlapping with this σS-promoter-like element (Figure 6E). Finally, expression of a transcriptional lacZ fusion to the flhDC promoter, which did not contain this FliZ binding site, did also not show regulation by FliZ (‘flhDC2::lacZ’ in Supplementary Figures S8C and D). We conclude that FliZ interferes with flhDC expression by binding to a sequence right downstream of the transcriptional start site, which resembles the −10 element of a σS-dependent promoter.
This raised the question whether this FliZ binding site might be a ‘cryptic’ σS-controlled promoter used under certain not yet known conditions, e.g. in a biofilm. With a lacZ reporter fusion devoid of the flhDC promoter, but containing the σS-promoter-like element as well as 40 nt upstream (‘flhDC3::lacZ’ in Figures 6C and F), we were unable to detect any β-galactosidase activity, no matter whether cells were grown for several days on plates (Figure 6C) or in liquid culture (data not shown). Thus, this element does not seem to constitute a σS-dependent promoter, indicating that in the control of flhDC, this sequence is just a regular operator for FliZ, which extends the regulatory influence of FliZ to a gene activated by a σ factor different from σS.
Together our data show that the overlapping DNA-binding specificity between FliZ and σS allows FliZ to directly bind to a subset of σS-dependent promoters and also to discriminate between these and vegetative promoters, which explains its global effect on many σS-dependent genes; yet, on the other hand, FliZ can also bind to alternative positions that are only similar to—but that do not function as—σS-dependent −10 promoter elements, and thereby exhibits also regulatory effects beyond the σS regulon.
Cellular concentration of FliZ during the growth cycle
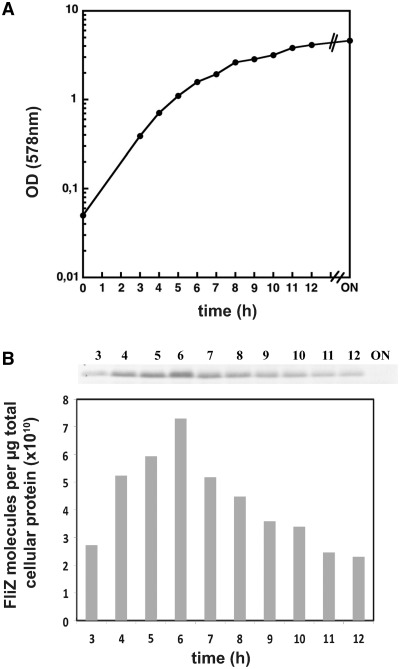
With FliZ acting as a global repressor that can directly target many σS-dependent promoter sequences as shown above, the observed global effect on σS-dependent gene expression (10) requires substantial cellular amounts of FliZ. Therefore we determined cellular FliZ levels throughout the growth cycle by immunoblot analysis (Figure 7 and Supplementary Figure S9). As expected for a FlhDC-activated gene product, FliZ levels increased during the post-exponential phase of a culture growing in LB, reached a peak at an OD578 of about 2, then steadily declined and could hardly be detected anymore in overnight cells (the limit of detection under our experimental conditions corresponded to approximately ≤5% of the maximal FliZ levels measured in the post-exponential phase; see Figure S9). During its peak phase, where FliZ-mediated repression of target genes such as mlrA is most pronounced (10), cellular levels of FliZ were found to be around 21.500 molecules per cell (Supplementary Fig. S10). In other words, FliZ is one of the most abundant DNA-binding proteins and therefore available to bind to numerous sites in the chromosome. FliZ can thus be regarded as an abundant nucleoid-binding protein.
Figure 7.
Cellular levels of FliZ during the growth cycle. OD578 (A) and cellular levels of FliZ (B) were determined in cells growing in LB medium at 28°C. FliZ levels were determined by immunoblot analysis, using defined amounts of purified FliZ as a reference for calculating absolute cellular protein levels (in molecules per μg total cellular protein; see Supplementary Figure S9). 6 µg of cellular protein was applied per lane. The experiment was done three times and a representative experiment is shown. FliZ levels in the ON sample were below detection (which corresponds to <5% of maximal levels determined). Based on direct measurements of cell numbers (as colony forming units), cellular FliZ contents were also calculated in molecules per cell (Supplementary Figure S10).
DISCUSSION
During the post-exponential growth phase of E. coli, where FliZ levels reach a maximum, FliZ globally interferes with σS-dependent gene expression and thereby gives priority to motility over σS-mediated functions which include curli fimbriae-mediated adhesion (10). In this study, we characterize FliZ as a global repressor with a DNA-binding specificity that overlaps with that of σS, with this overlap corresponding to an extended −10 region of σS-dependent promoters. This explains the mechanism by which FliZ antagonizes σS-activity. Moreover, we clarify the role of FliZ in motility regulation in E. coli. Our results may also shed some light on the mechanisms underlying the reported impact of FliZ in the regulation of virulence and motility in other bacteria.
FliZ is an abundant DNA binding protein which resembles σS in a specific DNA binding region and the DNA sequence recognized
FliZ directly binds to DNA regions that either correspond to known extended −10 promoter regions (consisting of nucleotides at positions −14 to −7) of σS-dependent genes such as mlrA and gadE (Figure 2) or to sequences that resemble such σS-dependent promoter elements but are not active as such, as analyzed here for the binding site right downstream of the flhDC promoter (Figure 6). In the yciR and hdeA promoters, FliZ seems to bind to sequences that also resemble the σS-dependent consensus −10 element but that overlap only with the actual promoter (Figure 2). It should also be noted that apart from the presence of sequences that constitute or resemble a σS-dependent −10 promoter element, we were unable to detect any other common features in the FliZ binding sites identified in this report. For example, mutating residues of a CAAAACTG motif that is found in the spacers of both the mlrA and yciR promoters did not affect FliZ binding to the mlrA promoter (Figure 3B).
The range of target genes affected (10) indicates that FliZ, just like σS itself, must be able to discriminate between the ‘stress’ promoters activated by σS and the very similar vegetative promoters activated by σ70. FliZ seems to achieve this in a way that is similar to that used by σS: FliZ contains a putative α-helix with sequence similarity to α-helix 3.0 of σS (Figure 4A), with R108 in FliZ corresponding to K173 in σS, which is involved in DNA recognition of the extended −10 promoter element (Figures 4B and 5) (26). This latter element, i.e. TC(−14/−13), which makes a direct contact to K173 in σS (26), also strongly contributes to recognition by FliZ; in addition, T(−12) and T(−7), which belong to the core −10 hexamer, are important for FliZ binding (Figure 3). Thus, the molecular mechanism of binding to an extended −10 region may be similar for σS and FliZ.
It should be noted, that while DNA binding by σS and FliZ overlaps in the extended −10 promoter region, additional DNA sequences or structural features that optimize DNA binding of either FliZ or σS are not identical. Using its domain 4, σS also binds to the −35 promoter region and for a subset of σS-controlled promoters, σS selectivity relies on the combination of a distal UP element half-site with a −35 region (25), while their −10 regions are similar to those of vegetative promoters. This is consistent with our previous finding that many but not all σS-dependent genes are targeted by FliZ (10).
FliZ-binding is not restricted to active σS-dependent promoters
In the flhDC promoter FliZ binds to a sequence, which is similar to—but not functional as a −10 element of a σS-dependent promoter and which is located right downstream of the known vegetative flhDC promoter. This shows that FliZ is not restricted to binding to ‘real’ σS-dependent promoters only and also extends the regulatory potential of FliZ to genes controlled by σ factors different from σS. In addition, FliZ binding sites may also occur overlapping with functional σS-dependent promoters without corresponding to the functional −10 promoter elements. The FliZ-binding site within the yciR promoter (Figure 2B) may represent an example for this kind of binding behavior.
Similar as discussed above for σS, FliZ could also make additional specific contacts to DNA that differ from the ones made by σS. Such contacts may account for FliZ binding to additional sites besides the extended −10(-like) elements as can be observed in the footprint experiments (Figure 2B). Moreover, in contrast to ‘directional’ promoter binding by EσS, FliZ could bind to the same or the opposite DNA strand. The latter might be the case for FliZ-binding to the hdeA promoter. The hdeA promoter does not contain a C(−13); however, the opposite strand features a sequence with similarity to a σS-dependent −10 promoter element (including an appropriate cytosine), which overlaps with the FliZ binding site (underlined in red in Figure 2B) and might therefore be recognized by the protein.
Together, these observations show that an overlapping binding-specificity between FliZ and σS directs FliZ towards—but does not restrict it to—binding at ‘typical’ σS-dependent promoters. These binding properties confer to FliZ a broad role in the global control of gene expression. This role is further emphasized by our finding that cellular FliZ levels rise up to 21 500 molecules per cell in the post-exponential phase (Figure 7 and Supplementary Figures S9 and S10), characterizing it as an abundant nucleoid-associated protein. Thus, FliZ is present in excess over RNAP (6), which is not only a prerequisite for effectively competing with σS-containing RNAP at σS-dependent promoters, but also allows it to bind to additional sites in the chromosome and thereby exert regulatory effects that go beyond antagonizing σS.
Similarities in DNA recognition by FliZ, domain 3 of σ factors and phage integrase family members
The alignment of FliZ with the core-binding domains of several phage intergrase family members reveals that the putative α-helix in FliZ, which contains R108 and looks like a mimic of the extended −10 recognition helix 3.0 in σS, corresponds to the DNA-binding α-helix 2 in phage integrase family members (Supplementary Figure S2A) (21,33–35). This α-helix also corresponds to one of two α-helices in the recombinase Cre, which make direct DNA contacts (33). Moreover, the residue corresponding to E109 in FliZ plays a role in determining DNA-binding specificity in lambda integrase (36,37). R108 and the two neighboring amino acids in FliZ are flanked by strongly conserved residues (Supplementary Figure S2A). The variable residues within these conserved structural elements were proposed to establish the unique function of each core binding domain (21). Overall, the DNA-binding helices in FliZ and in domain 3 of σS show the same pattern of conserved residues interspersed with variable residues used for specific DNA sequence recognition. This suggests that FliZ, domain 3 of σ factors and proteins of the phage integrase family may all use a similar mechanism of DNA interaction. This similarity may be the result of horizontal transfer of gene fragments, convergent evolution or may reflect homology, i.e. a common evolutionary ancestory.
The role of FliZ in controlling motility of E. coli
With FliZ being itself part of the flagellar gene hierarchy, several studies have implicated FliZ in the control of motility (23,28–31,38) but in E. coli the underlying molecular mechanisms had not been clarified. Here, we demonstrate that FliZ down-regulates flagellar gene expression by directly binding to a regulatory region of the flhDC operon which encodes the flagellar master regulator FlhDC (Figure 7). Being itself under positive FlhDC control, FliZ thus can exert a negative feedback control in motility. This feedback may have a homeostatic function in keeping motility gene expression at a level that can be shut down during entry into stationary phase. Since this shut-down is at least in part a consequence of the competition of σ70, σFliA and σS for RNAP core enzyme (10), and FliZ also globally affects σS-driven gene expression as discussed above, FliZ could be a key player in setting thresholds in the σ factor competition balance that controls switching between the motile ‘foraging’ life-style and a life-style characterized by slow or no growth, high stress resistance and cellular adhesion.
The role of FliZ in other bacterial species
Our study provides a comprehensive understanding of the molecular mechanism of FliZ in E. coli. How do our data relate to studies of FliZ and phenotypes reported in other bacterial species? In Salmonella, FliZ was suggested to post-transcriptionally regulate flagellar gene expression (31), the expression of pathogenicity island 1 (SPI1) genes (39–41) as well as type 1 fimbrial genes (42). However, a recent publication demonstrates that FliZ indirectly activates flagellar gene expression in Salmonella by directly binding to and repressing transcription from the nlpC promoter that also controls expression of the ydiV gene, which encodes the anti-FlhDC factor YdiV (38). In Xenorhabdus nematophila FliZ was also found to bind in vivo to unspecified regions upstream of flhD and two genes encoding hemolysins (23). These data support our finding that FliZ acts as a DNA-binding transcriptional regulator. Strong similarity of FliZ proteins from E. coli, Salmonella and X. nematophila (Supplementary Figure S2B) further supports that the molecular mechanism of FliZ action is similar in these organisms.
Moreover, FliZ-mediated regulation of motility through repression of ydiV in Salmonella (38) indicates that FliZ-mediated effects on processes other than transcription may be indirect. Indirect regulation by FliZ was also shown for the positive control of SPI1 virulence genes in Salmonella, where FliZ plays a not yet clarified role upstream of the HilD/HilC/RtsA/HilA transcriptional cascade (43). Species-specific differences with respect to factors involved in such indirect regulation may also explain discrepancies between FliZ-mediated effects in various species. Thus, FliZ acts as a positive regulator of motility in Salmonella (31) and X. nematophila (23), while we show here that in E. coli FliZ represses motility. Interestingly, differences in the expression of ydiV between E. coli and Salmonella have recently been reported (44). In contrast to Salmonella, expression of ydiV is below detection in E. coli cells grown in liquid culture (19). Thus, in the conditions under which motility and flagellar gene expression have been tested here, the direct negative effect of FliZ on flhDC expression (Figure 6) prevails over a potentially positive but indirect effect via YdiV. Finally, differences in FliZ-targeted genes in different bacteria are likely to reflect differences in the occurrence of FliZ binding sites in the chromosomes of these species. No matter whether these sites represent functional σS-regulated promoters or only resemble them, they can easily arise or be altered by minor mutations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Table 1, Supplementary Figures 1–10 and Supplementary References [2,10,18,21,45–59].
FUNDING
Deutsche Forschungsgemeinschaft (He 1556/13-2 to R.H.). Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Mika and A. Possling for performing the experiment shown in Supplementary Figure S4, and G. Klauck for performing the cfu measurements shown in Supplementary Figure S10.
REFERENCES
- 1.Ferenci T. Hungry bacteria-definition and properties of a nutritional state. Environ. Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- 2.Hengge R. The general stress response in Gram-negative bacteria. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, DC: ASM Press; 2010. pp. 25–289. [Google Scholar]
- 3.Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nyström T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 5.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 6.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl Acad. Sci. USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmann JD. Regulation by alternative sigma factors. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington DC: ASM Press; 2010. pp. 31–43. [Google Scholar]
- 8.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 9.Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes KT, Mathee K. The anti-sigma factors. Annu. Rev. Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 13.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 15.Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 18.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signalling within the σS network of Escherichia coli. Mol. Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 19.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology. 2009;155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- 20.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Swalla BM, Gumport RI, Gardner JF. Conservation of structure and function among tyrosine recombinases: homology-based modeling of the lambda integrase core-binding domain. Nucleic Acids Res. 2003;31:805–818. doi: 10.1093/nar/gkg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirumalai RS, Kwon HJ, Cardente EH, Ellenberger T, Landy A. Recognition of core-type DNA sites by lambda integrase. J. Mol. Biol. 1998;279:513–527. doi: 10.1006/jmbi.1998.1786. [DOI] [PubMed] [Google Scholar]
- 23.Lanois A, Jubelin G, Givaudan A. FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol. Microbiol. 2008;68:516–533. doi: 10.1111/j.1365-2958.2008.06168.x. [DOI] [PubMed] [Google Scholar]
- 24.Cairrao F, Chora A, Zilhao R, Carpousis AJ, Arraiano CM. RNase II levels change according to the growth conditions: characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II. Mol. Microbiol. 2001;39:1550–1561. doi: 10.1046/j.1365-2958.2001.02342.x. [DOI] [PubMed] [Google Scholar]
- 25.Typas A, Becker G, Hengge R. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- 26.Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter σS dependent? Role of the -13/-14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 2001;42:939–954. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 28.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikebe T, Iyoda S, Kutsukake K. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology. 1999;145(Pt 6):1389–1396. doi: 10.1099/13500872-145-6-1389. [DOI] [PubMed] [Google Scholar]
- 30.Kutsukake K, Ikebe T, Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- 31.Saini S, Brown JD, Aldridge PD, Rao CV. FliZ Is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J. Bacteriol. 2008;190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 34.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanya HS, Arciszewska LK, Baker RA, Bird LE, Sherratt DJ, Wigley DB. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorgai L, Yagil E, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J. Mol. Biol. 1995;252:178–188. doi: 10.1006/jmbi.1995.0486. [DOI] [PubMed] [Google Scholar]
- 37.Yagil E, Dorgai L, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J. Mol. Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 38.Wada T, Tanabe Y, Kutsukake K. FliZ Acts as a Repressor of the ydiV Gene, Which Encodes an Anti-FlhD4C2 Factor of the Flagellar Regulon in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2011;193:5191–5198. doi: 10.1128/JB.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kage H, Takaya A, Ohya M, Yamamoto T. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 2008;190:2470–2478. doi: 10.1128/JB.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin D, Rao CV, Slauch JM. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J. Bacteriol. 2008;190:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas RL, Lee CA. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2001;183:2733–2745. doi: 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini S, Slauch JM, Aldridge PD, Rao CV. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 2010;192:5767–5777. doi: 10.1128/JB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar typhimurium. J. Bacteriol. 2010;192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2011;193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006;2:2006 0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JH. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 48.Barembruch C, Hengge R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 2007;65:76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 49.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Germer J, Becker G, Metzner M, Hengge-Aronis R. Role of activator site position and a distal UP-element half-site for sigma factor selectivity at a CRP/H-NS-activated sigma(s)-dependent promoter in Escherichia coli. Mol. Microbiol. 2001;41:705–716. doi: 10.1046/j.1365-2958.2001.02548.x. [DOI] [PubMed] [Google Scholar]
- 51.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 52.Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5' mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 54.Lange R, Hengge-Aronis R. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 55.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma Z, Masuda N, Foster JW. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 2004;186:7378–7389. doi: 10.1128/JB.186.21.7378-7389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnqvist A, Olsen A, Normark S. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 62.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.