Abstract
Chemical inhibitors of histone deacetylase (HDAC) activity are used as experimental tools to induce histone hyperacetylation and deregulate gene transcription, but it is not known whether the inhibition of HDACs plays any part in the normal physiological regulation of transcription. Using both in vitro and in vivo assays, we show that lactate, which accumulates when glycolysis exceeds the cell’s aerobic metabolic capacity, is an endogenous HDAC inhibitor, deregulating transcription in an HDAC-dependent manner. Lactate is a relatively weak inhibitor (IC50 40 mM) compared to the established inhibitors trichostatin A and butyrate, but the genes deregulated overlap significantly with those affected by low concentrations of the more potent inhibitors. HDAC inhibition causes significant up and downregulation of genes, but genes that are associated with HDAC proteins are more likely to be upregulated and less likely to be downregulated than would be expected. Our results suggest that the primary effect of HDAC inhibition by endogenous short-chain fatty acids like lactate is to promote gene expression at genes associated with HDAC proteins. Therefore, we propose that lactate may be an important transcriptional regulator, linking the metabolic state of the cell to gene transcription.
INTRODUCTION
Histone modification by acetylation is an important process in transcriptional regulation. The chromatin of transcriptionally active genes is epsilon-amino (ε-NH2) acetylated on the N-terminal tails of histone H3 and histone H4, whereas that of inactive genes is hypoacetylated (1–4). The current paradigm is that histone modification and gene regulation arise from the differential recruitment of histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes to the chromatin through their association with protein complexes containing DNA binding and chromatin-modifying activities. These chromatin associated complexes are further stabilized by interactions with the modified histones, such as between the bromodomains of transcription factors and acetyl groups on histones H3 and H4 (5).
Normal mammalian cells possess 18 enzymes with HDAC activity (6). The classes I and II HDACs are simple hydrolases requiring no cofactors, whereas the class III enzymes are NAD dependent. Potent chemical inhibitors of the classes I and II HDAC activity, such as the aliphatic short-chain fatty acid butyrate and the hydroxamic acid trichostatin A, have been used experimentally to induce histone hyperacetylation in cells. After exposure to these agents, the acetylation of histone H4 increases rapidly to 70% of maximum within 3 h because HAT activity is no longer effectively opposed by HDAC activity. On removal of these inhibitors, acetylation levels return to normal within 3 h, illustrating the highly dynamic nature of histone acetylation and deacetylation (7). This dynamism of acetylation and deacetylation is further illustrated at the pS2 gene locus where activation by estrogen results in cyclical changes in histone acetylation levels associated with the periodic recruitment of HAT and HDAC activities (8).
Whilst HDAC activity can be inhibited experimentally using chemical inhibitors, there is no evidence that HDAC inhibition plays a role in the normal regulation of gene transcription. However, during the development of a Fourier transform mass spectrometry (FTMS) assay to measure histone H4 modifications, we observed that, in cell lines, histone acetylation levels increase during culture, reverting to baseline levels when the cell culture medium is refreshed. In this report, we demonstrate the mechanism of this effect, showing that two products of glycolytic metabolism, hydrogen ions and lactate ions, accumulate in the tissue culture medium and are directly inhibitory to HDAC activity. We show that lactate has global effects on gene transcription and that these effects overlap with the transcriptional changes induced by other HDAC inhibitors. We also show that the primary effect of HDAC inhibition is to increase gene transcription and propose that lactate may be an important transcriptional regulator, linking the metabolic state of the cell with transcription.
MATERIALS AND METHODS
Cell culture
HCT116 cells were grown in RPMI 10% FCS supplemented with penicillin and streptomycin. Phenol red free RPMI was used for Fluor de LysTM fluorimetric HDAC assays. For HDAC inhibitor experiments, RPMI solution was mixed 70:30 v:v with a balanced salt solution. This comprised 100 mg/L Ca(NO3)2.4H20, 400 mg/L KCl, 100 mg/L MgSO4.7H20, 2 g/L NaHCO3, 800 mg/L Na2HPO4 (anhydrous) and either 100 mM NaCl (control medium) or 100 mM sodium d- or sodium l-lactate. TSA (final concentration 10 nM), sodium butyrate (final concentration 0.2 mM) and zeocin (final concentration 20 µM) were added to control medium. Cells were incubated for 3 h in each treatment and harvested for RNA using TriZol reagent. All experiments were performed in triplicate.
Microarray analysis
RNA was extracted from cells (50% confluent) grown in T75 flasks using Trizol reagent and quality tested using an Agilent Bioanalyzer. RNA was amplified and biotin labeled using the Illumina Totalprep RNA amplification kit. The samples were hybridized onto Illumina-8 oligonucleotide expression arrays and detected using Cy3 streptavidin. Normalization of the raw data included a variance stabilizing transformation (VST) followed by robust spline normalization (RSN) using the R package Lumi (9). Background was not subtracted. An inverse of the VST was performed to convert the normalized data back to the original scale. Hierarchical clustering was performed using the ‘hclust’ function in R (2.7.1) with the Pearson’s correlation as the distance metric (1-R) and using the average linkage methodology.
The concordance of deregulation by different HDAC inhibitors—see Supplementary information
Fourier transform mass spectrometry
FTMS was performed on acid extracted histones according to a previously described method (7).
Western blotting
Western blots were performed on whole cell lysates using total anti-H4 (#07-108, rabbit polyclonal, Upstate) anti-H3 (#Ab1791, rabbit C-terminal polyclonal Abcam) antibodies. Affinity-purified acetylation specific rabbit polyclonal antibodies anti-acetyl-H4K5 (#07–327), anti-acetyl-H4K16 (#07–329) and anti-acetyl-H3K9 (#07–352) were all purchased from Upstate. The acetylation specificity of these antibodies was validated by the manufacturer using either peptide-specific binding assays or ChIP in yeast strains with specific histone H4 mutations.
HDAC assay
The effects of HDAC inhibitors on HDAC activity were assessed using the he Fluor de LysTM fluorimetric assay kit (BML AK500, Biomol) in strict accordance with the manufacturer’s protocol. The kit contains a protein extract from Hela cell nuclei which contains HDAC activity and a synthetic acetylated HDAC substrate (Fluor de Lys) which is fluorescent in the deacetylated state on the addition of the Fluor de Lys developer reagent. The fluorophore was excited at 360 nm and detected at 460 nm. An EnVision multilabel plate reader was used for quantification of fluorescence.
Nuclear magnetic resonance
Tissue culture media were harvested from cells in culture at the indicated times. The samples were centrifuged at 13 000 rpm for 3 min, and the supernatants were frozen and kept at −80°C until analysis. At analysis samples were diluted 50% in D2O. 1H NMR (nuclear magnetic resonance) spectra were obtained with a 360 MHz NMR (Bruker Biospin) operating at 360.1 MHz for 1H. Acquisition time was 2 s and a relaxation D1 of 3 s and phase-shift presaturation for water suppression was used. Sixteen scans were summed. Measurements were made at 25°C. XWin NMR software was used (Bruker Biospin).
RESULTS
A reversible increase in histone acetylation occurs during culture
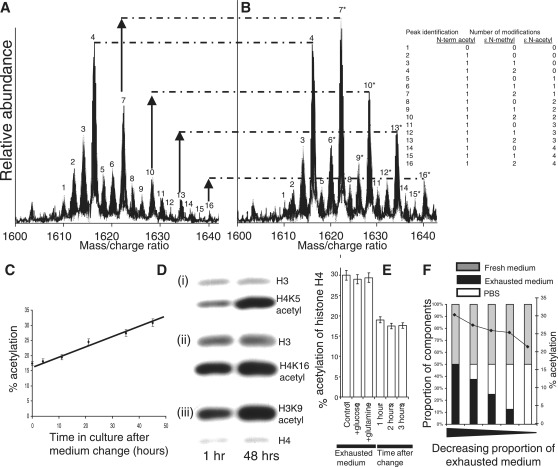
We previously developed a FTMS (9.4 Tesla) technique to quantify post-translational modifications on intact acid extracted histone H4 molecules (7). There are 16 peaks in the histone H4 mass/charge spectrum, and each peak corresponds to histone molecules with different numbers of additional methyl and acetyl groups, and potent HDAC inhibition causes a marked change in the spectrum towards maximal acetylation (Supplementary Figure S1). We previously identified all combinations of the most frequent histone H4 modifications occurring in cultured HCT116 cells (Figure 1A and B). Ninety-five percent of all histone H4 molecules lack the N-terminal methionine and are N-terminally acetylated on the first amino acid of the processed molecule, namely serine. This is a common form of primary protein processing in eukaryotes, occurring co-translationally. Histone H4 can be post-translationally modified on up to four lysine residues (lysine-5, 8, 12 and 16). The most abundant histone H4 molecule with a unique mass (28% of the total, peak 4 in Figure 1A) was unacetylated on all lysine residues and di-methylated on lysine-20 (7). We quantified the overall level of histone H4 acetylation as the percentage acetylation of all four lysines in the histone H4 spectrum.
Figure 1.
During mammalian cell culture compounds which promote histone hyperacetylation accumulate in the tissue culture medium. (A) The spectrum of post-translational modifications seen on histone H4 in HCT116 cells by FTMS. (B) Histone acetylation (peaks 7, 10, 13 and 16) increases in exhausted medium (after 48 h in culture). (C) Histone H4 acetylation levels increase during culture. (D) Confirmation (by Western blotting) of the changes in histone H4 acetylation detected by FTMS using modification-specific antibodies. Increased acetylation is also evident on lysine-9 of histone H3. (E) A complete fresh medium change is able to restore acetylation levels within 1 h. (F) Exhausted medium inhibits the restoration of acetylation levels after a medium change.
The level of histone H4 acetylation in fresh medium at neutral pH was 17% (Figure 1A). However, there was an increase in acetylation and a fall in the pH to 6.7 when HCT116 cells were grown to confluence over a period of 2 days (Figure 1B). The fall in pH during cell culture is a well-documented phenomenon. The accumulated hydrogen ions are a product of glycolysis. The proportions of histones with one, two, three and four post-translational acetylations were all seen to increase (Figure 1B, peaks 7 10, 13 and 16, respectively), and serial FTMS analysis showed that the overall increase in acetylation was gradual over this period (Figure 1C). The change in acetylation was confirmed by Western blotting using antibodies specific for acetylation of the histone H4 lysine-5 residue as well as the histone H4 lysine-16 residue (Figure 1D).
Exhausted medium contains factors that promote histone acetylation
We supplemented exhausted media with components that might potentially have been consumed during culture (such as glucose and glutamine) but were unable to correct the increase in acetylation. However, histone hyperacetylation was completely reversed within an hour of a complete change of medium (Figure 1E).
We set up an experiment to determine whether the correction of acetylation was due to the removal of factors that had accumulated in the medium during culture. At the time of medium change, we fed cells with medium that contained different proportions of fresh and exhausted medium. To control for the possibility that any effects observed might be due to nutrient replacement (rather than the removal of metabolic by-products), we ensured that the absolute quantity of fresh medium was the same (50% by volume) in all mixtures of used and fresh medium, while varying the amount of used medium by mixing it with phosphate-buffered saline (PBS) in different proportions (Figure 1F). We observed almost no correction of the hyperacetylation with a 50% mixture of fresh and used medium, but increasing correction as the amount of used medium was reduced. This result indicated that used medium contained metabolic products or factors that increased histone acetylation (Figure 1F).
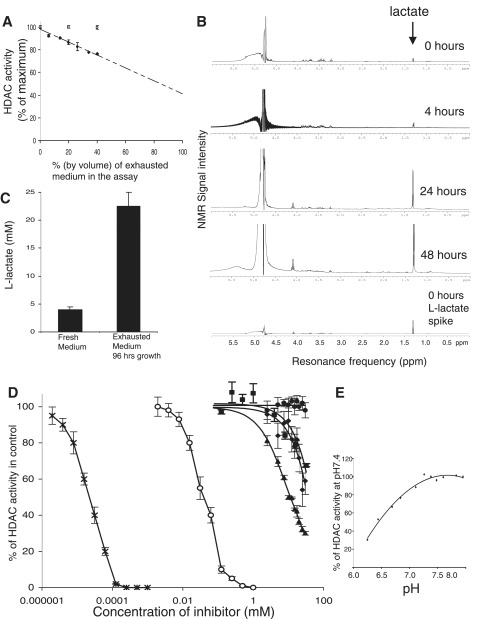
L-lactate anions accumulate progressively in culture medium and are weak inhibitors of histone deacetylases in vitro
We looked for inhibitory activity against type I and type II HDACs in Hela cell nuclei using a fluorescent HDAC substrate (Biomol HDAC assay, see ‘Materials and Methods’ section). We found that HDAC inhibition was proportionate to the amount of exhausted medium in the assay (Figure 2A). We then looked for compounds that might be responsible for HDAC inhibition by screening used medium by NMR spectroscopy. We identified a compound giving a double peak at a resonance of 1.3 ppm (Figure 2B). This signature is characteristic of lactate. We confirmed the compound’s identity by replicating the signature using fresh medium spiked with an l-lactate standard (Figure 2B, control).
Figure 2.
Exhausted tissue culture medium exhibits HDAC inhibitory activity due to the accumulation of l-lactate and increasing acidity. (A) In vitro HDAC assays to test the HDAC-inhibitory activities of fresh (open circles) and used tissue culture medium (black diamonds). All activities are percentages of the reaction buffer control. The media contributed 40% by volume to the total HDAC reaction mix. The pH values of the reaction mixtures are the same throughout (range pH 8.09–8.12). (B) NMR spectroscopy of complete tissue culture medium at various times in culture following a medium change. The control profile is of fresh medium spiked with an l-lactate standard. (C) Lactate measurements in fresh and used tissue culture medium. (D) l-Lactate, d-lactate and low-pH inhibit histone deacetylase activity in vitro. The graph shows HDAC activity in the presence of increasing concentrations of NaCl (control, filled circles), butyrate (open circles), TSA (black crosses), l-lactate (black diamonds), d-lactate (black triangles) and pyruvate (black squares). (E) In vitro HDAC activity is inhibited at low pH.
We also assayed l-lactate levels in exhausted medium using a Vitros 5,1 FS Bioanalyser. The medium was taken from a confluent culture of HCT116 cells that had been grown continuously for 96 h (Figure 2C). We compared this with the lactate level found in freshly made up medium (with 10% FCS). l-Lactate levels were greatly elevated (20–25 mM) in the used medium in comparison with fresh medium (4 mM). We therefore asked whether l-lactate and d-lactate could inhibit HDACs in vitro. We looked at the effects of purified lactate preparations (Sigma) on HDAC activity using the fluorescent reporter assay. We found that both l- and d-isomers of lactate were HDAC inhibitors at neutral pH, with d-lactate being a slightly more potent inhibitor than l-lactate (Figure 2D). The IC50 of l-lactate was 40 mM, whereas that of d-lactate was 10 mM. TSA and butyrate are much more potent inhibitors of HDAC activity in vitro, with IC50 of 2 nM and 50 µM, respectively. We also looked at the effects of pyruvate. In cells pyruvate is in chemical equilibrium with l-lactate, lactate dehydrogenase catalyzing the two-way conversion between pyruvate and lactate. Under normal conditions, the concentration of pyruvate is ∼10 times less than that of l-lactate (10). We confirmed that pyruvate is an inhibitor of HDAC activity in vitro, but contrary to a previous report (11) in our hands, the IC50 was similar to that of l-lactate (30 mM) with no discernible inhibition being observed at 1 mM. We also examined the effect of pH on HDAC activity in vitro using the HDAC reporter assay and found acid pH to be inhibitory, in agreement with a previous report (12) (Figure 2E).
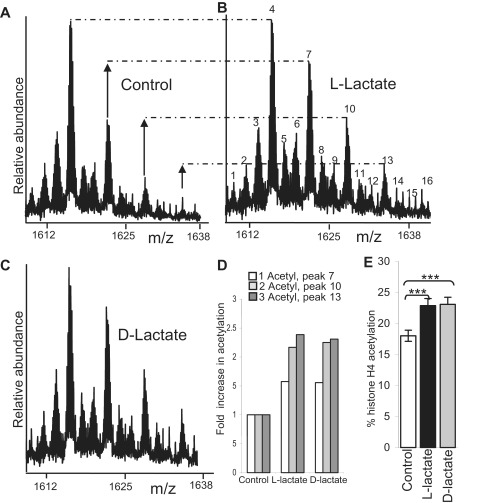
Histone hyperacetylation is induced by exogenous l-lactate and d-lactate in cultured HCT116 cells
As both lactate isomers inhibit HDACs directly in vitro, we determined whether they could induce hyperacetylation in cultured HCT116 cells in vivo. HCT116 cells were changed into fresh medium. After 2 h neutral solutions of either sodium chloride (Figure 3A, control), sodium l-lactate (Figure 3B) or sodium d-lactate (Figure 3C) were added to separate flasks, each to final concentrations of 25 mM. After a 3-h exposure to these agents, both l- and d-lactate were shown to induce histone acetylation as assayed by FTMS (Figure 3B and C). Bar graphs showing the relative increase in one-acetylated, two-acetylated and three-acetylated histone H4 molecules (compared with control) are shown in 3D, and the effects on overall histone H4 acetylation are shown in Figure 3E (***P < 0.001, unpaired t-test).
Figure 3.
l-Lactate and d-lactate induce histone hyperacetylation in vivo. Histone H4 FTMS profiles of HCT116 cells 3 h after exposure to mock (control, (A), 25 mM l-lactate (B) and d-lactate (C)). For peak identification refer to Figure 1A. Peaks 7 and 10 and 13 and are one and two and 3 ε-amino acetylated histone H4 molecules and are clearly increased in the lactate-treated cells relative to the corresponding molecules in the control. The profiles have been scaled to the zero acetylated (two methylated) peak (peak 4). (D) The fold increase in one, two and three acetylated histone H4 molecules compared with control. (E) Bar graphs showing the overall increase in total histone acetylation (***P < 0.001, unpaired test).
As HDACs are sensitive to pH, we considered the possibility that increasing the concentration of lactate in the medium might have increased acetylation by decreasing intracellular pH: high extra-cellular lactate would theoretically prevent the diffusion of monocarboxylates (such as lactate) and protons out of the cell through monocarboxylate transporters (13). We measured the intracellular pH using a flow cytometric method that utilizes the pH-sensitive fluorescent indicator BCECF-AM (Supplementary Figure S2 and Supplementary Information) (14). We found that neither high extra-cellular l-Lactate (30 mM) nor high extra-cellular d-lactate (30 mM) resulted in lower intracellular pH over the time course of this experiment (4 h).
Lactate, TSA and butyrate have similar effects on gene expression, suggesting a common mechanism of deregulation-dependent upon HDAC inhibition
We used Illumina-8 oligonucleotide arrays (22 177 probes) to investigate whether the expression changes induced by l-lactate and d-lactate were related to HDAC inhibition. We compared the gene expression changes induced by lactate to those induced by the established HDAC inhibitors, TSA and butyrate. The dosages used were those that had similar effects on histone H4 acetylation in vivo. The effects of the treatments were compared to control samples to which no drug had been applied. Sodium l-lactate and d-lactate were used at 30 mM, whereas the more potent inhibitors sodium butyrate and TSA were used at 0.2 mM and 10 nM, respectively. Parallel experiments established that these concentrations increased acetylation from 17% in control cells to 24% in l-lactate, 25% in TSA, 30% in d-lactate and 32% in butyrate. A sub-lethal dose of zeocin (20 μM) was used as a control for non-specific toxic effects. All drug exposures and controls were performed in triplicate, and an additional technical replicate was made of each of the control samples. Control set 1 (C1) consisted of three biological replicates of untreated cells, and control set 2 (C2) were the technical replicates of each of the C1 samples.
After normalization of the array data, we used t-tests to look for genes that were significantly deregulated in common by the different treatments compared with controls. Many more genes had changed in common than would be expected by chance (Table 1). Chi-squared tests demonstrated that all differences between the observed overlap, and the overlap expected by chance were highly significant (P < 0.0001). In contrast, with the exception of TSA and zeocin, there was no significant overlap between any HDAC inhibitor treatments and zeocin.
Table 1.
HDAC inhibitors deregulate more genes in common than would be expected by chance
| l-lac v C1 | d-Lac v C1 | TSA v C1 | But v C1 | Zeo v C1 | |
|---|---|---|---|---|---|
| l-lacvC2 | 912 (195)+ | 672 (174)+ | 678 (195)+ | 511 (187)+ | 62 (90)− |
| d-lacvC2 | 636 (152)+ | 1530 (195)+ | 932 (243)+ | 1098 (233)+ | 84 (112)0 |
| TSAvC2 | 515 (176)+ | 832 (266)+ | 1853 (195)+ | 963 (286)+ | 119 (137)0 |
| But.vC2 | 479 (165)+ | 1091 (234)+ | 1082 (263)+ | 1619 (195)+ | 78 (121)− |
| ZeovC2 | 61 (51)0 | 68 (73)0 | 134 (82)+ | 71 (78)0 | 308 (195)+ |
The number of genes deregulated by each treatment (bold) and co-deregulated by pairs of different treatments. The numbers that would be expected by chance (null hypothesis) are shown in brackets (see ‘Materials and Methods’ section). Chi-squared analysis with Yates correction was used to determine whether the differences were statistically significant: +significantly more than expected (P < 0.001); −significantly less than expected (P < 0.05); 0not significantly different (P > 0.05).
We next identified 338 genes whose expression had changed significantly and in the same direction in all four HDAC treatments compared with the C1 control set. Only 35 of these genes were also deregulated by zeocin in the same direction. Of the 338 genes identified, 185 had increased their expression compared with control set C1, and 153 had decreased their expression (Supplementary Tables S1 and S2). We generated the dataset 1000 times using random numbers (see Materials and Methods) and found that on average just 21 genes would be co-deregulated by all four treatments by chance (95% confidence limits 12–30).
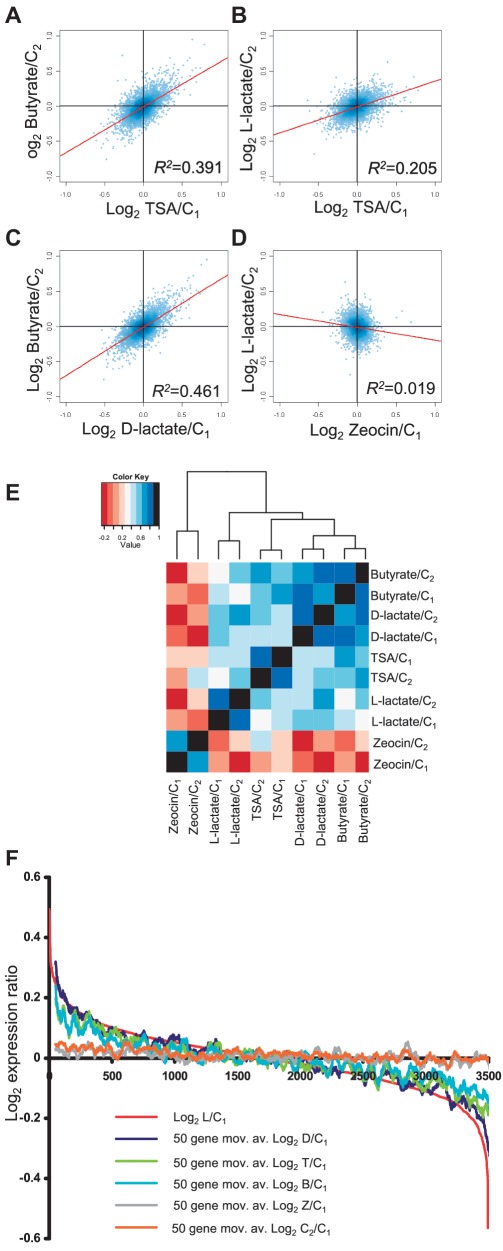
Gene expression changes induced by lactate correlate with changes induced by other HDAC inhibitors across the entire genome
Whilst the t-tests identified ∼1.5% (338) of the 22 177 genes as being significantly deregulated by all HDAC inhibitors, we also wanted to determine the extent to which the HDAC inhibitors affected the global pattern of gene expression. We therefore correlated changes in gene expression (compared with control) between pairs of treatments across the entire dataset. We found a clear tendency for gene expression changes induced by HDAC inhibitors to correlate positively and highly significantly, whereas, with the exception of zeocin and TSA, all correlations between HDAC inhibitors and zeocin were negative (Supplementary Table S3). The weak but significant positive correlation between zeocin, and TSA may indicate that TSA induces some toxic effects that are shared with zeocin.
We reasoned that the correlations observed might be even stronger (potentially demonstrating an even greater reliance of gene expression on normal HDAC activity) if we excluded all genes that were likely to be un-expressed or low-level expressed as these data points are known to have a high variance. We therefore excluded all probes with average expression values of <100 in control set C1. Of the remaining probes, we included only those that had a coefficient of variation within replicates of <10% in all of the treatments and control sets. Together these procedures eliminated without bias 18 681 of the 22 177 probes, leaving 3496 probes (Supplementary xls File 1). As predicted, the correlations between HDAC inhibitors were even higher within this refined set of probes (Table 2 and Figure 4 A–D). The established HDAC inhibitors TSA and butyrate showed strong positive correlations, but the strongest correlation was between d-lactate and butyrate (R2 0.461 and 0.483 l using different control sets). We performed hierarchical clustering of the treatment/control ratios of these 3496 genes using Pearson’s correlation R as the distance metric, the HDAC inhibitors clustered together separately from zeocin illustrating again the close relationship (shorter distance metric) between HDAC inhibitors as compared with zeocin (Figure 4E).
Table 2.
R2 values for the Pearson correlations of the expression changes observed in a refined dataset (n = 3496)
| l-lac/C1 | d-lac/C1 | TSA/C1 | But/C1 | Zeo/C1 | |
|---|---|---|---|---|---|
| l-lac/C2 | 0.248* | 0.205* | 0.101* | 0.019* | |
| d-lac/C2 | 0.283* | 0.231* | 0.483* | 0.064* | |
| TSA/C2 | 0.133* | 0.151* | 0.286* | 0.002# | |
| But/C2 | 0.107* | 0.461* | 0.391* | 0.020* | |
| Zeo/C2 | 0.001# | 0.034* | 0.057* | 0.003# |
Expression changes were expressed as ratios compared with the values in the control sets indicated. The numbers represent the R2 values for the indicated correlations. The significance of these correlations in indicated as follows: *P < 10-6; #P < 0.02. The P values for the Pearson correlations are 2-tailed significance tests of the t-value with n-2 degrees of freedom, where t = R.√[(n-2)/(1-R2)]. The shaded numbers indicate where correlations are negative.
Figure 4.
Gene expression changes induced by different HDAC inhibitors are positively correlated. (A–D) Correlations of the expression changes induced by different pairs of treatments. The data are taken from a refined dataset of 3496 probes. (A) Log2 butyrate/C2 versus TSA/C1; B, Log2 l-lactate/C2 versus TSA/C1; C, Log2 Butyrate/C2 versus d-Lactate/C1; (D) Log2, l-lactate/C2 versus Zeocin/C1. Regression lines are superimposed on the graphs together with their corresponding R2 values (also see Table 2). (E) Hierarchical cluster analysis of the expression changes induced by HDAC inhibitors and zeocin using the Pearson correlation R as the distance metric. (F) Probes ordered on the x-axis according to their expression ratios (log2, y-axis) in l-lactate (red line). Fifty-point moving averages of the log2 expression ratios of the same probes on exposure to the other HDAC inhibitor treatments (and controls) are over-laid. See key.
It was still formally possible that the gene expression changes that were identified as being positively correlated between any two HDAC inhibitors, for example TSA and butyrate, might be different from those identified as being correlated between different HDAC inhibitors, say d-lactate and l-lactate. To demonstrate that in general the same genes were being deregulated by HDAC inhibition, we aligned the 3496 genes in order of their expression changes in l-lactate and determined the magnitude of their expression changes when exposed to the other HDAC inhibitors (Figure 4F). The alignment showed a clear tendency for genes that are deregulated by l-lactate to also be deregulated by the other HDAC inhibitors. The effect of zeocin was completely different to that of the HDAC inhibitors, showing no tendency to follow the l-lactate-induced expression changes. As we had compared all expression to one control set (C1), we needed to control for the possibility that the trends observed for the HDAC inhibitors were not due to systematic errors in the C1 control measurements. To exclude this, we plotted changes in the second control set C2 relative to first control set C1 (C2/C1). There was no significant tendency for gene expression to vary to the extent that was observed with l-lactate (Figure 4F). Together these data demonstrate that different HDAC inhibitors, including l-lactate and d-lactate, cause global deregulation of the same genes.
HDAC-associated genes are more likely to be upregulated and less likely to be downregulated by HDAC inhibition
We next wanted to determine whether HDAC inhibition affected gene expression in any particular direction. The genes on the Illumina-8 arrays were both up and downregulated in our experiment, most likely because both the direct and the indirect effects are induced by HDAC inhibition. There is a general acceptance that promoter hypoacetylation is associated with reduced transcription. The inactive X chromosome is extensively hypoacetylated (3), and HDACs, which would induce hypoacetylation, are associated with transcriptional repressor complexes (15). However, a recent comprehensive study of HDAC binding to the promoters of genes in CD4 T cells demonstrated that HDAC1, HDAC2, HDAC3 and HDAC6 were bound with increased frequency to the promoters of genes with high-level expression (16). The authors (Wang et al.) speculated that HDAC activity might actually be required for the expression of active genes, as had previously been shown for the Hos2 deacetylase in yeast (17). Wang et al. used high-dose HDAC inhibitors (300 nM TSA and 2 mM butyrate) to investigate the effect of inducing hyperacetylation at quiescent genes. They found that histone acetylation could only be induced at genes which were also histone H3K4 methylated. This caused an increase in PolII binding, but gene expression could not be significantly induced at these genes.
We have used the Affymetrix gene expression and HDAC ChiP-sequencing data of Wang et al (16) to further dissect the function of HDACs on transcription. We asked whether genes that were significantly associated with the different HDACs in their study were more or less likely to be deregulated by HDAC inhibition than might be expected by chance.
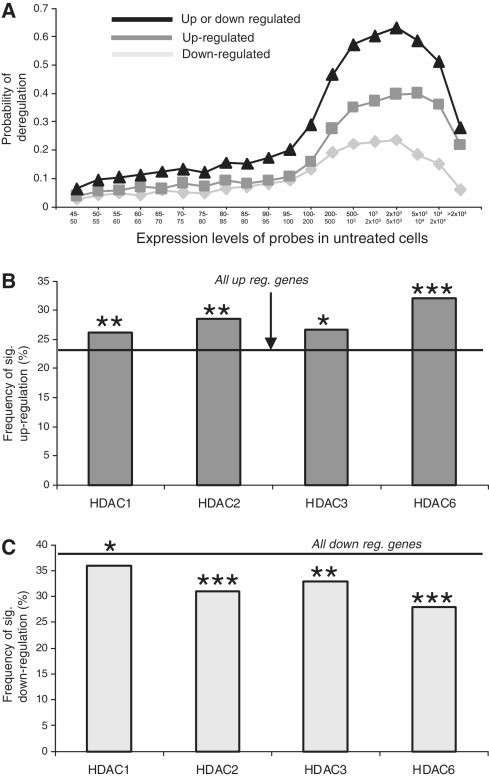
We used t-tests (P < 0.05) to assess significance of deregulation and found that the probability of detecting significant deregulation by HDAC inhibitors varied with the expression values of probes (Figure 5A). The greatest probability of deregulation occurred in probes with expression values between 1000 and 5000 in control cells. A similar pattern was seen with lactate and butyrate on our Illumina expression arrays (Supplementary Figure S2A and B). Interestingly in the Wang et al, dataset genes were more likely to be significantly downregulated than upregulated by HDAC inhibition (Figure 5A). However, when we looked specifically at the genes that were associated with HDAC proteins, we found highly significant effects on gene expression. Genes (probes) with expression values between 1000 and 5000 in control cells were significantly more likely to be upregulated by HDAC inhibition if they were associated with either of the HDACs (Figure 5B). Conversely the probability of being downregulated was less for probes that were associated with each of the HDACs (Figure 5C). Thus, while genes can be both upregulated and downregulated by exposure to HDAC inhibitors, the association of genes with HDAC proteins increases their likelihood of upregulation. This indicates that, rather than controlling the repression of non-expressed genes, HDACs exert a repressive effect on expressed genes and this effect would be modulated by endogenous HDAC inhibitors such as lactate.
Figure 5.
HDAC inhibition preferentially up-regulates HDAC-associated genes. (A) The probability of significant gene deregulation in CD4 cells after a 2 h exposure to high-dose butyrate and TSA. (B and C) Analysis of probes with expression values between 1000 and 5000 in CD4 cells. HDAC-associated genes/probes are more likely to be significantly upregulated (B) and less likely to be significantly downregulated (C) when compared with the overall probability of being up or downregulated on the array [horizontal lines (Chi-squared test)]. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
In this article, we show that, in addition to gene regulation caused by the differential recruitment of HDAC proteins to genes, genes can also be regulated by the in situ inhibition of these enzymes by an endogenous product of glycolytic metabolism, namely lactate. Furthermore, lactate induces HDAC-dependent gene deregulation at concentrations which arise in nature. Our study shows that pyruvate also has HDAC inhibitory activity but the concentration of pyruvate within cells is likely to be too low to inhibit HDAC activity in vivo. In cells, l-lactate and pyruvate are in chemical equilibrium. The levels of l-lactate and pyruvate in resting muscle cells have been measured as 6.6 mmol/kg dry weight and 0.5 mmol/kg dry, respectively (10). This equates to concentrations of 2.8 mM and 0.21 mM, respectively, assuming muscle is 70% water by weight. During intense exercise lactate levels rise to 130 mmol/kg dry weight (55.9 mM), whereas pyruvate levels rise to just 1.95 mmol/kg dry weight (0.83 mM). Thus l-lactate produced endogenously from glycolysis would be expected to inhibit HDAC activity during intense exercise, but pyruvate would not.
Structure studies show that the HDAC active site is lined by hydrophobic residues (18). The polar hydroxyl group on the second carbon of lactate (2-hydroxypropionate) would be predicted to diminish binding to the active site and probably explains why lactate is a less potent inhibitor than the aliphatic short-chain fatty acids (IC50 40 mM versus 50 µM). Our data predict that the HDAC-dependent transcriptional effects of l-lactate would be most significant when the intracellular lactate level rises, as it does when glycolysis is increased.
The concentrations of all HDAC inhibitors used in our experiments were very different (from 10 nM for TSA to 30 mM for lactate) but were calculated to induce similar effect on histone acetylation. The effects of lactate are likely to be complex, but the fact that lactate-induced effects on transcription that were similar to the effects induced by other established HDAC inhibitors (Figure 4F) is compelling evidence that a main mechanism of the effect is through the inhibition of histone deacetylation. However, transcriptional effects caused by the hyperacetylation of non-histone proteins cannot be ruled out.
The primary effect of HDAC activity on gene transcription is difficult to dissect on gene expression arrays because secondary changes are likely to cloud the primary effects. Interestingly, the most expressed genes in the genome appear to be those with the most HDAC activity bound (16). This is somewhat at variance with the generally held view that HDAC activity opposes gene transcription. However, we have shown that HDAC-associated active genes are more likely to be upregulated and less likely to be downregulated by HDAC inhibition. This indicates that HDAC activity most likely fine-tunes transcription at active genes, acting to dampen it. The purpose of HDAC inhibition by lactate may be to preserve transcription during temporary glycogen depletion brought about by increased glycolysis.
Our findings have relevance for human health and disease. Lactate levels rise very significantly (to 55 mM) in skeletal muscles during strenuous exertion (10) as glycolysis outstrips the capacity of mitochondria to metabolize the pyruvate produced. The effects of lactate may also be of relevance in tumour biology. Some tumours produce high levels of lactate due to an apparent over-reliance on glycolytic metabolism (19,20) and high lactate levels (up to 40 µmol/g wet weight, equivalent to ∼57 mM) have been reported to correlate with a tendency to metastasis (21). It remains to be determined whether lactate might regulate genes involved in tumour progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S3, Supplementary Tables S1–S3, Supplementary Methods and Supplementary File 1.
FUNDING
We thank the Melville Trust (T.L.), CRUK (T.L. and B.R.), Wellcome Trust (N.G.), Breakthrough Breast Cancer Charity (J.C., D.S., D.J.H., N.G. and B.R.), and RASOR Interdisciplinary Research Centre (L.M.) for support. Funding for open access charge: Division of Pathology, University of Edinburgh.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Meehan for critical reading of the manuscript.
REFERENCES
- 1.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 4.Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 6.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 7.Mackay CL, Ramsahoye B, Burgess K, Cook K, Weidt S, Creanor J, Harrison D, Langridge-Smith P, Hupp T, Hayward L. Sensitive, specific, and quantitative FTICR mass spectrometry of combinatorial post-translational modifications in intact histone H4. Anal. Chem. 2008;80:4147–4153. doi: 10.1021/ac702452d. [DOI] [PubMed] [Google Scholar]
- 8.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 9.Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 10.Putman CT, Jones NL, Lands LC, Bragg TM, Hollidge-Horvat MG, Heigenhauser GJ. Skeletal muscle pyruvate dehydrogenase activity during maximal exercise in humans. Am. J. Physiol. 1995;269:E458–E468. doi: 10.1152/ajpendo.1995.269.3.E458. [DOI] [PubMed] [Google Scholar]
- 11.Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, Sterneck E, Ganapathy V. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. 2006;66:11560–11564. doi: 10.1158/0008-5472.CAN-06-1950. [DOI] [PubMed] [Google Scholar]
- 12.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–11091. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 13.Halestrap AP, Meredith D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- 14.Franck P, Petitipain N, Cherlet M, Dardennes M, Maachi F, Schutz B, Poisson L, Nabet P. Measurement of intracellular pH in cultured cells by flow cytometry with BCECF-AM. J. Biotechnol. 1996;46:187–195. doi: 10.1016/0168-1656(95)00189-1. [DOI] [PubMed] [Google Scholar]
- 15.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 18.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl Acad. Sci. USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 20.Li X, Vigneron DB, Cha S, Graves EE, Crawford F, Chang SM, Nelson SJ. Relationship of MR-derived lactate, mobile lipids, and relative blood volume for gliomas in vivo. AJNR Am. J. Neuroradiol. 2005;26:760–769. [PMC free article] [PubMed] [Google Scholar]
- 21.Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfør K, Rofstad EK, Mueller-Klieser W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–921. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.