Abstract
The macrocyclic bis-naphthalene macrocycle (2,7-BisNP), belonging to the cyclobisintercalator family of DNA ligands, recognizes T–T mismatch sites in duplex DNA with high affinity and selectivity, as evidenced by thermal denaturation experiments and NMR titrations. The binding of this macrocycle to an 11-mer DNA oligonucleotide containing a T–T mismatch was studied using NMR spectroscopy and NMR-restrained molecular modeling. The ligand forms a single type of complex with the DNA, in which one of the naphthalene rings of the ligand occupies the place of one of the mismatched thymines, which is flipped out of the duplex. The second naphthalene unit of the ligand intercalates at the A-T base pair flanking the mismatch site, leading to encapsulation of its thymine residue via double stacking. The polyammonium linking chains of the macrocycle are located in the minor and the major grooves of the oligonucleotide and participate in the stabilization of the complex by formation of hydrogen bonds with the encapsulated thymine base and the mismatched thymine remaining inside the helix. The study highlights the uniqueness of this cyclobisintercalation binding mode and its importance for recognition of DNA lesion sites by small molecules.
INTRODUCTION
Mismatched, or non-complementary, base pairs in DNA are permanently formed in living cells due to misincorporation of nucleotides during DNA synthesis, inclusion of chemically damaged nucleotides, or inclusion of normal nucleotides opposite damaged bases in the DNA template strand (1). The rate of generation of DNA mismatches is greatly increased by exogenous factors, such as genotoxic chemicals or UV radiation. DNA mismatches disrupt the genetic information and are therefore highly toxic for the organism. As a consequence, all organisms possess enzyme systems responsible for the detection and repair of damaged DNA (DNA repair systems) (2,3). The primary task of the DNA repair system, i.e. detection and excision of damaged and mismatched bases from the DNA, is met by the DNA glycosylases that locate the damaged or ‘wrong’ nucleobases embedded in the massive excess of undamaged genome much like a needle in a haystack. The mechanism, by which DNA glycosylases accomplish this formidable task, extremely efficiently and rapidly, is still a matter of debate (1,4–9), but ultimately its deeper understanding will allow us to control the DNA repair processes in normal and tumor tissues.
Notably, modern cancer therapy is largely based on the agents (drugs or radiation) killing cancer cells more efficiently than normal cells, and DNA-damaging drugs represent the primary therapy in most cases. However, due to the activity of the DNA repair system, a large part of the drug-induced lesions is repaired before they cause cell death and stop proliferation. Therefore, the efficacy of the DNA damage-based cancer therapy can be increased by inhibition of such repair pathways, and DNA repair enzymes represent a topic of an immense interest in this context (10–15). Apart from a few known inhibitors of DNA repair enzymes such as APE1 (16–20), PARP (21–23) or UDG (24,25), it may be assumed that small molecules, interfering with repair enzymes by binding to their substrate, i.e. DNA lesion sites, could lead to modulation of DNA repair pathways and enhance of the anti-cancer activity of DNA-damaging agents.
Along these lines, a few families of small molecules that selectively bind to DNA lesions, e.g. to mismatched base pairs in the DNA, have been described during the last decade. Among them, derivatives of naphthyridine have been shown to bind to G–G, G–A, A–A or C–C mismatched base pairs by insertion into the duplex and formation of hydrogen bonds with the mismatched and neighboring bases (26–30). The solution structure of a complex of naphthyridine–azaquinolone dimer with an oligonucleotide having A–A mismatch in the CAG/CAG context was characterized by NMR; it features the binding of two molecules of ligand in the mismatch site, accompanied by ejection of two flanking cytosine residues from the duplex (31). The second important class of mismatch binders is represented by bulky metal complexes, such as [Rh(bpy)2(chrysi)]3+ and [Co(phen)2(HPIP)]3+, which do not form hydrogen bonds with the nitrogenous bases, but insert into mismatched and abasic sites taking advantage of their higher accessibility, as compared to the Watson–Crick base pairs (32–37). Structures of [Rh(bpy)2(chrysi)]3+ bound to A–C and A–A mismatches were determined in the solid state using X-ray diffraction analysis (38,39), whereas the structure of a complex with a C–C mismatch was recently characterized in solution using NMR spectroscopy (40). Notably, in all three cases, the binding of the metal complex to the mismatch site leads to ejection of both mispaired bases out of the stack. However, intercalation of [Rh(bpy)2(chrysi)]3+ between two well-matched base pairs was also observed in the solid state (38). Most importantly from the therapeutic point of view, mismatch-selective metal complexes inhibit the growth of mismatch-repair-deficient cancer cell lines, and not that of mismatch repair-proficient ones, which supports the hypothesis that recognition of base mismatches by small molecules and interference with the DNA repair enzymes is possible in cells as well (41,42).
In another approach, we have shown that certain macrocyclic ligands containing two aromatic planes connected by two flexible polyamine chains (also termed cyclobisintercalators, or CBI) (43) can selectively bind to DNA lesions such as abasic sites (44,45) and homopyrimidine mismatches (46,47). The initial studies were performed with the 2,7–substituted bis-acridine (BisA) and 2,6-substitued bis-naphthalene (2,6-BisNP) macrocycles. Later, however, screening of a panel of 20 homo- and heterodimeric macrocyclic ligands led to the identification of the 2,7–substituted bis-naphthalene isomer (2,7-BisNP, Scheme 1) as the ligand with the highest binding selectivity for mismatched versus fully matched DNA duplexes (48). The selective binding of this family of ligands to DNA lesion sites is, presumably, due to their peculiar constrained conformation and the short distance between the two aromatic planes, which does not allow their simultaneous intercalation into regular DNA, as it would violate the neighbor-exclusion principle (49). However, it appears that at sites of weaker thermodynamic stability, such as mismatches and abasic sites, this principle can be violated, allowing the highly efficient binding of cyclobisintercalators. In fact, the binding of the cyclobisacridine BisA to a DNA duplex with an abasic site has been characterized by a high-resolution NMR and molecular modeling study, which showed that one of the acridine units of BisA occupies the abasic pocket, while the second one intercalates at the neighboring base pair step, in a threading bisintercalation fashion (45). Thus, we speculated that binding of 2,7-BisNP to T–T mismatch may proceed in a similar fashion. In addition, it was postulated that one of the mismatched thymines may be extruded (flipped out) from the helix, as this would be in agreement with their higher susceptibility to permanganate oxidation observed upon binding of CBI macrocycles (46). However, the structural details of the recognition event have not been available, so far.
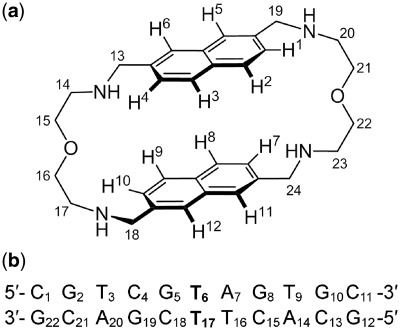
Scheme 1.
(a) Chemical structure and proton numbering scheme of 2,7-BisNP. (b) Sequence and numbering of the TT-DNA duplex. The T-T mismatch is highlighted in boldface.
In this study, we report the NMR characterization and molecular modeling of 2,7-BisNP binding to an 11-mer oligonucleotide duplex containing a thymine–thymine mismatch target (TT-DNA, Scheme 1). The central part of TT-DNA (5′-TCG T AGT-3′/5′-ACT T CGA-3′) corresponds to that of 17-mer duplex, used in our previous studies (46,46); the terminal GC base pairs were added to increase the thermal stability of the short duplex. Moreover, the asymmetry of the binding site with respect to the neighboring base pairs (G–C versus A–T) allowed us to hope that one type of ligand–DNA complex would be present exclusively, unlike in the case of BisA–abasic site recognition, where the symmetry of the binding site (two flanking G–C pairs) gave rise to two complexes (45). In addition, the solid-state conformation of 2,7-BisNP alone was obtained from X-ray diffraction analysis and served for comparison with the one observed upon binding to the DNA mismatch in solution.
MATERIALS AND METHODS
Solid-state structure of 2,7-BisNP
The synthesis and characterization data of 2,7-BisNP × 4 HCl were reported previously (48). Crystals suitable for X-ray diffraction analysis were obtained by vapor diffusion of Et2O or t-BuOMe into solutions of 2,7-BisNP × 4 HCl in MeOH. A satisfactory dataset was obtained with a crystal grown form a MeOH–Et2O solvent pair. Diffraction data were collected by using a Bruker Kappa X8 APEX II diffractometer with graphite-monochromated Mo Kα radiation (λ = 0.71073 Å) at 100 ± 1 K. The structure was solved by direct methods using SHELXS-97 and refined by full-matrix least-squares techniques using SHELXL–97 with anisotropic displacement parameters for all non-hydrogen atoms. X-ray data for 2,7-BisNP × 4 HCl × 4 MeOH (CCDC-847671): C36H60Cl4N4O6, M = 786.68 g mol−1, space group P21/c (no. 14), a = 32.786 6(9), b = 5.524 80(10), c = 23.056 6 (6) Å, α = γ = 90°, β = 105.943 0(10)°, V = 4015.81(17) Å3, Z = 4, μ = 0.342 mm−1, F(000) = 1680, crystal size 0.20 × 0.11 × 0.06 mm3. A total of 98 941 reflections (1.29° ≤ θ ≤ 33.30°) were collected, of which 15 469 were unique (Rint = 0.0470) and 10 758 observed [I > 2σ (I)] were used in calculations. The structure was refined on F2 to R = 0.0820 [I > 2σ(I)] for 483 refined parameters and 6 restraints. Largest difference peak and hole 1.741 and −1.264 e Å−3.
NMR studies
Sample preparation
The oligonucleotides (purified by HPLC and ion exchange chromatography) were purchased from Eurogentec (Belgium). The two strands d(CGTCGTAGTGC) and d(GCACTTCGACG) were dissolved and mixed in 500 µl of 10 mM sodium phosphate buffer (pH 6) containing 50 mM NaCl, heated to 80°C and slowly cooled to room temperature to form the duplex. The sample was lyophilized once from water, twice from 99.9% D2O and then redissolved in 500 µl of 99.99% D2O. For NMR experiments involving exchangeable protons, the oligomers were dissolved in 90% H2O/10% D2O (v/v). The final duplex concentration was about 1.5 mM. For the 2,7-BisNP–TT-DNA complex, the ligand was dissolved in the NMR buffer at 19 mM and added progressively to the free DNA in the NMR tube until a 1:1 complex was obtained, as confirmed by 1D NMR spectroscopy.
NMR experiments
1H NMR experiments were carried out with a Varian Unity Plus 500 MHz spectrometer or a Varian Unity 800 MHz spectrometer equipped with a cryo-probe. Chemical shifts were calibrated relative to sodium 3–(trimethylsilyl)propionate-2,2,3,3-d4 (TSP-d4). Data were processed with VNMR software (Varian) and displayed and analyzed with the CCPNMR program (50).
The NOESY spectra of the free DNA and of the ligand–TT-DNA complex were recorded in D2O buffer at 10°C using mixing times of 75, 100, 150 and 250 ms. The TOCSY spectra were recorded with a spin-lock time of 40 and 60 ms using a MLEV17 pulse train of 8 kHz for the free DNA and with a spin-lock time of 60 or 110 ms for the complex. The experiments were recorded with 512 (t1) × 2048 (t2) complex data points. Ninety-six scans were acquired over a spectral width of 5000 Hz (for experiments at 500 MHz) or 64 scans were acquired over a spectral width of 8000 Hz (for experiments at 800 MHz) in both dimensions. Water resonance was suppressed by a standard presaturation during the relaxation delay and during the mixing time for NOESY. The DQFCOSY spectrum was run with 2048 points in t2, 256 points in t1, and 96 scans for each t1 value. For a sample in 90% H2O, NOESY spectra were acquired at 10 or 5°C using the sculpting gradients scheme with selective pulses of 3–4 ms for water suppression. The mixing time was set to 350 ms. The experiments were recorded with 340 (t1) × 4092 (t2) complex data points and 160 scans for each FID. 1D proton-decoupled 31P spectra and 2D 1H-31P HSQC were recorded at 10°C at 202 MHz; chemical shifts were referenced to trimethyl phosphate.
For the free 2,7-BisNP, 1D NMR spectra, 2D NOESY with mixing time of 0.8 s and DQFCOSY were performed in a D2O buffer containing 10 mM sodium phosphate (pH 6) and 50 mM NaCl at 10°C.
Distance restraints and dihedral angles
The NOE intensities measured from the NOESY spectra in D2O (mixing time of 150 ms) were converted into distance restraints in the CCPNMR program and calibrated to the fixed reference cytosine H5–H6 = 2.45 Å. The lower and upper boundaries were set to 0.8 and 1.2 multiples of calculated distances, respectively. For dihedral angles restraints, a qualitative analysis of the DQFCOSY cross-peaks intensities was used to evaluate the approximate values of the JH1′–H2′, JH2″–H3′ and JH3′–H4′ coupling constants and deduce the pseudorotation angles P. The P-values were then used in the Antechamber module (AMBER 10.1 software, University of California, San Francisco) to generate the sugar pucker restraints. For the ligand–DNA complex, intermolecular NOEs between the ligand and DNA were measured and classified semi-quantitatively as distances into three categories: strong (2.0–3.0 Å), medium (3.0–4.0 Å) and weak (4.0–5.0 Å).
Molecular modeling
The B-DNA duplex was built using the Biopolymer module of Biosym Insight II software, saved as a pdb file and exported into xLEaP module of the AMBER program. Sodium counterions were added. Structure calculations of the free DNA and of the ligand–TT-DNA complex were performed with the SANDER module of AMBER10.1 using the ff99SB force field. A 2 fs time step was used for all the calculations, and the SHAKE algorithm was applied to remove bond stretching. The tetraprotonated 2,7-BisNP ligand was constructed in Insight II and minimized with the AMBER force field, which lead to a U-shaped conformation with a near-parallel orientation of naphthalene units. The partial charges were calculated using HF/6-31G* (Gaussian 03 Rev. A.1, Gaussian, Inc., Pittsburgh, PA, USA) and the RESP module in AMBER. A new AMBER library containing the ligand parameters was then created.
Structure of the free TT-DNA
Five random starting structures were prepared using 3–5 ps unrestrained molecular dynamics at elevated temperature ranging from 600 to 1000 K. Each of these structures was subjected to 100 ps restrained simulated annealing protocol with implicit solvent (Born solvation model) using 232 experimental interproton distances, 100 sugar dihedral angles, 91 backbone torsion angles which maintained the right-handed character of the helix and 27 angles which preserved base pairing as restraints. The temperature was rapidly increased to 400 K and maintained over the first 20 ps. The system was then gradually cooled from 400 to 100 K for 70 ps and finally to 0 K for the last 10 ps. The TAUPT parameter was set to 0.4 ps for the first 20 ps, and then during cooling it was increased to 4.0 ps. From 90 to 95 ps, TAUPT was decreased to 1 ps and to 0.1 ps for the last 5 ps. The weight of NMR restraints (REST parameter) increases from 0.1 to 1 during the first 10 ps (3.2 to 32 kcal mol−1 Å−2 for distance restraints and 50 kcal mol−1 rad−2 for torsion angle restraints) and kept at this value until the end of the simulation. The five final structures were averaged and solvated in a periodic TIP3 water box that extended to a distance of 5 Å from any solute atom (approximate dimensions 35 × 35 × 50 Å, 1766 water molecules). One thousand steps of minimization of the solvent and counterions only were applied; the whole system was then subjected to 5000 steps of minimization. A 20 ps NVT dynamics were then performed at 300 K on the water only (with the DNA and sodium ions weakly constrained) before 100 ps of NPT dynamics at 300 K were performed on the whole system without NMR restraints, and then 100 ps of NPT dynamics at 300 K with NMR restraints. The structures over the last 5 ps were analyzed. The structures have been deposited to the Protein Data Bank (code 2LL9).
Structure of the 2,7-BisNP–TT-DNA complex
The structure of the free TT-DNA obtained from the above protocol was modified in the Biopolymer module of Biosym Insight II software to create an intercalation site at the A7pG8 and C15pT16 steps. The backbone angles of the T6pA7pG8 and C15pT16pT17 steps were manually changed, and T6 and T17 were pushed toward the major groove. 2,7-BisNP was manually docked into the DNA. The complex was solvated with a new periodic TIP3 water box extended to a distance of 4.5 Å from any solute atom (approximate dimensions 40 × 40 × 55 Å, 2400 water molecules). One thousand steps of minimization of the solvent and counterions only were applied, and then the whole system was subjected to 5000 steps of minimization using NMR restraints with a force constant of 32 kcal mol−1 Å−2. Then 20 ps of equilibration followed by 40 ps of simulated annealing molecular dynamics were performed. No force constants were introduced to ensure the A7–T16 base pairing. The final structure has been deposited to the Protein data bank (code 2LLJ).
DNA melting experiments
Thermal denaturation studies were performed in triplicate according to the previously publishing protocol in 10 mM sodium cacodylate buffer containing 50 mM NaCl, pH 6 (48). The concentration of duplex TT-DNA or TA-DNA (5′-CGT CGT AGT GC-3′/5-GCA CTA CGA CG-3′) was 2 µM, and the concentration of ligand was varied from 0 to 8 µM.
RESULTS
Structure of 2,7-BisNP in the solid state
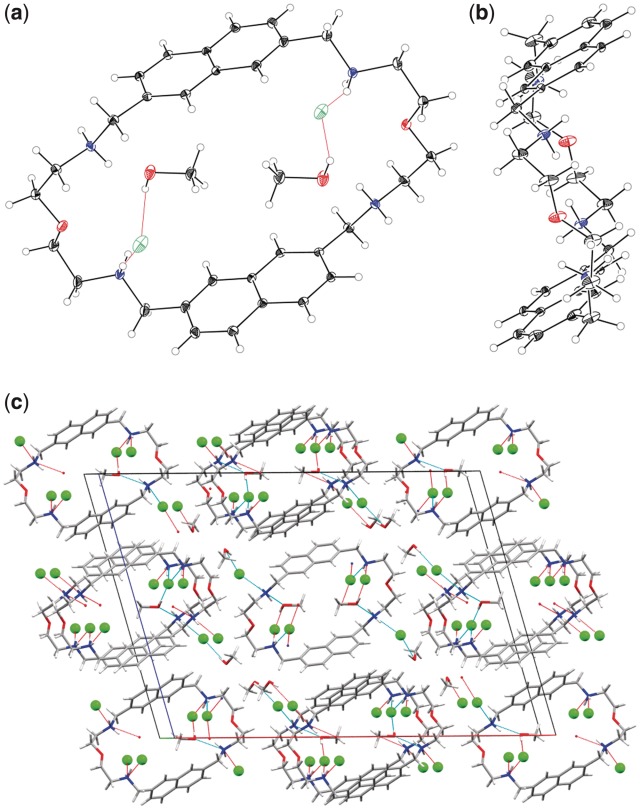
The preliminary X-ray diffraction data obtained with several crystals grown form MeOH–Et2O or MeOH–t-BuOMe solvent pairs indicated the same space group and unit cell volume, which provided evidence of the identical structure. A detailed analysis was performed with a crystal of 2,7-BisNP × 4 HCl × 4 MeOH grown from MeOH–Et2O. Two crystallographically non-equivalent cations of the macrocycle with similar geometric parameters were observed in the asymmetric unit. The structure (Figure 1a and b) reveals the non-overlapping, anti-parallel orientation of the naphthalene units within each cation. The planes of the aromatic units are separated by a distance of 6.6–6.7 Å. Two molecules of MeOH are inscribed in the macrocycle, hold by the hydrogen bonds between the OH groups, chloride ions, and the protonated amino groups of the side chains of 2,7-BisNP, as well as by van der Waals contacts between the methyl groups and naphthalene units. Two other solvent molecules and chloride ions reside in the intermolecular cavities (Figure 1c). Importantly, the naphthalene units do not participate in intra- or intermolecular π-stacking interactions. The structure is hold together by a network of hydrogen bonds between chloride anions, methanol molecules and protonated amino groups of the side chains.
Figure 1.
Structure of 2,7-BisNP × 4 HCl in the solid state. (a) ORTEP view along the crystallographic b axis of 2,7-BisNP with the enclosed solvent molecules and anions. The thermal ellipsoids of the non-hydrogen atoms are shown with 30% probability. The oxygen atoms of the side chains occupy two alternate positions; the ones with the higher occupancy factor (0.83) are shown. (b) ORTEP view along the crystallographic a axis without solvent molecules and anions. (c) Crystal packing (CPK coloring scheme).
Although the anti-parallel conformation of 2,7-BisNP in the solid state does not feature a confined space between the naphthalene units, it may be expected that in solution the macrocycle is more dynamic giving rise to numerous possible conformers. Indeed, a molecular dynamics study of 2,7-BisNP in a water box identified several conformers differing mainly by a more or less open orientation of their naphthalene units (48). In addition, in a previous X-ray structural study, the amphi isomer (2,6-BisNP) was found to adopt a semi-closed conformation with a parallel orientation of the two aromatic units upon binding of an aromatic dicarboxylate guest (51). It is therefore clear that the conformation of the macrocycle, free or in a complex, is influenced both by the attachment position of the two linkers on the aromatic moieties (48) and by the nature of the guest trapped inside the macrocyclic scaffold, which points out to the ability of this class of macrocycles to adapt to a large variety of guests.
DNA melting experiments
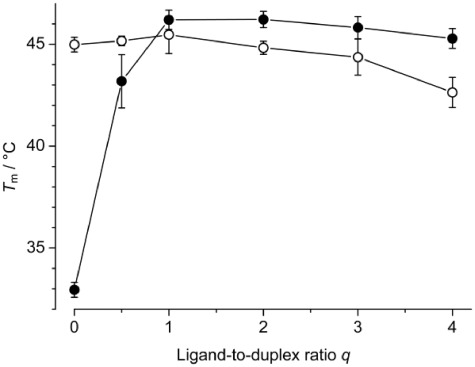
To confirm the selective binding of 2,7-BisNP to the 11-mer TT-DNA and get a rough idea of the binding stoichiometry and affinity, thermal denaturation experiments were performed at variable ligand-to-duplex ratios. In conditions comparable to those of the NMR experiments ([Na+] = 60 mM, pH 6), the duplex TT-DNA alone denatured at Tm = 33.0°C. The addition of 2,7-BisNP led to significant stabilization of TT-DNA, as demonstrated by large increase of the melting temperature (Figure 2). The maximal effect was observed at a 1:1 ligand-to-DNA ratio (ΔTm = 13.3°C), whereas at higher ligand loadings, the stabilization effect leveled off and slightly decreased to 12.3°C. This minor decrease is attributable to weak binding of the excess ligand to the single-stranded form, which may slightly counterbalance the strong stabilizing effect induced by duplex binding. At a ligand-to-DNA of 0.5:1, a broad melting curve was observed, indicative of incomplete saturation of the binding site (52).
Figure 2.
Melting temperatures of TT-DNA (filled circles) and TA-DNA (empty cirlces) in the absence and in the presence of 2,7-BisNP (0–4 equivalents). Conditions: cDNA = 2 µM in cacodylate buffer, pH 6.0, [Na+] = 60 mM.
To confirm the selective binding of 2,7-BisNP to mismatched DNA, the same experiment was performed with the fully paired duplex TA-DNA. The denaturation profile of TA-DNA was hardly influenced by the presence of 2,7-BisNP (Figure 2), and only at high ligand loadings, a decrease in melting temperature was observed (ΔTm = −2.4°C at a 4:1 ligand-to-DNA ratio), indicative of weak binding to the single DNA strands as described for analogous macrocycles (53,54). Interestingly, melting temperature of the free duplex TA-DNA (Tm = 45.0°C) is very close, within the experimental error, to the melting temperature of TT-DNA in the presence of one molar equivalent of ligand (46.2°C), indicating that binding of 2,7-BisNP leads to stabilization of the T–T-mismatched duplex to the level of a regular T–A duplex. Altogether, thermal denaturation experiments speak for the preferential binding of the macrocycle at the T–T base pair site.
Structure of TT-DNA in the absence of ligand
Proton and 31P assignments
The base and sugar protons of TT-DNA were identified on the basis of analysis of 2D NOESY, TOCSY and DQFCOSY spectra using the sequential assignment strategy, well described for right-handed DNA duplexes (55). All cross-peak intensities were consistent with a B-like DNA conformation and the bases adopting an anti-conformation. Qualitative analysis of the proton–proton coupling constants in the DQFCOSY spectra (no detectable H3′–H4′ and H2″–H3′ cross-peaks corresponding to very small JH3′–H4′ and JH2″–H3′ coupling constants, but characteristic H1′–H2′ pattern) along with intensities in the NOESY spectra indicate that all the sugars, except C1, C15 and G22, are in a C2′-endo conformation. For these sugars, a P-value in the range 162–180° was used. C1, C15 and G22 showed large JH3′-H4′ coupling constant and small but detectable JH2″–H3′, corresponding to a pseudorotation phase angle P ranging from 90 to 120°. For the others sugars, a P-value in the range 162–180° was used.
Around the mismatched base pair T6–T17, the bases are well-stacked and both T6 and T17 remain inside the helix, as confirmed by the sequential NOE connectivities observed between T6H6 and G5H1′, G5H2′ and G5H2″, A7H8 and T6H1′, T6H2′ and T6H2″, T17H6 and T16H1′, T16H2′ and T16H2″, C18H6 and T17H1′, T17H2′ and T17H2″, as well as characteristic H8/H6 cross-peaks (G5H8/T6H6, T6H6/A7H8, T16H6/T17H6, T17H6/C18H6).
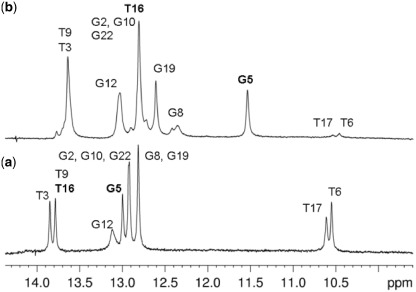
The exchangeable imino protons were assigned with the 1D and 2D NOESY spectrum recorded in 90% H2O at 10 or 5°C. The 1D spectrum shows three main sets of imino proton: thymine imino protons in the range 13.5–14.0 ppm, guanine imino proton in the range 12.8–13.1 ppm and two resonances at 10.56 ppm and 10.61 ppm, corresponding to the mismatched thymines (Figure 3a). It is known that non-hydrogen-bound imino protons, such as those of bulged bases (56), bases facing abasic sites (57), or non-Watson–Crick base pairs like base mismatches (58–62) or wobble base pairs (63,64) resonate in this range. These resonances were assigned to the imino protons of T6 and T17, respectively, based on the cross-peaks observed in the 2D spectra. However, it was not possible to deduce the hydrogen bonding pattern between T6 and T17, as the imino protons of these bases were almost coincident. Nevertheless, we can conclude that the T–T mismatch does not introduce dramatic distortions to the duplex structure, as chemical shifts of the DNA protons are in the common range observed for regular DNA.
Figure 3.
1D imino spectra of (a) free TT-DNA at 10°C and (b) 2,7-BisNP–TT-DNA complex at 5°C. T16 and G5, which undergo most significant shifts, are highlighted in bold.
31P chemical shifts were used to obtain information about the conformation of the phosphodiester backbone, as they are sensitive to the local distortions. The 1D 31P spectrum was recorded at 10°C (Supplementary Figure S1), and the signals were assigned from a 1H-31P HSQC experiment (Supplementary Table S1). The 31P chemical shifts were observed in the range from −3.72 to −5.14 ppm, clearly indicating that all torsion angles are in the BI conformation (65).
Structural modeling of the free TT-DNA
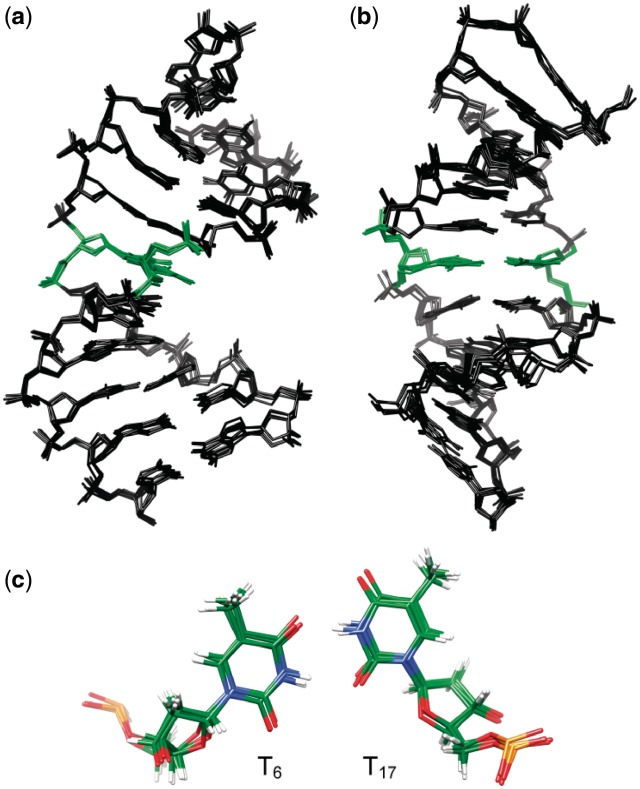
The structural analysis of the free TT-DNA was carried out using a simulated annealing protocol within the AMBER 10.1 program. A set of five random conformations was generated to ensure that each of the starting structures was in different regions of the conformational space and that the final conformation is independent of the initial structure. A 100 ps restrained dynamics in implicit water were then performed followed by 100 ps of restrained dynamics in explicit solvent (see Materials and Methods). An ensemble of the structures obtained over the last 5 ps of the dynamics is shown in Figure 4. The DNA adopts a regular B-DNA-like structure with well-formed Watson–Crick alignments of all canonical base pairs. The bases T6 and T17 remain inside the helix, and the major and minor groove size are normal around the mismatch. Thus, the T–T mismatch does not introduce dramatic distortion to the structure, in accordance with what has been observed in earlier NMR studies (58–62). A good agreement with the NMR experimental restraints was obtained: 13 restraints were violated by 0.2–0.3Å, 14 by 0.3–0.5 Å and four by more than 0.5 Å. The mutual root mean square deviation (rmsd on all atoms except the terminal base pairs) between the last five structures from the dynamic are in the range 0.32–0.39 Å, evidencing no major fluctuations between the structures and a good convergence. This successful convergence to similar conformation is indicative of the performance of the restrained molecular dynamics. Most significant differences are observed at the terminal base pairs and can probably be attributed to end fraying.
Figure 4.
Structure of TT-DNA: superposition of the five last structures emergent from the restraint molecular dynamics viewed (a) from the side and (b) into the major groove. The mismatched thymine residues T6 and T17 are depicted in green, hydrogen atoms are not shown. (c) Close-up down view of the T6–T17 mismatch (O: red, N: blue, H: white, P: orange).
Structure of the 2,7-BisNP–TT-DNA complex
The titration of 2,7-BisNP to TT-DNA was monitored by 1D 1H NMR spectroscopy (Supplementary Figure S2). Upon the addition of the ligand new resonances appeared and increased while the resonances of the free oligonucleotide decreased, disappearing completely when a 1:1 ratio was reached. This indicates a slow exchange regime on NMR timescale. Line broadening was also observed, and some of the aromatic protons of the ligand were significantly shifted upfield. These observations are consistent with a specific binding and insertion of the ligand inside the DNA.
Proton and 31P assignments of the 2,7-BisNP–TT-DNA complex
Most of the non-exchangeable protons of the oligonucleotide were assigned using a combination of 2D NOESY, TOCSY and DQFCOSY. In the NOESY, the observed sequential internucleotide connectivities H6/8 and H1′ as well as H6/8 and H2′/H2″ allowed the assignment of most of the protons. However, this sequential walk was interrupted: no NOEs were observed between G8H8 and A7-sugar protons and between T16H6 and C15-sugar protons, which gives a clear evidence of intercalation of an aromatic unit between the base pairs A7–T16 and G8-C15, as these NOEs were clearly present in the free oligonucleotide. Adenine A7 was difficult to identify, as its signal was not observed in the typical range of chemical shifts normally observed for adenine (downfield), but was significantly shifted upfield (δ = 7.88 ppm) and overlapped with five H8 or H6 base protons. This also undoubtedly points out to binding of the ligand in the neighborhood of A7.
Weak NOE signals were observed between T6H6 and G5-sugar protons, but not between T17H6 and T16-sugar protons. Similarly, no NOE were present between C18H8 and T17-sugar protons. Unfortunately, it was not possible to unambiguously determine if A7H8 gave NOE with T6-sugar proton, as the peak of A7H8 was strongly overlapped with other protons. These observations suggest ejection of the mismatched thymine T17 from the double helix, with the opposite T6 remaining closer to its flanking bases G5 and A7 than T17 does to C18 and T16. The intranucleotide NOEs indicate that even after binding of 2,7-BisNP to DNA, the bases maintain their anti-conformation. The DQFCOSY cross peak pattern suggests that the sugars mainly adopt the C2′-endo puckering, even the thymines T6 and T17, as it was observed for the oligonucleotide in the absence of the ligand.
The imino and amino protons were assigned on the basis of the 2D-NOESY spectrum of the complex recorded in H2O at 10 or 5°C. Usual correlations for Watson–Crick base pair were observed between G-NH imino protons and C-NH2 amino protons and between T-NH and A-H2 protons. Imino–imino NOEs between flanking base pairs were also detected throughout the duplex. G5NH and T16NH were markedly shifted upfield (1.5 ppm and 1.0 ppm, respectively), which gave evidence for intercalation of 2,7-BisNP at the G5pT6 and C15pT16 steps. The T6NH and T17NH were easily detected in the 1D spectrum as broad signals at 10.49 and 10.56 ppm (Figure 3b), but could not be unambiguously assigned even in the 2D spectra due to the absence of correlation peaks. This is consistent with the fact that they are extruded from the duplex and more exposed to the solvent.
The protons of 2,7-BisNP were assigned following the analysis of the NOESY, TOCSY and DQFCOSY datasets in D2O solution. The aromatic region of the DQFCOSY spectrum displayed four sets of vicinal coupling connectivities corresponding to H1/H2, H3/H4 and H7/H8, H9/H10. The protons H1, H4, H7 and H10 were identified on the basis of their long-range coupling constant with H5, H6, H11 and H12, respectively, and their NOE with the methylene proton of side chains (H19, H13, H24 and H18). The other methylene protons of the linker chains were identified by combined use of TOCSY and NOESY cross-peak. Unfortunately, unambiguous assignment of all the protons of the side chains was not possible due to broad signals and severe overlaps. Upon binding to the DNA, the methylene protons become non-equivalent and have different, but very similar, chemicals shifts leading to numerous overlapping signals which were difficult to assign.
Finally, the 31P spectra of the 2,7-BisNP–TT-DNA complex (Supplementary Figure S1) show similar chemical shift dispersion compared to the free TT-DNA, indicating no major backbone deformation upon binding of the ligand.
Chemical shift mapping upon binding of 2,7-BisNP to TT-DNA
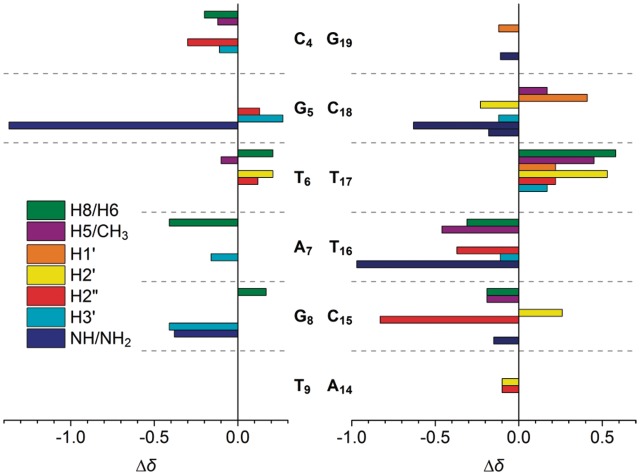
Changes in the chemical shift upon binding represent a valuable tool for the localization of the binding site of the ligand. The interaction of 2,7-BisNP with the TT-DNA duplex resulted in line broadening and significant variation of chemical shifts relative to the free DNA (Figure 5). As can be seen, the central part of the sequence T6pA7pG8 previously identified as the possible binding site undergoes the most pronounced changes. Indeed, most of the protons of the base pairs, which flank the T6–T17 mismatch are shifted upfield due to the intercalation of the ligand and the associated π-stacking effect. For example, A7H8 is shifted upfield by 0.41 ppm, T16H6 is shifted by 0.31 ppm, T6CH3 is shifted by 0.46 ppm and almost all the protons of C15 are shifted upfield. The most striking effect is observed for the exchangeable protons: G5NH is shifted by −1.37 ppm, T16NH by −0.97 ppm, G8NH by −0.38 ppm and C18NH2 protons are shifted by −0.63 and −0.18 ppm. This provides a strong evidence of the insertion of the ligand into DNA. The large shift experienced by G5NH and, to a lesser extent, by C18NH2 together with the shift of the non-exchangeable protons of A7–T16 pair clearly reveals the position of one of the aromatic ring of 2,7-BisNP inside the helix in the plane of T6–T17. Indeed, such shift would be inconsistent with an intercalation occurring between the base pairs T6–T17 and A7–T16.
Figure 5.
Changes in 1H chemical shifts of TT-DNA (Δδ = δbound − δfree) upon binding of 2,7-BisNP. Only significant changes (|Δδ| ≥ 0.10 ppm) are shown.
The signals of some protons were, instead, shifted downfield. Notably, most protons of the mismatched thymines T6 and especially T17 exhibited higher chemical shifts compared with the free TT-DNA, providing evidence of their ejection from the base stack. No significant changes (|Δδ| ≥ 0.10 ppm) were detected for the base pairs other than C4–G19, G5–C18, T6–T17, A7–T16, G8–C15 and T9–A14, which speaks for the localization of the binding site in the central part of the duplex. The signals of the protons of 2,7-BisNP also undergo significant upfield shifts (Δδ up to −1.36 ppm; Table 1) which are characteristic of its intercalation into the DNA.
Table 1.
1H chemical shifts (ppm) of 2,7-BisNP in the free form and in the 2,7-BisNP–TT-DNA complex and observed intermolecular NOEsa
| Protonb | δfree | δbound | Δδc | 2,7-BisNP–TT-DNA NOEs |
|---|---|---|---|---|
| H1 | 7.43 | 6.86 | −0.57 | T16H1′ (m), T16CH3 (w), T17H1′ (m), T17H2′ (m), T17H2″ (m) |
| H2 | 7.81 | 6.72 | −1.09 | T16H1′ (w), T16CH3 (m), T17H1′ (s), T17H2′ (s), T17H2″ (m) |
| H3 | 7.81 | 7.15 | −0.66 | |
| H4 | 7.43 | 7.25 | −0.18 | |
| H5 | 7.78 | 6.65 | −1.19 | G5NH1 (m), C18H5 (w) |
| H6 | 7.78 | 6.59 | −1.13 | G5NH1 (m) |
| H7 | 7.43 | 6.59 | −0.84 | T16H1′ (s), T16H2′ (s), T16H2″ (s), T16CH3 (m) |
| H8 | 7.81 | 6.67 | −1.14 | T16H1′ (m), T16H2′ (m), T16H2″ (m), T16CH3 (m) |
| H9 | 7.81 | 7.15 | −0.66 | |
| H10 | 7.43 | 7.12 | −0.31 | |
| H11 | 7.78 | 6.86 | −0.92 | |
| H12 | 7.78 | 6.42 | −1.36 | |
| CH2-13 | 4.22 | 4.07/4.16 | −0.15/−0.06 | |
| CH2-14 | 3.40 | NA | NA | |
| CH2-15 | 3.88 | 3.44/3.42 | −0.44/−0.46 | |
| CH2-16 | 3.88 | NA | NA | |
| CH2-17 | 3.40 | NA | NA | |
| CH2-18 | 4.22 | 4.07 | −0.15 | T16CH3 (w) |
| CH2-19 | 4.22 | 3.93 | −0.29 | |
| CH2-20 | 3.40 | NA | NA | |
| CH2-21 | 3.88 | NA | NA | |
| CH2-22 | 3.88 | 3.22 | −0.66 | |
| CH2-23 | 3.40 | 3.43 | 0.03 | |
| CH2-24 | 4.22 | 3.89/4.07 | −0.33/−0.15 | A7H1′ (m) |
aChemical shifts at 10°C.
bFor the (arbitrary) proton numbering of 2,7-BisNP, see Scheme 1.
cΔδ = δbound − δfree.
s, strong; m, medium; w, weak; NA, Not assigned.
(3) Intermolecular NOEs between 2,7-BisNP and TT-DNA
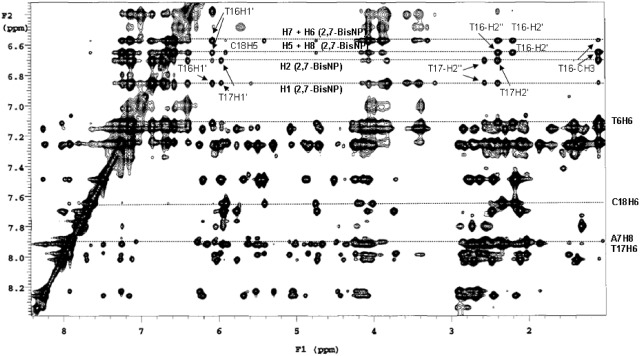
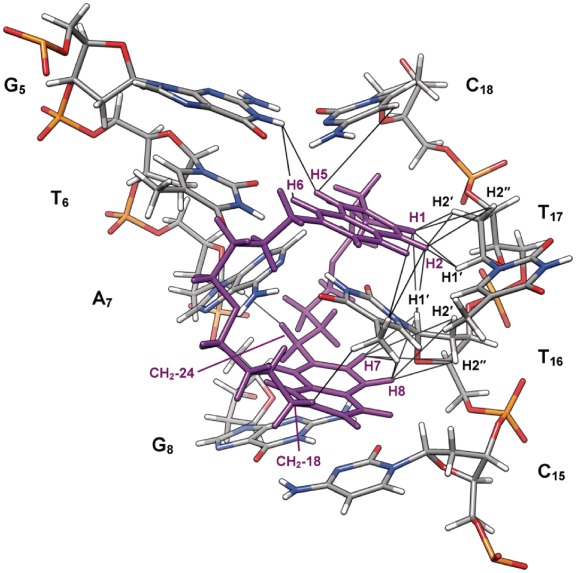
The assignment of the ligand and DNA protons provided the basis for identification of the ligand–DNA contacts, which gave insight into the mode of binding of 2,7-BisNP. Twenty-three specific intermolecular ligand–TT-DNA contacts (Table 1) were detected in the 2D NOESY spectrum (Figure 6), which allowed, in combination with the observations described previously, to identify more precisely the location of the binding site and the binding mode. Numerous NOE contacts were observed between the two aromatics cores of 2,7-BisNP and the T16 base, consistent with the ‘sandwiching’ of T16 between the aromatic planes of the ligand (Figure 7). Some NOEs were also detected between one naphthalene ring (protons H1 and H2) and the sugar protons of T17. NOEs between A7H1′and the side-chain protons CH2-24 indicate that this chain lies in the minor groove. On the contrary, the NOE observed with the other side chain, namely between CH2-18 and T16CH3, requires the second chain to be placed in the major groove. This is only possible if the binding of 2,7-BisNP occurs via a threading intercalation mode, with the linking chains occupying both the minor and the major grooves. The NOE cross-peaks between H5 and H6 of the ligand and the imino proton of the guanine (G5NH1) as well as between H5 and the cytosine (C18H5) flanking the mismatch site give another evidence of insertion of the ligand at the level of T6–T17 base pair.
Figure 6.
Expansion of the 250 ms NOESY spectrum of the 2,7-BisNP–TT-DNA complex in D2O at 10°C. Correlations are between the aromatic protons and the sugar protons. Selected intermolecular 2,7-BisNP–TT-DNA cross-peaks are indicated.
Figure 7.
Structure of the complex of 2,7-BisNP (purple) with TT-DNA (CPK colors) from NMR-restrained molecular dynamics calculations showing the binding site and the NOEs observed in NMR spectra (black lines).
Structure of the 2,7-BisNP–TT-DNA complex from restrained molecular dynamics calculations
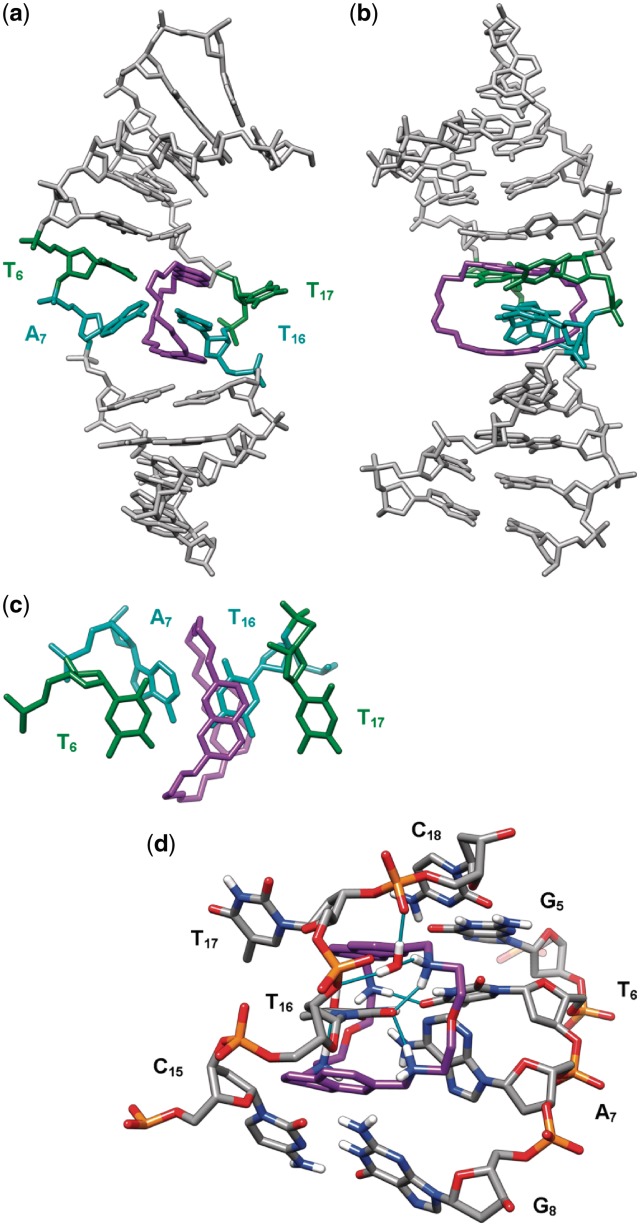
The 23 intermolecular distance restraints determined from the NOESY spectra were used for molecular dynamics calculation in order to obtain a model of 2,7-BisNP binding to TT-DNA that would agree with NMR data. The structure obtained after 40 ps of dynamics in explicit water is displayed in Figures 7 and 8. It shows the threading of the macrocyclic ligand through TT-DNA and the specific binding around the central thymine–thymine mismatch site. One of the naphthalene rings of 2,7-BisNP faces T6 and occupies the place of T17, which is extruded toward the major groove. This naphthalene is almost perpendicular to the main axis of A7–T16 base pair, which allows a good stacking with T16, but not with A7 (Figure 8c). The T6 remains stacked with the flanking G5 residue. The sandwiched base pair A7–T16 forms a buckle of around 38° to allow the ligand to snugly fit inside the helix. The second naphthalene ring of 2,7-BisNP also stacks with T16, leading to the ‘sandwiching’ of the latter in the cavity delineated by the two naphthalene units. The side chains of 2,7-BisNP are located in the middle of each groove. Stabilization of the complex occurs, in addition to the aforementioned π–stacking interactions, due to formation of the hydrogen bonds (Figure 8d) between the protonated amino groups of the ligand and carbonyl groups of thymines T6 and T16 (C13-NH2+ and T6O4, C19-NH2+ and T16O2, C24-NH2+ and T16O2). In particular, the hydrogen bond with T6 may explain the specificity of the ligand for T-X mismatch, while the two hydrogen bonds with the ‘encapsulated’ thymine T16 largely contribute to the stabilization of the complex. In addition, four hydrogen bonds between the positively charged amino groups and water molecules were detected (66). One of the water molecules mediates the bonding between C18–NH2+ and T16O4, while the other one mediates the interaction between C19–NH2 and two phosphate groups (pT16p).
Figure 8.
Threading intercalation binding mode of 2,7-BisNP (purple) to TT-DNA from NMR-restrained molecular dynamics calculations, viewed (a) from the major groove and (b) from the side of the flipped-out T17. (c) Top view of the naphthalene rings of 2,7-BisNP stacking with the encapsulated thymine T16. (d) Close-up view of the binding site (view from the minor groove, only heteroatom-bound hydrogen atoms are shown) showing direct and water-mediated hydrogen bonds (cyan sticks) between 2,7-BisNP and TT-DNA.
DISCUSSION
Thermal denaturation data strongly suggest the formation of a complex with a 1:1 stoichiometry upon binding of the bis-naphthalene macrocycle to the 11-mer duplex containing a TT mismatch. Accordingly in the NMR-monitored titration, clean formation of the ligand–DNA complex was observed at a 1:1 ratio and no excess of the ligand was necessary. Finally, 1D and 2D NMR data give evidence that only one type of complex between 2,7-BisNP and TT-DNA is formed.
The binding model, deduced from the numerous ligand–DNA NOE interactions, indicates that the macrocycle is located inside the double helix and sandwiches the thymine T16 of the A–T base pair in the vicinity of the T6–T17 mismatched base pair. Presumably, the weaker stability of the A7–T16 base pair compared with the G5–C18 pair which flanks the T–T mismatch on the other side, leads to formation of a single complex. It is likely that the hydrogen bonding between A7 and T16 is weakened due to the strong kink (38°) observed between the planes of these bases. The two polyammonium linkers of the macrocyclic scaffold lie both in the minor and the major grooves in a threading mode. In addition, the binding features two hydrogen bonds with the encapsulated thymine (T16O2) and one hydrogen bond with one of the mismatched thymines (T6O4). Both mismatched thymines are strongly displaced as compared to their position in the free duplex, especially T17 which is flipped out of the duplex toward the major groove. Thus, binding of the macrocycle induces a local distortion of the mismatch site, and interactions are observed over the four base pairs from C5–G18 to G8–C15. Altogether, the ligand–DNA complex results from a complex array of multiple interactions involving the mismatched base pair and the three base pairs in the vicinity, which is based on hydrogen bonding, electrostatic interactions and π–stacking. It is clear that the macrocycle takes benefit of the local thermodynamic instability of the mismatch site and of the neighboring base pairs to enter inside the DNA and accommodate a thymine ring in its cavity. The binding of the ligand necessitates firstly the disruption of the Watson–Crick base pairing between A7 and T16 and of the wobble pairing between T6 and T17. These events should be energetically compensated by the final stability of the complex, as evidenced by the large increase in melting temperature. In this regard, the three hydrogen bonds established between the ammonium groups of the ligand linkers and the carbonyl groups of the two thymine bases (T6 and T16) should provide significant contribution. Additionally, partial restoration of hydrogen bonding between A7 and T16, which is likely (although not evidenced by NMR), may contribute to stabilization of the complex. Another important contribution is due to strong electrostatic interaction of the highly charged (+4) ligand and DNA phosphate backbone, which is highly favorable since both ligand linkers occupy the grooves of the DNA. We can also anticipate that the entropic factors also largely participate to the stabilization effect, since, first, binding of the ligand leads to release of water molecules from the DNA grooves and from the solvation sphere of the ligand and, second, the ligand is preorganized due to its macrocyclic nature, which means low entropy cost for adaptation to the binding site.
The cyclobisintercalation or pseudocatenation binding mode observed in the present study is remarkable in many aspects.
This very particular and unusual DNA binding behavior is evidenced here for the second time with a CBI macrocycle and a DNA duplex containing a single-base pairing defect (abasic site or a mismatch). Indeed, in a previous study, we have shown that a macrocyclic bis-acridine BisA binds via a similar threading double intercalation mode to an apurinic site with encapsulation of a pyrimidine base (cytosine) of the G–C base pair flanking the lesion (45). However, in that case, two complexes in an 85:15 ratio were observed: in the major complex, both acridine units were intercalated, sandwiching the G–C base pair flanking the lesion, whereas in the minor complex only one acridine was inserted in the abasic pocket, the location of the second one being not defined. This is different in the present study since the asymmetry of the base pairs flanking the mismatch site (A–T versus G–C) led to formation of only one complex, through the encapsulation of the more labile A7–T16 base pair. Thus, this very particular binding mode involving double threading of the double helix at a base pairing defect with encapsulation of the neighboring base pair seems to be a unique feature of the CBI macrocycles. Nonetheless, it should be noted that threading bisintercalation has also been characterized in the case of a macrocyclic bis-naphthalenediimide (67) or proposed in the case of a macrocyclic bis-acridine (68,69). However, both derivatives were built with two long linker chains (> 10 Å) and were found to bind to regular double-stranded DNA with spanning over 2–4 bp. This binding behavior is in stark contrast with that of our macrocyclic bis-naphthalene that does not bind fully paired duplex DNA.
As we anticipated (46–48), the characteristic DNA binding behavior of cyclobisintercalators studied by our group is obviously determined by the short length of the polyammonium linkers that imposes a constrained and short distance between the two aromatic moieties. Indeed, an interchromophoric distance of ∼6.7–6.8 Å can be measured for 2,7-BisNP bound to the T-T mismatched duplex. On the one hand, this short distance prevents intercalation at contiguous Watson–Crick base pairs for steric reasons (neighbor-exclusion principle) (49); on the other hand, it is ideally suited for the encapsulation of aromatic guests, particularly nucleosides and nucleotides (70). This structural particularity governs both the binding preference for the T–T mismatch over the regular duplex and the sandwiching of the neighboring pyrimidine base (T16) by π-stacking interactions. Interestingly, the short distance between the two naphthalene planes is roughly the same in the solid state (6.6–6.7 Å). Conversely, the X-ray analysis indicates a Z-shaped conformation with antiparallel orientation of the two aromatic units, which differs from the almost parallel orientation of these units (U-shaped conformation) in the DNA-bound macrocycle as shown by the NMR analysis. This indicates a certain degree of conformational flexibility of the macrocyclic scaffold, resting mainly on the free rotation about the Car−C and C−N bonds between the aromatic units and the spacers, and is consistent with previous data obtained from the molecular dynamics study of the free 2,7-BisNP in a water box (48). This is an evidence of the conformational adaptability of the bis-naphthalene macrocycle that enables fitting with its DNA target and is likely a key factor of the binding interaction.
Finally, the observed binding mode sheds light on the high structural deformability (plasticity) of duplex DNA which, albeit well-known, is often underestimated when designing DNA-interactive agents. In this regard, a comparison with the two other classes of mismatch-binding agents, i.e. naphthyridine derivatives (31) and bulky metal complexes (38–40), shows that in all cases binding proceeds via ejection of one of the mismatched bases, as in the present study. Thus, all three families of ligands take advantage of the local thermodynamic instability of the mismatch sites, compared to Watson–Crick base pairs. However, the uniqueness of the CBI series is represented by their two-point binding mode (mismatched base pair and intercalation pocket at the neighboring base pair step) and their conformational restrictions, which do not allow binding to fully paired DNA regions. In summary, the observed threading interaction is thus the result of a subtle interplay between the conformational flexibility of the macrocyclic ligand and the local dynamics of the DNA mismatched site.
CONCLUSIONS
In the present study, we characterized the structural details of a bis-naphthalene macrocycle bound to a T–T DNA mismatch via a rare cyclobisintercalation binding mode, which involves encapsulation of the neighboring thymine base between the aromatic planes of the macrocycle and ejection of one of the mispaired thymines. This binding mode is unique since it provides high specificity for DNA base pairing defects with respect to regularly paired DNA, and allows recognition of unpaired thymine due to the hydrogen bonding pattern of the 2,7-BisNP ligand. Interestingly, the structure of the ligand–DNA complex obtained in the present study shows that the macrocyclic scaffold of the ligand allows the introduction of additional chemical functionalities, either at the linking chains or in the positions of the naphthalene units which are not in close contact with the DNA, without perturbing the binding specificity. This might enable us to endow the macrocycle with reporter groups (fluorescent, redox units) (71) for detection of the pairing defect. It may be expected that this pseudocatenated interaction can result in a slow dissociation of the ligand–DNA complex, which may dramatically increase the residence time of the macrocycle inside the DNA, as compared with classical intercalators or minor groove binders and interfere with the processing of mismatch repair by enzymes. Altogether, the present work highlights that bis-naphthalene 2,7-BisNP and, more largely, bisintercalators of the CBI type hold a great potential for therapeutic and diagnostic applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–3.
FUNDING
The Centre national de la recherche scientifique [TGE RMN THC Fr3050 (NMR studies at 800 MHz) and the interdisciplinary action ‘Interface physique, biologie et chimie: soutien à la prise de risque’ (PIR051 ‘DNA Lesion Binders’)]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marie-Louise Dheu-Andries and Sebastien Morin for their help with the AMBER 10 program, Dr. Adrien Favier for his help with NMR experiments, and Dr. Hélène Jamet (UJF Grenoble) for Gaussian calculations. Prof. Elmar Weinhold (RWTH Aachen) is greatly acknowledged for helpful discussions. The figures in this manuscript were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
REFERENCES
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukui K. DNA mismatch repair in eukaryotes and bacteria. J. Nucleic Acids. 2010 doi: 10.4061/2010/260512. Article ID 260512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdine GL, Bruner SD. How do DNA repair proteins locate damaged bases in the genome? Chem. Biol. 1997;4:329–334. doi: 10.1016/s1074-5521(97)90123-x. [DOI] [PubMed] [Google Scholar]
- 5.Rajski SR, Jackson BA, Barton JK. DNA repair: models for damage and mismatch recognition. Mutat. Res. 2000;447:49–72. doi: 10.1016/s0027-5107(99)00195-5. [DOI] [PubMed] [Google Scholar]
- 6.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 7.Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 9.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 11.Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med. Chem. 2008;8:417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Hu J, Hu Y, Liu W. Targeting DNA repair pathways: a novel approach to reduce cancer therapeutic resistance. Cancer Treat. Rev. 2009;35:590–596. doi: 10.1016/j.ctrv.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen TJ. Enhancing radiosensitivity: targeting the DNA repair pathways. Cancer Biol. Ther. 2009;8:665–670. doi: 10.4161/cbt.8.8.8304. [DOI] [PubMed] [Google Scholar]
- 14.Tell G, Wilson DM., III Targeting DNA repair proteins for cancer treatment. Cell. Mol. Life Sci. 2010;67:3569–3572. doi: 10.1007/s00018-010-0484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Bapat A, Glass LS, Luo M, Fishel ML, Long EC, Georgiadis MM, Kelley MR. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J. Pharmacol. Exp. Ther. 2010;334:988–998. doi: 10.1124/jpet.110.169128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiple LA, Cardellina JH, II, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol. Pharmacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM., III Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS One. 2009;4:e5740. doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson DM, III, Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci. 2010;67:3621–3631. doi: 10.1007/s00018-010-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol. Med. 2005;11:456–463. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J. Med. Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 23.Underhill C, Toulmonde M, Bonnefoi H. A review of PARP inhibitors: from bench to bedside. Ann. Oncol. 2011;22:268–279. doi: 10.1093/annonc/mdq322. [DOI] [PubMed] [Google Scholar]
- 24.Jiang YL, Ichikawa Y, Stivers JT. Inhibition of uracil DNA glycosylase by an oxacarbenium ion mimic. Biochemistry. 2002;41:7116–7124. doi: 10.1021/bi025694y. [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Stivers JT, Greenberg MM. Competitive inhibition of uracil DNA glycosylase by a modified nucleotide whose triphosphate is a substrate for DNA polymerase. J. Am. Chem. Soc. 2009;131:1344–1345. doi: 10.1021/ja807705z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatani K, Sando S, Kumasawa H, Kikuchi J, Saito I. Recognition of guanine-guanine mismatches by the dimeric form of 2-amino-1,8-naphthyridine. J. Am. Chem. Soc. 2001;123:12650–12657. doi: 10.1021/ja0109186. [DOI] [PubMed] [Google Scholar]
- 27.Nakatani K, Sando S, Saito I. Scanning of guanine-guanine mismatches in DNA by synthetic ligands using surface plasmon resonance. Nat. Biotechnol. 2001;19:51–55. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]
- 28.Kobori A, Horie S, Suda H, Saito I, Nakatani K. The SPR sensor detecting cytosine[bond]cytosine mismatches. J. Am. Chem. Soc. 2004;126:557–562. doi: 10.1021/ja037947w. [DOI] [PubMed] [Google Scholar]
- 29.Hagihara S, Kumasawa H, Goto Y, Hayashi G, Kobori A, Saito I, Nakatani K. Detection of guanine-adenine mismatches by surface plasmon resonance sensor carrying naphthyridine-azaquinolone hybrid on the surface. Nucleic Acids Res. 2004;32:278–286. doi: 10.1093/nar/gkh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatani K. Recognition of mismatched base pairs in DNA. Bull. Chem. Soc. Jpn. 2009;82:1055–1069. [Google Scholar]
- 31.Nakatani K, Hagihara S, Goto Y, Kobori A, Hagihara M, Hayashi G, Kyo M, Nomura M, Mishima M, Kojima C. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat. Chem. Biol. 2005;1:39–43. doi: 10.1038/nchembio708. [DOI] [PubMed] [Google Scholar]
- 32.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J. Am. Chem. Soc. 1997;119:12986–12987. [Google Scholar]
- 33.Jackson BA, Barton JK. Recognition of base mismatches in DNA by 5,6-chrysenequinone diimine complexes of rhodium(iii): a proposed mechanism for preferential binding in destabilized regions of the double helix. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 34.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. A rhodium(iii) complex for high-affinity DNA base-pair mismatch recognition. Proc. Natl Acad. Sci. USA. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeglis BM, Pierre VC, Barton JK. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Yang P, Yuan C, Pu X. Study on the binding of base-mismatched oligonucleotide d(GCGAGC)2 by cobalt(iii) Complexes. Eur. J. Inorg. Chem. 2005:3141–3148. [Google Scholar]
- 37.Chen H, Dou C, Wu Y, Li H, Xi X, Yang P. Structure-specific binding of [Co(phen)2(HPIP)]3+ to a DNA duplex containing sheared G:A mismatch base pairs. J. Inorg. Biochem. 2009;103:827–832. doi: 10.1016/j.jinorgbio.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl Acad. Sci. USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode. Biochemistry. 2009;48:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordier C, Pierre VC, Barton JK. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: an NMR solution study. J. Am. Chem. Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. DNA mismatch-specific targeting and hypersensitivity of mismatch-repair-deficient cells to bulky rhodium(iii) intercalators. Proc. Natl Acad. Sci. USA. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernst RJ, Song H, Barton JK. DNA mismatch binding and antiproliferative activity of rhodium metalloinsertors. J. Am. Chem. Soc. 2009;131:2359–2366. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teulade-Fichou M-P, Vigneron J-P. Interactions of macrocyclic compounds with nucleic acids. In: Demeunynck M, Bailly C, Wilson WD, editors. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes. Weinheim: Wiley-VCH; 2003. pp. 278–314. [Google Scholar]
- 44.Berthet N, Michon J, Lhomme J, Teulade-Fichou MP, Vigneron J-P, Lehn J-M. Recognition of abasic sites in DNA by a cyclobisacridine molecule. Chem. Eur. J. 1999;5:3625–3630. [Google Scholar]
- 45.Jourdan M, Garcia J, Lhomme J, Teulade-Fichou M-P, Vigneron J-P, Lehn JM. Threading bis-intercalation of a macrocyclic bisacridine at abasic sites in DNA: nuclear magnetic resonance and molecular modeling study. Biochemistry. 1999;38:14205–14213. doi: 10.1021/bi991111h. [DOI] [PubMed] [Google Scholar]
- 46.David A, Bleimling N, Beuck C, Lehn JM, Weinhold E, Teulade-Fichou M-P. DNA mismatch-specific base flipping by a bisacridine macrocycle. ChemBioChem. 2003;4: 1326–1331. doi: 10.1002/cbic.200300693. [DOI] [PubMed] [Google Scholar]
- 47.Bahr M, Gabelica V, Granzhan A, Teulade-Fichou MP, Weinhold E. Selective recognition of pyrimidine-pyrimidine DNA mismatches by distance-constrained macrocyclic bis-intercalators. Nucleic Acids Res. 2008;36:5000–5012. doi: 10.1093/nar/gkn392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Granzhan A, Largy E, Saettel N, Teulade-Fichou M-P. Macrocyclic DNA-mismatch-binding ligands: structural determinants of selectivity. Chemistry. 2010;16:878–889. doi: 10.1002/chem.200901989. [DOI] [PubMed] [Google Scholar]
- 49.Crothers DM. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968;6:575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- 50.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 51.Paris T, Vigneron J-P, Lehn J-M, Cesario M, Guilheim J, Pascard C. Molecular recognition of anionic substrates. Crystal structures of the supramolecular inclusion complexes of terephthalate and isophthalate dianions with a bis-intercaland receptor molecule. J. Inclusion Phenom. Macrocyclic Chem. 1999;33:191–202. [Google Scholar]
- 52.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 53.Slama-Schwok A, Peronnet F, Hantz-Brachet E, Taillandier E, Teulade-Fichou MP, Vigneron JP, Best-Belpomme M, Lehn J-M. A macrocyclic bis-acridine shifts the equilibrium from duplexes towards DNA hairpins. Nucleic Acids Res. 1997;25:2574–2581. doi: 10.1093/nar/25.13.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teulade-Fichou MP, Fauquet M, Baudoin O, Vigneron JP, Lehn J-M. DNA double helix destabilizing properties of cyclobisintercaland compounds and competition with a single strand binding protein. Bioorg. Med. Chem. 2000;8:215–222. doi: 10.1016/s0968-0896(99)00283-7. [DOI] [PubMed] [Google Scholar]
- 55.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 56.Kalnik MW, Norman DG, Li BF, Swann PF, Patel DJ. Conformational transitions in thymidine bulge-containing deoxytridecanucleotide duplexes. Role of flanking sequence and temperature in modulating the equilibrium between looped out and stacked thymidine bulge states. J. Biol. Chem. 1990;265:636–647. [PubMed] [Google Scholar]
- 57.Coppel Y, Berthet N, Coulombeau C, Coulombeau C, Garcia J, Lhomme J. Solution conformation of an abasic DNA undecamer duplex d(CGCACXCACGC) × d(GCGTGTGTGCG): the unpaired thymine stacks inside the helix. Biochemistry. 1997;36:4817–4830. doi: 10.1021/bi962677y. [DOI] [PubMed] [Google Scholar]
- 58.Arnold FH, Wolk S, Cruz P, Tinoco I., Jr Structure, dynamics, and thermodynamics of mismatched DNA oligonucleotide duplexes d(CCCAGGG)2 and d(CCCTGGG)2. Biochemistry. 1987;26:4068–4075. doi: 10.1021/bi00387a049. [DOI] [PubMed] [Google Scholar]
- 59.Kouchakdjian M, Li BF, Swann PF, Patel DJ. Pyrimidine · pyrimidine base-pair mismatches in DNA. A nuclear magnetic resonance study of T·T pairing at neutral pH and C·C pairing at acidic pH in dodecanucleotide duplexes. J. Mol. Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 60.Gervais V, Cognet JAH, Le Bret M, Sowers LC, Fazakerley GV. Solution structure of two mismatches A·A and T·T in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Eur. J. Biochem. 1995;228:279–290. [PubMed] [Google Scholar]
- 61.Trotta E, Paci M. Solution structure of DAPI selectively bound in the minor groove of a DNA T.T mismatch-containing site: NMR and molecular dynamics studies. Nucleic Acids Res. 1998;26:4706–4713. doi: 10.1093/nar/26.20.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharya PK, Cha J, Barton JK. 1H NMR determination of base-pair lifetimes in oligonucleotides containing single base mismatches. Nucleic Acids Res. 2002;30:4740–4750. doi: 10.1093/nar/gkf601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel DJ, Kozlowski SA, Marky LA, Rice JA, Broka C, Dallas J, Itakura K, Breslauer KJ. Structure, dynamics, and energetics of deoxyguanosine. thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982;21:437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- 64.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 65.Gorenstein DG. Conformation and dynamics of DNA and protein–DNA complexes by 31P NMR. Chem. Rev. 1994;94:1315–1338. [Google Scholar]
- 66.Mills JE, Dean PM. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput.-Aided Mol. Des. 1996;10:607–622. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 67.Chu Y, Hoffman DW, Iverson BL. A pseudocatenane structure formed between DNA and a cyclic bisintercalator. J. Am. Chem. Soc. 2009;131:3499–3508. doi: 10.1021/ja805676w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmerman SC, Lamberson CR, Cory M, Fairley TA. Topologically constrained bifunctional intercalators: DNA intercalation by a macrocyclic bisacridine. J. Am. Chem. Soc. 1989;111:6805–6809. [Google Scholar]
- 69.Veal JM, Li Y, Zimmerman SC, Lamberson CR, Cory M, Zon G, Wilson DM. Interaction of a macrocyclic bisacridine with DNA. Biochemistry. 1990;29:10918–10927. doi: 10.1021/bi00501a009. [DOI] [PubMed] [Google Scholar]
- 70.Teulade-Fichou M-P, Vigneron J-P, Lehn J-M. Molecular recognition of nucleosides and nucleotides by a water-soluble cyclo-bis-intercaland receptor based on acridine subunits. Supramol. Chem. 1995;5:139–147. [Google Scholar]
- 71.Granzhan A, Teulade-Fichou M-P. Synthesis of mono- and bibrachial naphthalene-based macrocycles with pyrene or ferrocene units for anion detection. Tetrahedron. 2009;65:1349–1360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.