Abstract
While functional roles of several long non-coding RNAs (lncRNAs) have been determined, the molecular mechanisms are not well understood. Here, we report the first experimentally derived secondary structure of a human lncRNA, the steroid receptor RNA activator (SRA), 0.87 kB in size. The SRA RNA is a non-coding RNA that coactivates several human sex hormone receptors and is strongly associated with breast cancer. Coding isoforms of SRA are also expressed to produce proteins, making the SRA gene a unique bifunctional system. Our experimental findings (SHAPE, in-line, DMS and RNase V1 probing) reveal that this lncRNA has a complex structural organization, consisting of four domains, with a variety of secondary structure elements. We examine the coevolution of the SRA gene at the RNA structure and protein structure levels using comparative sequence analysis across vertebrates. Rapid evolutionary stabilization of RNA structure, combined with frame-disrupting mutations in conserved regions, suggests that evolutionary pressure preserves the RNA structural core rather than its translational product. We perform similar experiments on alternatively spliced SRA isoforms to assess their structural features.
INTRODUCTION
Completion of the mouse transcriptome project demonstrated that a large fraction of the mammalian genome is transcribed into RNA molecules with no potential for protein coding (1). In light of their poor evolutionary conservation, these non-coding RNAs (ncRNAs) were initially thought to have little or no functional capability (2). However, studies over the past decade have shown that ncRNAs play critical roles in the cell (3,4). While the functional capabilities of short ncRNAs (e.g. miRNAs and siRNAs) are well understood, long non-coding RNA molecules (lncRNAs) represent a largely unexplored area of the transcriptome. Long ncRNAs often possess cell-specific expression profiles (5), show intracellular localization patterns, and are linked to various diseases including cancer (6). To date, the functional roles of lncRNA transcripts have been uncovered in signaling sensors (7), embryonic stem cell differentiation (8), brain function (5,9), subcellular compartmentalization and chromatin remodeling (10).
While the functional studies of some lncRNAs have been performed (11–14), the molecular basis for function is poorly understood. Two basic mechanistic questions have yet to be answered: (i) Is the functional performance dominated by primary sequence or specific secondary elements (6)? (ii) Does the RNA exist as an intermixed RNA–protein complex [e.g. ribosome (15)], or, is the structure dominated by RNA [e.g. bacterial group II intron (16)]. Because RNA functional performance is typically driven by its secondary and tertiary organization, determining the structural and functional domains of lncRNAs, as well as sequence specific requirements, will lay the foundation for a detailed mechanistic understanding of long non-coding RNAs.
In this study, we report the first experimental characterization of the secondary structure of the entire steroid receptor RNA activator (SRA). This lncRNA is a coactivator for several nuclear receptors (17) and is associated with breast cancer (18–21). Among lncRNAs (200 nt–100 kb), SRA has the advantage of being short enough (∼1 kb) for mechanistic studies, but long enough to encapsulate many of the characteristics of long ncRNAs. While this RNA was originally characterized to act as a regulatory non-coding RNA (17), subsequently, coding isoforms of the SRA RNA were found to exist and code for the SRA protein (SRAP) (22,23).
SRA has been found to participate extensively in nuclear coactivation for many hormone-related systems, including the estrogen receptor (17,24–28), androgen receptor (29), progesterone receptor (17,30), retinoic acid receptor (31), thyroid hormone receptor (32), dosage sensitive sex reversal protein (DAX1) and steroidogenic factor-1 protein (SF1) (33), as well as myogenic differentiation factors (34,35). In addition, it has been recently shown that SRA together with the dead box protein p68 mediates insulation function of the CCCTC-binding factor (CTCF) (36). Coimmunoprecipitation studies place SRA in multicomponent nuclear receptor complexes (17,31,37). Direct binding to the SRA has been observed for the following proteins: pseudouridylases Pus1p and Pus3p (38), RNA helicases p68/p72 (37), nuclear receptor coactivator SRC-1 (17), and nuclear repressors such as SHARP (26) and SLIRP (28). The estrogen-signaling pathway is involved in breast tumorigenesis, as a subset of breast cancer cells express the estrogen receptor and require estrogen for their growth and proliferation. Elevated SRA expression profiles have been shown during tumor progression (18–21). Additionally, the relative ratio of SRA/SRAP expression differs in breast cancer tumor cells, with higher recovery rates for patients for whom SRAP is overexpressed (39). Alterations in SRA expression levels in estrogen-dependent breast tumorigenesis make this RNA a promising new tumor-control target (18–21).
A diverse range of coding and non-coding SRA RNA isoforms is observed in human and mouse organisms. This expression appears to be tissue-specific (17,40). The ratio between coding and non-coding SRA RNA transcripts in human muscle cells has been shown to be important in myogenic differentiation (40). To date, up to 20 SRA transcripts have been identified (40–42). These transcripts primarily differ in (1) their 5′- and 3′-extensions, (2) point mutations or (3) possession of either a full intron or partial portion of the intron. These differences dictate the coding versus non-coding potential of SRA (23,43). For example, possession of the 5′-extension introduces initiation codons for protein synthesis, while the intron insertion causes an amino acid frame-shift or the introduction of premature stop codons that abort protein synthesis (43). With the aim of gaining structural insights into lncRNA function, we performed probing experiments on three reported RNA transcripts, which constitute major representatives of the SRA isoform spectrum in humans: (i) the non-coding isoform (ncSRA), 0.87 kb in length, (ii) the coding isoform (cSRA) with a 5′-extension possessing two start codons, both utilized for the synthesis of 224 and 236 amino acid SRAPs, and (iii) a non-coding isoform of SRA (intSRA) that is an alternatively spliced transcript possessing a partial intron insertion.
In previous in vivo studies, serial deletions of the SRA sequence negatively affected the steroid receptor transcriptional activity. In addition, certain fragments of SRA were shown to lack the functional performance of the full sequence (30). These results suggested that SRA functional capabilities are not limited to one particular substructure, but may require a complex structural organization of the lncRNA. To assess the secondary structure of this RNA, we employ several chemical probing tools along with covariance analysis across multiple species. These techniques have proven to be indispensable in building the initial ribosomal RNA secondary structures, elucidating important structural motifs and secondary structures in many other RNAs, including introns, riboswitches and short non-coding RNAs (44–49). Leontis and coworkers used chemical probing to validate a variety of designed self-assembled RNA nano-objects (49). In this work, we utilize the SHAPE methodology developed by Weeks and coworkers that has been recently used to map the secondary structure of the HIV genome (50). In addition, we support these investigations with DMS probing, in-line (51) and RNase V1 digestions. We present the experimentally derived secondary structure model of the lncRNA, describe its structural architecture and discuss its evolution.
MATERIALS AND METHODS
RNA synthesis
Double-stranded DNA templates for RNA isoform synthesis were generated using multiple cycles of PCR from smaller DNA fragments (∼ultramers of 150–200 nt in size, IDTDNA). Five to seven DNA pieces, depending on the desired RNA, were engineered to have overlapping regions. Two DNA fragments were first annealed and extended by Taq polymerase to generate pre-dsDNA templates. This was followed by the addition of a third fragment, accompanied by additional rounds of PCR to obtain a larger dsDNA template. This stepwise protocol was continued until all DNA pieces have been utilized. Additional PCR reactions were performed to (i) amplify the final dsDNA product using reverse and forward primers and (ii) incorporate the T7 promoter region. These templates were used in run-off transcription using a high yield AmpliScribe T7 synthesis kit from Epicentre Biotechnologies. The RNA products were extracted with phenol–chlorophorm and further precipitated with the addition of one volume of 5 M ammonium acetate. The integrity of RNA was checked on agarose and polyacrylamide gels.
Chemical probing
Prior to probing, RNA was denatured in water at 94°C for 2 min and snap-cooled on ice. Folding was carried out in 1 × HMK buffer (50 mM HEPES–NaOH pH 8.0, 100 mM KCl, 6 mM MgCl2,) for 30 min at 37°C. An exception in the folding protocol was made for in-line probing, which is described below. In all probing reactions, the final concentration of RNA was adjusted to 250 nM.
SHAPE
SHAPE probing was performed as recommended by Weeks and coworkers (52) using the fast-acting 1M7 reagent (53). 1M7 was synthesized from 4-nitroisatoic anhydride using a protocol developed by Mortimer and Weeks (53). Folded RNA was adjusted with 1M7 (dissolved in DMSO) to a final concentration of 3 mM and incubated at 25°C for 5 min. Parallel RNA samples were treated with the same amount of pure DMSO to obtain the blank. Modified RNAs were collected using the standard sodium acetate/ethanol precipitation technique.
DMS
An amount of one-twentieth volume of 10% DMS in ethanol (or pure ethanol for blank trace) was added to the folded RNA followed by incubation for 1 h on ice. The reactions were quenched by the addition of one volume of stop solution (1 M Tris–HCl pH 8.0; 1 M B-mercaptoethanol, 1 M sodium acetate). To precipitate the alkylated RNAs, 2.5 volumes of ethanol were added to the mixture followed by incubation at −80°C and centrifugation.
In-line
In-line probing reactions were performed as described previously (51). After the RNA denaturation step, they were carried out in a 1× in-line probing buffer (50 mM Tris–HCl pH 8.3; 20 mM MgCl2, 100 mM KCl) for 46 h at 25°C. The products of the in-line cleavage were precipitated using the sodium acetate/ethanol procedure.
RNase V1
Serial dilutions of RNase V1 (Ambion) were tested to optimize the conditions of the cleavage. The optimal cleavage pattern was obtained with the final amount of 0.000125 U/µl of RNase V1. The digestion reactions were carried out for 20 min at 25°C, followed by the addition of the precipitation/inactivation buffer (supplied with the enzyme) as outlined in the manufacture’s protocol.
Analysis of chemical probing reactions
Reverse transcription/primer design
The modification/cleavage sites of the RNA were analyzed by primer extension of fluorophore-labeled primers with the SuperScript III reverse transcriptase from Invitrogen. DNA primers of 25 nt were designed to target the regions of SRA separated by ∼150–200 nt. Fluorophore-labeling of primers was achieved using DNA oligos synthesized with an amino moiety on their 5′-end (IDTDNA) and Alexa Fluor 488 amine reactive ester purchased from Invitrogen. The fluorophore-labeled primers were further purified on reverse phase HPLC. Reverse transcription reactions were performed in the following manner: 6 pmol of RNA (2 µl) were mixed with 2 pmol of site-specific primer (1 µl), 1 µl of water and 1 µl of dNTP mix (2.5 mM). The mixture was heated up for 5 min at 65°C and placed on ice. This was followed by the addition of 2 µl of 4× Reverse Transcription buffer and 1 µl of Superscript III (200 U/µl). The 4× Reverse Transcription buffer was prepared by combining four parts of 5× First Strand buffer and one part of 0.1 M DTT supplied with the enzyme. The mixture was incubated for 1 h at 55°C followed by additional 15 min at 70°C for enzyme inactivation. The mixture was diluted with water to a final volume of 40 µl and desalted with micro Bio-Spin columns filled with Bio-gel P6 (Bio-Rad Life Science). One quarter of the mixture (10 µl) was dried under the vacuum and resuspended in 20 µl of deionized formamide. Two dideoxy sequencing reactions were performed in parallel (A-sequencing and C-sequencing). The reverse transcription protocol is similar as outlined above except that the 1 µl of water was substituted with 1 µl of 1 mM ddNTP (ddTTP for A-sequencing and ddGTP for C-sequencing).
Capillary electrophoresis and trace processing
The products of reverse transcription were resolved by capillary electrophoresis on an ABI PRISM 3100-Avant genetic analyzer. Prior to loading, the samples were denatured for 3–4 min at 95°C. The sequencing and probing primer extension reactions were run on either 50 or 80 cm capillaries loaded with POP-6 polymer. Traces were manually aligned and then Gaussian integrated. Probing reactivity traces were further corrected for exponential decay using the statistical model implemented in the ShapeFinder software package (54). Reverse transcription stops observed in the blank traces were subtracted from the probing traces. The SHAPE traces were further normalized by the average reactivities for highly reactive nucleotides such that the range extends from 0 to 1.5, as recommended by Vasa and coworkers (54). In-line, DMS and RNase V1 probing traces were normalized in a similar fashion.
Comparative sequence analysis (conservation and covariance analysis)
Multiple sequence alignments of 45 eukaryotic sequences comprising the full or partial SRA gene were obtained from the ENCODE project (55). Covariant base pairs were determined with in-house code that monitors changes in a given base pair across species, in a manner similar to Hofacker and coworkers (56). Alignment figures were prepared using the Jalview program (57).
RESULTS
Agreement between SHAPE, in-line, DMS and RNase V1 probing experiments across the entire length of the lncRNA
The secondary structure of the non-coding SRA transcript has been assessed by extensive probing investigations using chemical (SHAPE, in-line and DMS) and enzymatic probes (RNase V1). These methods utilize different chemical mechanisms, resulting in either modification or cleavage of RNA molecules depending on their structural folds. SHAPE, in-line and DMS probe single-stranded nucleotides, while RNase V1 digests base paired regions. Probing sites have been analyzed by reverse transcription using multiple site-specific primers and capillary electrophoresis.
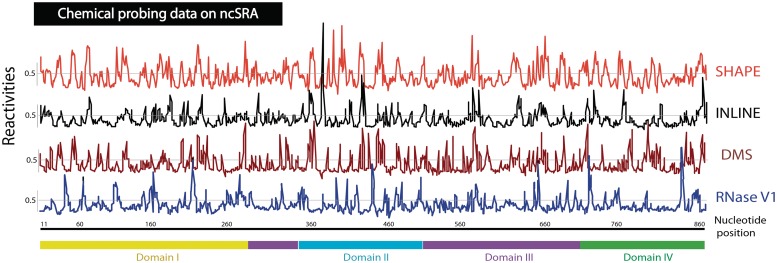
The collection of processed data for the entire length of the lncRNA is summarized in Figure 1. It shows the processed reactivities of each nucleotide from SHAPE, in-line, DMS or RNase V1 probing methods plotted against their position in the sequence. High-intensity values in SHAPE (red), in-line (grey) and DMS (brown) plots define single-stranded nucleotides of RNA. High-intensity regions show a high degree of overlap, suggesting that the results from the three methods are highly consistent with each other. High-intensity values in RNase V1 (blue) indicate base-paired nucleotides, which, as expected, are primarily positioned in the regions showing lower SHAPE, in-line and DMS reactivities. Enlarged regions of these plots can be found in Supplementary Figure S1. Overall, the results from the four methods are consistent with each other and have been used in aggregate to determine the structure of the lncRNA.
Figure 1.
Processed probing reactivities versus nucleotide position for entire SRA lncRNA spanning 874 nt. Examples of raw traces are shown in Figures 2–3. Red, plot of SHAPE-probing reactivities for SRA using multiple site-specific primers; grey, in-line probing reactivities; brown, DMS reactivities; blue, RNase V1 digestion. SHAPE probing reactivities were normalized by the average reactivities for highly reactive nucleotides. In-line, DMS and RNase V1 probing reactivities were normalized in a similar fashion. Global cutoff value of 0.5 is drawn for each probing profile.
Figures 2 and 3 summarize our experimental findings and display our experimentally derived secondary structure, annotated with either SHAPE and in-line reactivities or DMS and RNase V1 reactivities, respectively. The lncRNA appears to possess a complex secondary structure organization consisting of four major domains, comprising 25 helices in total. To simplify the following discussion, we also outline the helix nomenclature of the lncRNA based on several conventions previously applied to the ribosomal RNA secondary structure. First, the numbering of the helices was sequential, starting from the 5′-end to the 3′-end. Second, helices were differentiated when they were separated by either (i) a junction, (ii) a large internal loop (>12 nt in total), or (iii) a highly asymmetric internal loop with zero residues present on one side and a large number of single-stranded nucleotides (>6 nt) on the other side.
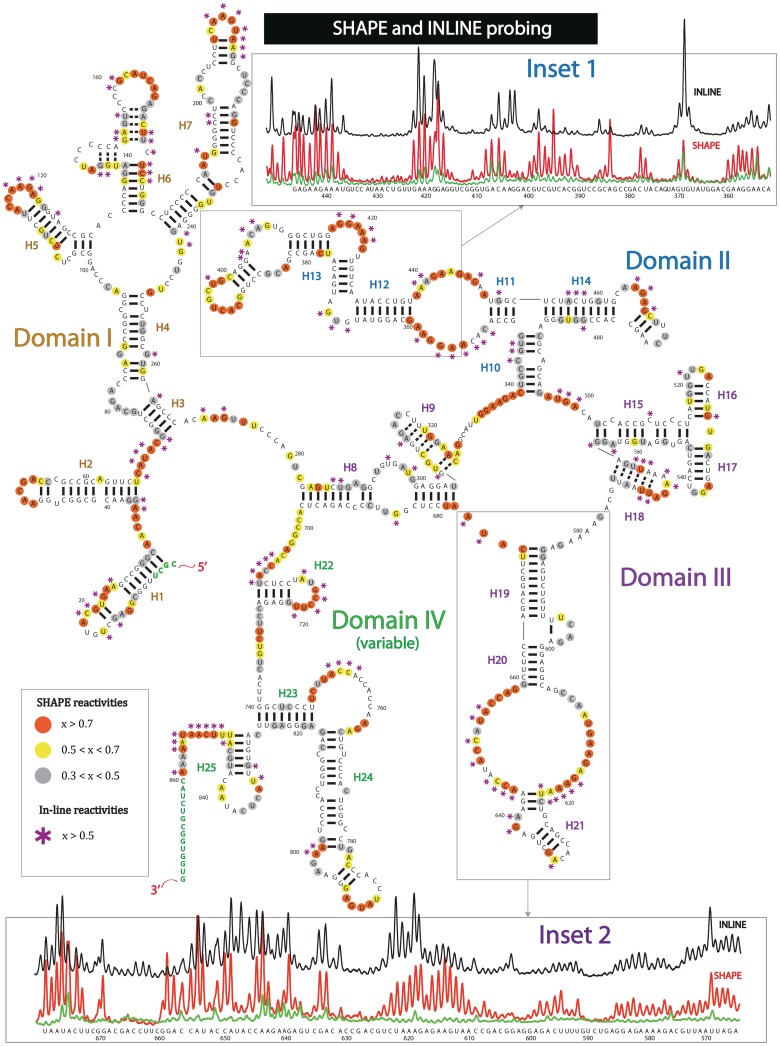
Figure 2.
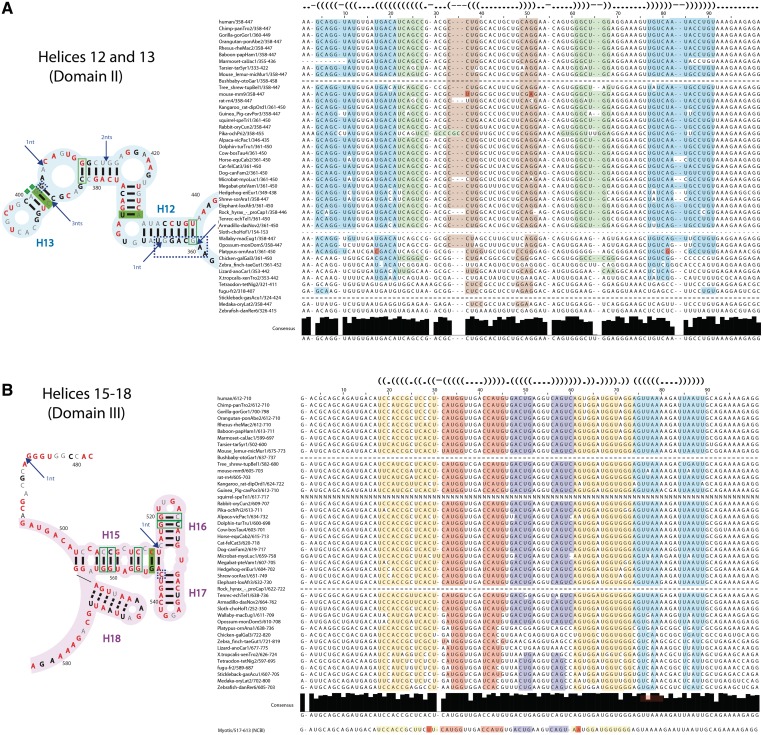
Secondary structure of entire SRA lncRNA, based on SHAPE and in-line probing experiments. Both experimental techniques detect single-stranded regions of RNA. Uncircled nucleotides, normalized SHAPE reactivity < 0.3; grey circled nucleotides, 0.3 < normalized SHAPE reactivity < 0.5; yellow circled nucleotides, 0.5 < normalized SHAPE reactivity < 0.7; orange circled nucleotides, normalized SHAPE reactivity > 0.7; purple asterisks, normalized in-line reactivity > 0.5; green nucleotides, no probing data. Helices are indicated by H1,…,H25. INSET 1: Raw capillary electropherograms of RNA region containing helices H12-H13 of domain II. Red, SHAPE reactivity; black, in-line reactivity; green, raw blank trace. INSET 2: Raw capillary electropherograms of RNA region containing helices H19–H21 of domain III. Red, SHAPE reactivity; black, in-line reactivity; green, raw blank trace.
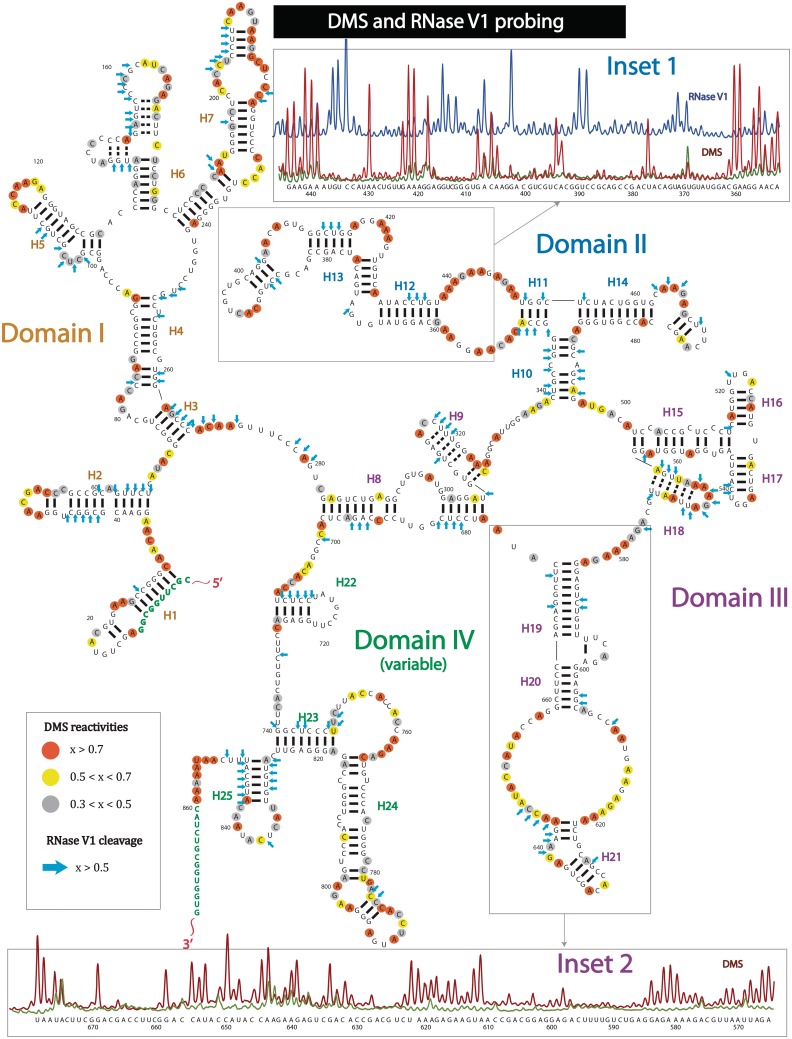
Figure 3.
Secondary structure of entire SRA lncRNA, based on DMS and RNase V1 experiments. DMS probing detects single-stranded regions; RNase V1 digestion detects double stranded helical regions. Uncircled nucleotides, normalized DMS reactivity < 0.3; grey circled nucleotides, 0.3 < normalized DMS reactivity < 0.5; yellow circled nucleotides, 0.5 < normalized DMS reactivity < 0.7; orange circled nucleotides, normalized DMS reactivity > 0.7; blue arrows, normalized RNase V1 cleavage > 0.5; green nucleotides, no probing data. Helices are indicated by H1, … , H25. INSET 1: Raw capillary electropherograms of RNA region containing helices H12–H13 of domain II. Brown, DMS reactivity; blue, RNase V1 cleavage; green, raw blank trace. INSET 2: Raw capillary electropherograms of RNA region containing helices H19-H21 of domain III. Brown, DMS reactivity; blue, RNase V1 cleavage; green, raw blank trace.
The proposed secondary structure model is consistent with the probing results: single-stranded regions are extensively modified or cleaved by the chemical reagents targeting single-stranded nucleotides, while base paired nucleotides are cleaved by RNase V1. A detailed discussion of the probing data and the resulting secondary structure is presented below.
Highly mobile and flexible nucleotides of the lncRNA are mapped by SHAPE and in-line probing
SHAPE probing targets flexible nucleotides of RNA molecules and does not suffer from solvent accessibility issues (58). Nucleotides that are extensively acylated at the 2′-OH position by the SHAPE reagent and have a normalized reactivity >0.5 are considered to be highly flexible and likely to be single-stranded (Figure 1, red curve; Supplementary Figure S1, red chart). Nucleotides that undergo no or relatively little modification are likely to be constrained either by base pairing or other non-canonical interactions (SHAPE reactivities <0.5) (59).
Nucleotides, which exhibit high SHAPE reactivities, are mainly located in the terminal loops, internal loops and junction regions (outlined in yellow and red dots—Figure 2). This indicates that these regions are highly mobile and likely to be single-stranded. Some examples of such regions include residues belonging to the terminal loops of H2 and H7, internal loops separating H11–H12 and H20–H21, and junction regions such as the multi-way junction connecting helices H8, H9, H10, H15, H18 and H19. Nucleotides restrained by base pairing interactions generally show a much lower tendency toward modification (outlined in grey dots or not outlined). The vast majority of our helices demonstrate this behavior. There are a few select instances where nucleotides involved in base pairing are also reactive towards the SHAPE reagent. These tend to be located close to single-stranded regions or bulges. Specific examples occur in H2, H4 and H13. This is also a common observation from the SHAPE probing of the rRNA, the secondary structure of which is well known (60). In addition, minor instances of SHAPE-reactive nucleotides positioned in the central part of a helix have also been previously observed in rRNA (60). In certain cases, it appears that the presence of the GU wobble base pairs might bring some flexibility to the helical segment. Exceptions to the above examples are helices H6, H9 and H18, where excessive modifications have been observed. These local regions of RNA have the potential to form helices (potential base pairs are indicated with dashes); however, these same nucleotides can be relatively mobile. For example, H9 can potentially form a 9-bp stem with three GU pairs and one GA mismatch. The following helical composition can explain the relative low stability of such a secondary structure element. We note that SRA is known to bind multiple proteins (17,26,28,37,38). Thus, we do not exclude the involvement of additional stabilizing factors such as proteins.
To verify the SHAPE probing results, we have also performed in-line probing, developed to study RNA structure by Breaker and coworkers (51). This method does not require any specific chemical reagents. Instead, the technique utilizes magnesium-catalyzed transesterification of RNA in flexible regions. In general, in-line digestion corresponds very closely to the SHAPE results (Figure 1 and Supplementary Figure S1). For detailed comparison, we present an example in inset 2 of Figure 2. This case shows raw SHAPE and in-line capillary electropherograms collected for the region of SRA from positions 565–680 (helices H19-H21), where in-line and SHAPE reactivities are extremely similar in intensities. Examination of the probing results of the entire transcript shows isolated instances of significantly reduced in-line cleavage in the regions of highly-reactive nucleotides towards the SHAPE reagent. The following suggests that these residues do not sample the optimal reaction geometry for transesterification by magnesium ions. An example of this case is included in inset 1 of Figure 2, which shows SHAPE (red) and in-line (black) capillary traces for the SRA region between positions 355–445. In particular, the intensity of cleavage in the region between positions 370 and 400 is suppressed significantly compared to SHAPE.
RNase V1 maps base paired nucleotides, while DMS verifies single-stranded nucleotides
To gain more insight into the structural organization of the lncRNA, we also employ dimethyl sulfate (DMS) and enzymatic RNase V1 probing on the entire length of the lncRNA transcript (Figure 1 and Supplementary Figure S1). DMS targets adenosines and cytosines, which do not participate in base pairing or other tertiary contacts (61). RNase V1 generally digests base-paired regions and stacked single-stranded nucleotides (62–64). While DMS data helped us to verify the information obtained through SHAPE experiments, RNase V1 probing contributed to our understanding of helix formation. In addition, due to bulkiness of the enzymatic probes, RNase V1 has the potential to reveal solvent-exposed sites of RNA molecules. DMS reactivities were analyzed and normalized in a similar manner as the SHAPE data for easy comparison. The DMS probing profile (Figure 3) and the SHAPE results (Figure 2) clearly demonstrate a strong overlap of DMS-modified nucleotides with SHAPE-reactive residues (see also Supplementary Figure S1). Adenosines and cytosines, which are strongly methylated by DMS (yellow and red), appear in the majority of single-stranded regions previously captured by the SHAPE reagent. Detailed examples of this correspondence can be seen by comparing raw DMS capillary traces of Figure 3 with SHAPE traces in Figure 2. However, DMS probing alone was not sufficient to capture all single-stranded elements (e.g. loop between H19 and H20, and terminal loop of H22). Nevertheless, more extensive methylation was observed for the single-stranded regions of H7 and the junction region connecting helices H18 and H19.
The complete RNase V1 cleavage profile is designated with blue arrows in Figure 3. Due to the large size of the enzyme, it is limited to the cleavage of solvent-exposed sites of RNA molecules (63). For example, the five base pair portion of helix H12 was not digested by RNase V1 (see also raw RNase V1 trace in the inset 1 of Figure 3). Interestingly, the most extensively digested sequences by RNase V1 belong to helices H2, H3, H7 (near the terminal loop), H13, H22 and H25. This preference of digestion sites might indicate that these RNA regions are more solvent exposed and, therefore, accessible to RNase V1. Additionally, RNase V1 is known to cleave stacked single-stranded nucleotides (62). We observe several instances of such action in the internal loop of H14, the terminal loop of H18 and in the internal loop located between H20 and H21. The single-stranded nature of these regions is well supported by SHAPE, in-line and DMS probing.
LncRNA SRA has a complex structural architecture, organized into four distinct domains
Based on the experimental data, the derived secondary structure of the lncRNA appears to be organized in four major domains. Domain I contains helices H1, H2, H3, H4, H5, H6 and H7. Domain II contains helices H10, H11, H12, H13 and H14. Domain III contains H8, H9, H15, H16, H17, H18, H19, H20 and H21. Domain IV contains helices H22, H23, H24 and H25 (Figures 2–3). The overall helical composition of the lncRNA accounts for 48% of the total number of SRA nucleotides with a helical density of 1 helix per 34 residues. This RNA is roughly half the size of the ribosomal subunit (874 nt versus 1542 nt) and contains 25 helical segments, 16 terminal loops, 15 internal loops and 5 junction regions. The current secondary structure of 16S rRNA comprises 45 helices, 31 terminal loops, 26 internal loops and 18 junction regions. Thus, the relative number of secondary structure elements in SRA is similar to that in 16S rRNA. The only exception is the relatively low number of junction regions in SRA. Junctions, however, are the most difficult RNA structural elements to define. For example, only eight junctions of 16S rRNA were initially determined (44). Interestingly, the majority of purine-rich sequences are located in the single-stranded regions of SRA, including junctions, internal loops and certain terminal loops. This is consistent with the secondary structures of ribosomal RNAs and riboswitch RNAs, which both show a similar structural trend of placing purine-rich stretches of sequence in single-stranded locations. Several CU and CA non-Watson–Crick pairs are observed in the lncRNA. This phenomenon is also observed in the human 18S rRNA secondary structure. Finally, we observe an ACC tri-loop, also found in the human 18S rRNA secondary structure.
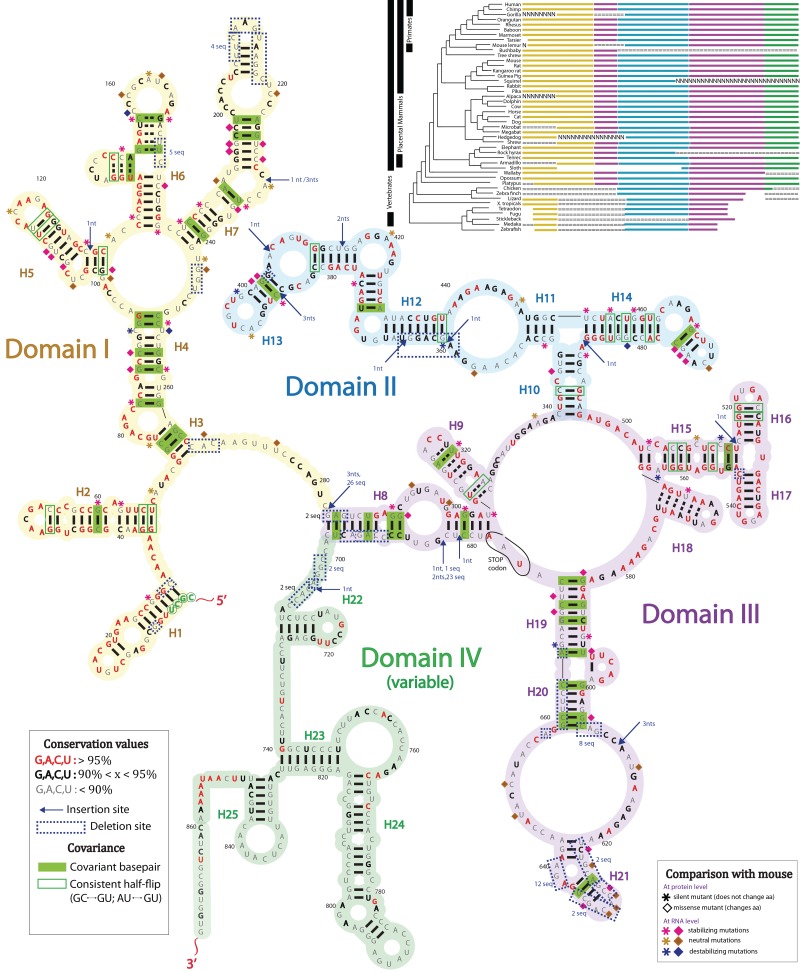
Domains I–III represent the core region of SRA and are the most conserved across species (Figure 4). Specifically, domain I (outlined in yellow) consists of two independent helices, H1 and H2, followed by a larger subdomain region with helices H3–H7. Helix H6 contains three CCCC or CCCCC stretches. These regions exhibit no reactivity towards chemical reagents, but appear to cause reverse transcriptase pausing at the nucleotide positions located in between the C-rich regions (Figures 2 and 3). This could be indicative of a complex tertiary structure that cannot be easily assessed via chemical probing. We do not guarantee the accuracy of the H6 fold.
Figure 4.
Conservation diagram of SRA lncRNA across wallaby, opossum, platypus and placental mammals (36 sequences total). Dark grey nucleotide letters, <90% conserved; bold black nucleotide letters, 90–95% conserved; red nucleotide letters, >95% conserved. Yellow highlighting, domain I; blue highlighting, domain II; purple/grey highlighting, domain III; green highlighting, domain IV. Dashed boxes, deletions that occurred in at least one of 36 vertebrate sequences. If deletions occur in more than one species, then the number of species undergoing deletion at this position is specified. Arrows, insertion positions. Number of nucleotides, x, incorporated at insertion site is indicated by ‘x nt’. If insertion occurs in more than one species, number of species undergoing insertion at this position is specified. Green filled boxes, covariant base pairs. Green outlined boxes (not filled), base pairs undergoing a change from a Watson–Crick base pair to a GU or UG, which do not have any instances of mismatches across all the organisms. Pink asterisks, mouse-to-human mutation that stabilizes human RNA helix and is silent with respect to amino acid sequence; brown asterisks, mouse-to-human mutation that is neutral with respect human RNA structure and is silent with respect to amino acid sequence; blue asterisks, mouse-to-human mutation that destabilizes human RNA helix and is silent with respect to amino acid sequence. Pink diamonds, mouse-to-human mutation that stabilizes human RNA helix and changes amino acid sequence; brown diamonds, mouse-to-human mutation that is neutral with respect human RNA structure and changes amino acid sequence; blue diamonds, mouse-to-human mutation that destabilizes human RNA helix and changes amino acid sequence. Mutations from mouse-to-human tend to stabilize the RNA structure of the lncRNA. Inset: Phylogenetic tree displays evolution of RNA structural domains across 45 vertebrates possessing the SRA gene. Yellow bars, domain I; cyan bars, domain II; purple/magenta bars, domain III; green bars, domain IV; ‘===’ and ‘NNN’ denote uncertainty in sequence alignment according to ENCODE database conventions.
Domain II (outlined in blue in Figure 4) is defined by a three-way junction, branching helices H10, H11 and H14. Subsequently, helix H11 gives rise to the helices H12 and H13. Domain III (outlined in purple) is the largest subdomain of SRA. It is composed of several individual secondary structure elements including H9, H15, H16, H17, H18, H19, H20 and H21, which are all locked in a globular fold by helical stem H8.
Validation of secondary structure by comparing SHAPE experiments on smaller stretches of RNA sequence to the full RNA
To validate our structural fold and eliminate alternatives, we performed SHAPE experiments on smaller fragments of SRA sequence. If the structural elements are formed via close-range base pairing in the context of the entire SRA, we expect them to also base pair in the context of smaller fragments. We note that the deletions in RNA sequence may expose secondary elements that were previously hidden, or affect the flexibility of previously rigid structural elements. These effects could modify signals introduced by the chemical reagents. Nevertheless, in order to test our strategy, we randomly chose two sequences ∼220 nt in size that overlap two regions of SRA sequence between positions 260–479 and 480–693 (Supplementary Figure S2). The first stretch of RNA (260–479) has significant overlap in SHAPE reactivity with the SHAPE reactivity profile of the full RNA for positions 360–445. The relative ratio in reactivities changed slightly; however, the positions of base paired nucleotides remained the same. The overlap in SHAPE data suggests that the region occupying positions 360–445 forms an autonomous secondary structure and comprises the well-defined helices H12 and H13 of Domain II. The second fragment (positions 480–693) allowed us to resolve the nucleotides from positions 495 to 665. The SHAPE reactivities of this stretch of RNA are very similar to the SHAPE reactivities of this sequence in the context of the entire SRA. Helices H15, H16, H17, H18, H19, H20 and H21 of domain III, which belong to this region, do indeed base pair in a close-range and are limited to this sequence. Therefore, the proposed secondary structure of entire SRA is in a good agreement with fragment analysis, which was quite useful in the validation of a number of our substructures in domains II and III.
Due to high sequence variability, domain IV was not expected to be highly structured. Surprisingly, this domain, which undergoes many insertions and deletions among mammals, is well organized into chain of smaller helical regions with a slightly lower helical density relative to the rest of the lncRNA. Because of its low sequence conservation, we refer to this domain as ‘variable’. Interestingly, eukaryotic ribosomal RNAs also contain highly variable regions called expansion segments (65). In the X-ray structure, these regions are well-defined RNA helices and participate in the formation of complex tertiary motifs, necessary for maintaining ribosome functionality.
Conservation and covariance analysis of SRA secondary structure
Due to the emergence of both non-coding and coding RNA transcripts from the SRA gene, we cannot eliminate the possibility that two evolutionary constraints are placed on its sequence—first to preserve the RNA structure and second to maintain a protein fold. We note that no antisense RNA has been reported to arise from this gene. Therefore, conservation was assessed only for the sense strand. To date, the conservation of the SRA gene has been examined only in terms of its translational product across certain vertebrates and species from lower taxa (42). Our new secondary structure of human SRA allows us to examine the coevolution of 45 vertebrate SRA sequences at the RNA structure and protein structure levels, which are available through the ENCODE project (55). We have calculated the conservation values for each nucleotide of the SRA sequence from 36 of these sequences, ranging from platypus to human. The remaining nine sequences extracted from chicken, zebrafish and other phylogenetically distant vertebrates were excluded from the conservation comparison because they contain only small portions of the SRA gene (see upper right corner in Figure 4); however, they were not neglected in further analysis. Nucleotides with the conservation values >95% are highlighted in red (Figure 4). Thus, regions abundant in red are the most conserved regions. These include helices H1, H2, partial regions of H3, H4 and H5 of domain I, helices H12 and H13 of domain II and helices H15, H16, H17 and H18 of domain III. With regard to RNA structure, we note that terminal loops, bulges and other looping regions in mammalian SRA are generally more highly conserved relative to base paired regions.
The most striking feature of the lncRNA secondary structure is the local region of domain III, occupying positions 493–586 and comprising a three-way junction branching helices H15, H16 and H17 (detailed alignment can be found in Figure 5). This RNA segment could be important functionally, as 57% of the nucleotides in this region are 100% conserved across vertebrates, from platypus to human.
Figure 5.
Sequence alignment for the highly conserved SRA regions. (A) SRA conservation diagram for helices H12 and H13 of domain II (nucleotide positions 357–440). Left, secondary structure. Annotation is as in Figure 4. Right, sequence alignment across 45 vertebrates. Top line shows dot-bracket notation of secondary structure. Light blue, complementarity of helix H12; light brown and light green, complementarity of helix H13. Red, covariant base pairs. (B) Same as (A) for H15–H18 of domain III (nucleotide positions 478–583).
We compared the H15–H17 region with nine sequences of lower vertebrates, including lizard and several species of fish (pufferfish, stickleback, Japanese killifish and zebrafish) that were initially excluded from the conservation calculations (Figure 5B). Interestingly, one of the lowest organisms in the comparison (zebrafish) still contains portions of H15. Helices H16 and H17 become more stabilized throughout evolution, extending from 3 to 5 bp. For comparison, we also show in detail the phylogenetic alignment of the region comprising H12 and H13 located in Domain II (Figure 5A). This region is highly conserved across primates and placental mammals. We note that Helix H13 is associated with structure-7 (STR-7) of SRA previously proposed by Lanz (30). There are a number of studies that show the functional importance of the H13 region, which is directly involved in protein binding (28). However, H13 is significantly less conserved relative to H15–H18 for a large number of lower species including chicken, zebra finch, lizard, frog, and several species of fish (pufferfish, stickleback, Japanese killifish and zebrafish). Juxtaposition of this lack of conservation of H13 with the high conservation of H15–H18 suggests that the H15–H18 region in domain III may be a very important element of the lncRNA structure.
The high level of sequence conservation allowed us to easily determine the positions of covariant base pairs across 36 organisms. Watson–Crick base pairs, mutated to other Watson–Crick base pairs, are considered covariant and are denoted by solid green boxes in Figure 4. Mutations that interconvert a Watson–Crick base pair and a GU wobble pair are considered partially covariant and are denoted by an empty green box in Figure 4. Helices H2, H3, H4, H6, H7, H8, H9, H12, H13, H14, H15, H19, H20 and H21 possess at least one covariant base pair. Covariance analysis was not performed on the variable domain due to poor sequence alignment in this region.
Amino acid frame disruptions in the conserved regions of mammalian SRA suggest the loss of protein-coding potential for SRAP synthesis
We also examine the impact of nucleotide insertions and deletions across 36 vertebrates. Insertion sites are denoted with blue arrows and deletion sites are presented as dashed boxes in Figure 4. There is a high occurrence of these events after position 700 in domain IV. Therefore, due to space limitations, these sites were not outlined in this figure.
Insertions and deletions of integral codon length (i.e. insertions and deletions that are multiples of 3 nt) occur in the regions that show reduced or a complete lack of sequence conservation between vertebrates. These insertions and deletions are located primarily in the single-stranded regions or close to the terminal loops such as H7 in domain I and H21 in domain III. Insertions and deletions of integral codon length represent the majority of all cases and are very common amongst the various vertebrates.
An interesting feature of the insertion/deletion map concerns protein coding frame disruption for the SRAP encoded by the coding isoform of SRA, which is identical to the non-coding isoform, apart from a short region at the 5′-end. Frame disrupting insertions and deletions are non-integral codon insertions and deletions (i.e. insertions and deletions that are not multiples of 3). We observe such insertions and deletions in well-conserved regions of SRA such as H10, H12 and H13 of domain II and H15, H16 and H17 of domain III. These insertions and deletions constitute a small fraction of the total number of the insertion/deletion points and are mainly introduced by three eukaryotes: pika, microbat and rock hyrax. Interestingly, these three organisms are not descendants of each other and belong to different branches of the mammalian phylogenetic tree. Thus, it is likely that each of these three species express only the non-coding form of SRA RNA, while their ancestors may have expressed both the coding and non-coding isoforms. The SRA gene of rock hyrax has a single nucleotide deletion in the H13 of domain II (also see detailed alignments in Figure 5A). The pika gene does not have deletions, but has the highest number of insertions, which primary occur in the helices H12 and H13. The microbat gene has accumulated a 1-nt insertion in the junction connecting H10, H11 and H14 of domain II. This is followed by an additional 1-nt insertion and 1-nt deletion in the junction of H15, H16 and H17 of domain III (Figure 5B). All three above mentioned organisms introduce changes that negatively affect translation, by compromising the amino acid reading frame. This disruption will have no obvious effect on the RNA structure. One possible exception may be an insertion in H13 of pika, where the formation of an alternative 6-bp stem might be the case. Despite high conservation and limited number of sequences, the formation of H12 and H13 is validated by the covariant base pairs outlined in green in Figure 4. H15 also possesses a covariant base pair from Myotis lucifugus (Figure 5B). Frame-disruptions in the conserved regions of the SRA gene suggest that evolutionary pressure in mammals preserves the RNA structural/functional core rather than its translational product.
The sequence of RNA helix H2 is the most highly conserved at the protein level and shows a reduced rate of evolution relative to other regions of the lncRNA
Helix H2 in domain I is highly conserved from the point of view of RNA and protein structures (Supplementary Figure S3). Lanz and coworkers show that this RNA region is important in the coactivation performance of the lncRNA (30). Site-directed mutagenesis of this RNA substructure reduced the coactivation performance of this RNA by 40%. In addition, the formation of well-defined RNA helical composition is supported by our SHAPE, in-line, DMS and RNase V1 probing experiments (Figures 2 and 3). Surprisingly, it was found recently that the sequence belonging to this helix is the most conserved at the protein level between human and lower organisms, including trichoplax. This suggests that the H2 sequence may play an important role in the translated protein (42). We believe that while this region is functionally important at the RNA structure level, it has a significantly reduced rate of evolution relative to the remainder of the lncRNA due to strong constraints imposed by coding requirements of the SRA gene. This example provides evidence that different evolutionary rates are placed on the same sequence. In contrast to H2, the functionally important H13 (STR-7) evolved later and is absent in lizard, pufferfish and other vertebrates.
Nucleotide mutations from mouse to human produce more stable RNA helices in human
With the RNA secondary structure of human SRA (Figure 4) and recent NMR structure of mouse SRA protein (PDB ID: 2YRU), we can simultaneously assess the mutational effects of the SRA gene at the RNA structure and protein structure levels. The solved NMR structure of mouse SRAP contains amino acids corresponding to the positions 93–216 (Figure 6). The IUPred package (66,67) predicts that human SRAP residues 1–90 and 220–236 are highly disordered. We note that while amino acid positions 13–21 (RGWNDPPQF) of SRAP are predicted to be disordered, this region corresponds to the highly structured H2 in Domain I and is highly conserved at the protein and RNA levels.
Figure 6.
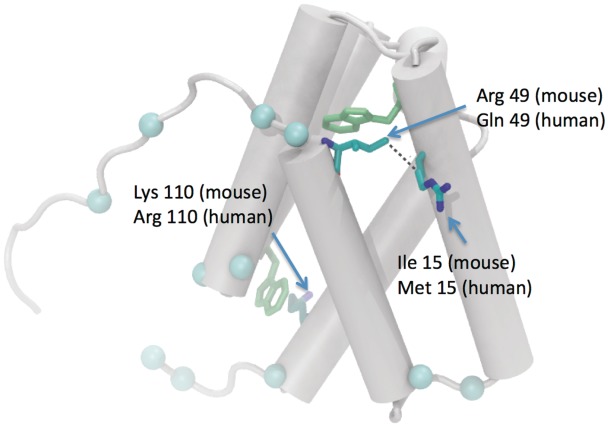
SRAP protein corresponding to coding isoform of SRA. NMR structure of mouse SRAP protein (PDB ID: 2YRU) corresponding to positions 271–609 of SRA RNA transcript. Grey rods, α helices of the protein. Blue spheres, C-α atoms of residues that differ between mouse and human. Bonds representation is used for mutations from mouse to human located in α helices. The majority of mutations occur in linkers connecting helices.
Based on the ENCODE SRA multiple sequence alignment (55), there are 99 positions in the SRA gene that have mutated between mouse and human (Figure 4). A detailed map at the nucleotide level can be also found in the Supplementary Figure S4. At the level of protein sequence, 48 mutations are silent (asterisks) and do not change the amino acid (Figure 4). The remaining positions are missense mutations (diamonds), resulting in the coding of a different amino acid. We also map the locations of missense mutations on the NMR structure of mouse SRAP (Figure 6). In summary, the majority of amino acid mutations (blue spheres) are positioned in inter-helical linker regions. In general, these loops lack secondary structure and generally possess the highest number of amino acid insertion/deletion sites (68). However, the possibility of their involvement in active site formation should not be neglected. We observe only three amino acid mutants positioned in α helices, which could alter the helical packing of SRAP: Ile 15, Arg 49 and Lys 110 (amino acid numbers correspond to protein databank accession code 2YRU). Cooper and coworkers have constructed a homology model of human SRAP based on the NMR structure of mouse SRAP (42), which adopts a similar five-helix conformation to that of mouse. This group has also noticed that mouse SRAP shares strong structural similarities with yeast splicing factor prp18, suggesting that SRAP could possess similar RNA-binding motifs (42).
At the RNA structure level, 58 of 99 mutations have a stabilizing effect on RNA helix formation. These include (i) changes from AU base pairs in mouse to GC base pairs in human, (ii) changes from GU pairs in mouse to GC base pairs in human, and (iii) changes from non-Watson–Crick mismatches in mouse to Watson–Crick base pairs in human. Of the 99 mutations, 32 are neutral and do not affect the secondary structure fold. These mutations are positioned in single-stranded regions. We note that it is possible that these neutral mutations may have positive or negative effects on tertiary interactions because we do not know all possible canonical and non-canonical tertiary interactions involved in the structure. In total, 59% of all mutations contribute to the stabilization of the secondary structure, while only 9% have a negative effect on RNA helix stability. While the mutational effects at the protein level are not easily assessed, they appear to be minimal. The mutational contributions towards stabilization of the RNA structure are profound.
RMDetect identifies several secondary structure elements consistent with SHAPE, in-line, DMS and RNase V1 probing
The RMDetect computational tool for structural RNA module searching was applied to the human ncSRA isoform alone and to the ENCODE multiple sequence alignment across 36 organisms (69). In the single sequence run of RMDetect, the code identified the C-loop in H24 of domain IV at positions 769–776 on the 5′-side and 805–813 on the 3′-side. Interestingly, RMDetect also predicts a kink-turn motif in H24 at 779–784 on the 5′-side (CUGACC) and 794–802 on the 3′-side (GGGAAGAAG). While this differs slightly from the configuration depicted in Figures 2–4, we emphasize that this kink-turn predicted by RMDetect is consistent with all four probing techniques used in this study. This kink-turn is defined by an AGA bulge and a four pair stem with AG, GA, GC and GC pairs. When running RMDetect using the multiple sequence alignment provided by the ENCODE genome server, RMDetect identifies a tandem-GA loop at 780–783 on the 5′-side and 795–798 on the 3′-side, capped by Watson–Crick base pairs on each end. We emphasize that domain IV is highly variable and has poor sequence alignment.
Structural changes in H1 and the H4/H5 junction region are manifested between coding and non-coding SRA RNA isoforms
As mentioned previously, a number of SRA transcripts have been determined to date (40–42). These transcripts are alternatively spliced variants of the SRA1 gene of human chromosome 5q31.3 comprising 5 exons and 4 intronic regions. The majority of SRA isoforms are fully spliced (all four introns are removed), while a few transcripts retain full or partial intron sequences. We decided to chemically assess the differences between the various isoforms in order to understand the possible structural alterations that can be caused by these alternative splicing events.
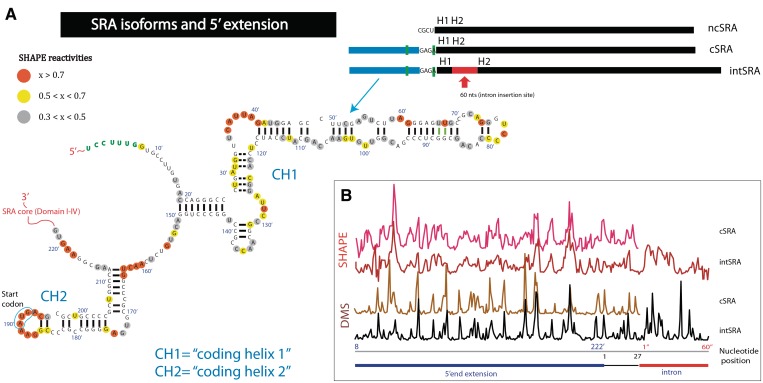
We chose three key SRA systems, which represent the majority of transcripts determined to date in humans: the original non-coding form, the coding form, and a second, longer non-coding form (Figure 7). System 1 (Figures 1–4) is the non-coding form (ncSRA, NCBI ID: AF092038), which is lacking initiation codons for the translational machinery and is 222 nt shorter than the coding form. ncSRA is the first SRA transcript determined and proven to act as a non-coding RNA (17). This transcript possesses an interesting 4-nt mutation relative to the coding sequence: positions 1–4 of ncSRA have the sequence CGCU rather than the GAGA sequence present in the coding form cSRA (Figures 7–8). This nucleotide substitution silences the initiation codon AUG located between positions 4–6 by modifying its sequence to UUG. In addition, the substituted nucleotides are able to participate in the formation of helix H1. System 2 is the coding isoform of SRA (cSRA, NCBI id: AF293024). In this isoform, exon 1 is extended by 222 nt on the 5′-side relative to the ncSRA isoform. This 5′-extension contains two initiation AUG codons, both utilized in the production of 224 and 236 amino acid SRAP proteins (29,70). System 3 is an alternatively spliced non-coding transcript of SRA. This isoform (intSRA, NCBI id: DQ286291) contains a portion of an intron that is normally spliced out of ncSRA. This intron portion in intSRA exists between positions 27 and 28 in ncSRA numbering (Figure 7, upper right). This SRA variant has the same 5′-extension as coding isoform (cSRA); however, a pre-mature stop codon in the intron sequence aborts translation (43).
Figure 7.
Alternatively spliced isoforms of SRA. ncSRA, non-coding isoform of SRA; cSRA, coding isoform of SRA; intSRA, isoform of SRA that contains an intron not present in ncSRA and cSRA. (A) Experimentally determined secondary structure of the region of the coding isoform of SRA (cSRA) that differs from the non-coding isoform (ncSRA). This region is called the 5′-extension. Annotation is same as Figure 2. The cSRA and intSRA have identical secondary structures in this region. Upper right, schematic of alternatively spliced isoforms of SRA. The ncSRA comprises fully spliced core of SRA gene (black line). The cSRA possesses an extended exon-1 (blue line) with two initiation codons for SRAP synthesis (green bars). The intSRA retains a portion of an intron that is spliced out of ncSRA, located between exon-1 and exon-2 of SRA gene (red). (B) Processed SHAPE and DMS reactivities for cSRA and intSRA 5′-end extensions.
Figure 8.
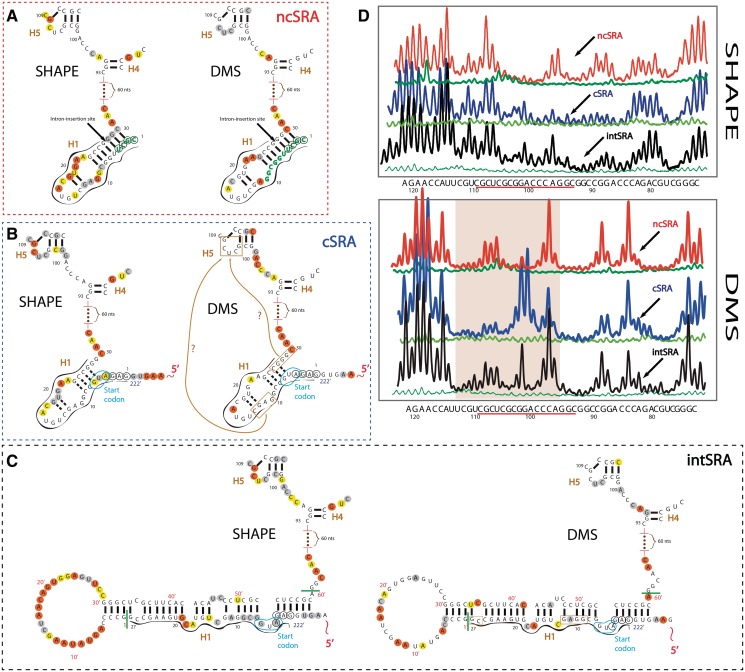
Differences in the secondary structures of alternatively spliced SRA RNA isoforms. (A) Secondary structure of non-coding isoform ncSRA H1 and H4/H5 junction region annotated with SHAPE (left) and DMS (right) reactivities. Annotation is similar to Figures 2–3. Circled green nucleotides, first 4 nt (CGCU) differ with respect to the coding isoform (cSRA). The position of the splicing site between exon-1 and exon-2 is indicated by black arrow. (B) Same as (A), but for coding isoform cSRA. Nucleotides circled in black (GAGA) differ from non-coding isoform ncSRA. These nucleotides are preceded by an extended region. Additional extension of 222 nt is not fully shown due to space limits. Brown arrows with ‘question marks’ in the DMS-annotated structure (right) show the potential base pair interaction sites between the nucleotides in brown boxes. (C) Secondary structure model of intron-comprising isoform of SRA (intSRA) H1 and H4/H5 junction region. Structure contains the intron present in intSRA and displays the changes caused by the intron retention between exon-1 and exon-2. Start and end positions of the intron sequence are pinpointed by green bars located after position 27 and before 28. Nucleotides of the intronic region are numbered separately (red primed numbers). Other annotation is as in (A). (D) Raw SHAPE and DMS capillary traces for ncSRA (red), cSRA (blue) and intSRA (black) for the H4/H5 junction region. Green, blank traces for unmodified RNA. Nucleotide positions are shown below the traces.
We have performed probing experiments (SHAPE and DMS) on the entire sequences of cSRA and intSRA transcripts and compared them with the ncSRA probing results. SHAPE and DMS probing traces collected for the 5′-end extension region in the context of the cSRA and intSRA sequences appear identical (Figure 7 and Supplementary Figure S5). This suggests that they share the same structural fold and are not affected by the intron sequence. The secondary structure of the 5′-extended exon appears to be an autonomous unit of the sequence, consisting of one relatively large domain followed by a smaller helical segment. Interestingly, one of the AUG initiation codons is located in the terminal loop of helix CH2. Possession of the 5′-extension did not appear to interfere with the SRA core sequence: the core sequence remains almost entirely undisturbed across all non-coding and coding isoforms. As shown by Hube and coworkers, cSRA lacks the coactivation performance of ncSRA transcript (40). However, when its coding features are disrupted via mutation of the AUG codon, this transcript has been shown to perform regulatory activities. This correlates well with our observation that the structural fold of the core sequence across isoforms remains largely unchanged. The only significant changes in SHAPE and DMS probing occur in the region containing helix H1 and in the junction region connecting H4 and H5, distant from the intron insertion site and 5′-extension. Raw SHAPE and DMS capillary traces for the H4/H5 junction region are shown in Figure 8D. The changes are more pronounced in the DMS profile.
In order to fully describe the structural changes, Figure 8 shows probing annotations for these regions in the ncSRA, cSRA and intSRA transcripts. Figure 8A depicts the SHAPE-annotated (left) and DMS-annotated (right) helix H1 and the H4/H5 junction region for the ncSRA transcript. Figure 8B presents the probing annotations for the cSRA transcript. Figure 8C shows the intSRA, which includes a 60 nt intron inserted into helix H1. The first four mutated nucleotides of ncSRA, CGCU, participate in the formation of H1 (Figure 8A). The coding SRA isoform in Figure 8B lacks these nucleotides. Therefore, the SHAPE and DMS reactivity profiles of H1 are changed. Likewise, significant changes in the DMS profile of the H4/H5 junction region are also observed relative to ncSRA (see also raw DMS capillary traces in Figure 8D). Specifically, low-reactivity CUC nucleotides occupying positions 105–107 are almost completely unreactive, while C99 and A100, located in close proximity, are highly exposed to DMS methylation. In addition, A96 shows reduced intensity.
In the intSRA (Figure 8C), DMS reactivities of H4/H5 junction region seem to correspond closely to that of ncSRA. While the H4/H5 junction region appears to be similar in both ncSRA and intSRA transcripts (non-coding forms of SRA), this region displays different reactivities for cSRA (Figure 8D). Since the probing results of the 5′-end extensions present in cSRA and intSRA are very similar, we excluded the possibility that 5′-end extension has any effect on this structural change. As the remainder of the SRA core sequence remains identical across isoforms, the only possible cause for this structural change is the region of RNA corresponding to H1. From the probing results, it is not obvious which nucleotides from H1 of cSRA may interact with the H4/H5 region. This will require future site-directed mutagenesis studies. However, there are two low-reactive and complementary sequences in H1 of cSRA that could pair with the internal loop of H5 (outlined in brown boxes and connected with question mark labeled curves in Figure 8B). Interestingly, in intSRA, these two sequence sites are blocked via close-range base pairing with the intron sequence of intSRA.
Overall, we find it intriguing that non-coding isoforms share a similar fold in the H4/H5 junction region, distinct from that of the coding transcript. We also note that the GAGA sequence (positions 1–4 in cSRA, Figure 8B) contributes to the formation of a much stronger Kozak signal context: GagAUGG, as opposed to GaaAUGa, located upstream in CH2 of the 5′-end extension.
DISCUSSION
Structural architecture plays a key role in understanding the mechanism of functional RNAs. Before mechanistic understanding of many functional RNAs (e.g. ribosomal RNAs, tRNAs, group I and II introns) could be achieved, extensive secondary structure studies, along with comparative sequence analysis, were performed to lay the foundation for mechanistic studies (44,45,71,72). RNAs originating from the SRA gene act as regulatory non-coding RNAs and as coding transcripts, which produce the protein SRAP. The functions of the SRA RNA and SRAP often overlap in the estrogen-signaling pathway (22,73). We used a variety of tools to produce a secondary structure consistent with SHAPE, in-line, DMS, RNase V1, and covariance analysis across 36 species. Each probing technique utilizes various mechanisms to target nucleotides. We find that using the combination of these tools gives the most comprehensive picture. DMS probing alone was not sufficient to capture all single-stranded nucleotides; however, it was indispensable in the determination of the internal loop of H7 and the junction region H18/H19, which were not well captured by SHAPE and in-line probing. The combined assembly of these complementary data represents convincing experimental evidence supporting the structural organization depicted in Figure 4.
Previous studies have investigated the functional performance (i.e. coactivation) of SRA and variants of SRA, including site-directed mutagenesis and deletions of stretches of sequence. These studies suggested that the SRA function is not limited to one sub-structure, but rather requires the full sequence (30). Our new experimental probing results reveal that the lncRNA is organized into four domains, with various secondary elements ranging from small, autonomous helical stems (e.g. H1 and H2 of domain I) to larger structures formed via long-range base pairing (e.g. H10 of domain II and H19 of domain III). Previous deletion studies showed that removal of the 3′-end (after position 634) results in a 49% decline in the coactivation efficiency (30). This is consistent with our proposed model where such a deletion would disrupt the formation of three helices (H19–H21) and the globular fold of domain III (by disrupting H8). Earlier studies also showed that the removal of the 5′-end (positions 1–142) resulted in a 32% decline of coactivation activity. In our new secondary structure, a deletion of positions 1–142 leaves domains II–IV intact.
Site-directed mutagenesis of SRA also appears consistent with our secondary structure. Lanz and coworkers observed no effect on the coactivation performance when nucleotides G78 and A96 were simultaneously mutated to C78 and G96. These residues are positioned in the looping regions of our secondary structure. A single-point mutation of G123 to A123 also did not affect the SRA function, consistent with our secondary fold. Mutations of the UCU622–624 region and C630 resulted in a 40% decline. This data supports our structure, as these changes result in disruption of H21. We note that mutants of A246 and G249 result in a 60% reduction in SRA functional abilities. This could be a result of the disruption of key tertiary interactions in the junction region connecting helices H4–H7; however, further studies are required to test this hypothesis.
The RNA motifs STR-1 (helix H2) and STR-7 (helix H13), identified previously by site-directed mutagenesis (30), are consistent with the proposed secondary structure model. For example, it has been suggested that STR-1 (H2) forms a stem loop helix interrupted by an A-bulge and a UC mismatch. Indeed, strong SHAPE and DMS modification sites are observed in the terminal loop region and in the junction region preceding the helix, suggesting that it is a completely autonomous unit of the lncRNA. Similarly, STR-7 (H13) has been proposed to have a stem-loop structure comprising an asymmetric internal loop. We have probing results consistent with the overall shape of this RNA motif.
It is sometimes assumed that RNA sequences with poor conservation lack functional relevance (74), and therefore, exist as disordered regions with no defined structural organization. Despite low conservation of domain IV, this domain is highly structured, containing well-defined helices and secondary structure motifs. Extensions on the 5′-end in the cSRA and intSRA isoforms also exhibit a well-defined secondary structure.
Additionally, this lncRNA transcript is known to interact with a variety of proteins, including its own translation product SRAP, suggesting a possible formation of a multicomponent RNA–protein complex (17,26,28,37,40,75). We observe large internal loops positioned in domain II (between H11 and H12) and domain III (between H20 and H21), which may become more structured upon protein binding. This may also be the case for helices H9 and H18 of domain III. Interestingly, X-ray crystallography has recently revealed that the expansion segments specific for eukaryotic ribosomal RNAs contain many single-stranded stretches of RNA that interact with proteins to form highly ordered non-helical elements (65). For example, expansion segment ES39L in the large subunit contains three long single-stranded stretches of RNA that form a platform for the binding of six ribosomal proteins. This region also serves to bind the signal recognition particle, which facilitates recruitment of the ribosome to the endoplasmic reticulum.
We employ analysis of conserved regions to address the following two questions: (i) are these nucleotides primarily conserved to maintain structure at the RNA level or at the protein level, or both simultaneously? and (ii) does the evolutionary pressure apply uniformly across the entire sequence or should different evolutionary trends be considered? Comparison across vertebrates shows that the evolution of the lncRNA structure towards the human structure occurs in a stepwise manner. For example, the highly conserved helix H2 of domain I is already present early in vertebrate evolution, existing in lizard, frog and several species of fish (pufferfish, stickleback, Japanese killifish and zebrafish). This helix remains largely unchanged for the rest of vertebrate evolution. H2 is followed by the appearance of helices H15, H16 and H17 of domain III in opossum, wallaby and sloth. H12 and H13 of domain II appear later in evolution, present in armadillo and tenrec. Comparative structural analysis between mouse and human strongly suggests that a large number of evolutionary changes occur to stabilize the RNA structural core, while the mutational effects at protein level are relatively minimal. Multiple frame-disrupting insertions and deletions in other mammal sequences indicate a significant disruption of coding potential at late times in evolution, compromising many well-conserved elements at the protein level.
Moreover, it has been previously proposed that alternative SRA splicing maintains the coding/non-coding transcript balance (40,43). The coding/non-coding potential of SRA transcripts appears to be encoded in its primary sequence via the presence or absence of initiation codons or through the introduction of premature stop codons in the intron. In addition to differences in primary sequence, we find differences in structure between the coding and non-coding forms in H1 and the H4/H5 junction region.
Several theories on the origins of non-coding RNA have been discussed previously (76). One of them relies on the degeneration of the ability to code for protein (‘pseudogenization’). An example is Xist RNA, a long non-coding transcript involved in X chromosome inactivation in mammals that originates from the protein-coding Lnx3 gene (77,78). Interestingly, the Lnx3 gene was still a coding gene in opossum; however, later vertebrates gave rise to the non-coding Xist RNA, with the help of frame-shifting mutations in the exon regions (77). It is not clear whether the transformation of the protein-coding gene to the non-coding transcript occurs in a gradual, stepwise mechanism or via a sudden change. It has been proposed that, at some point in evolution, the non-coding Xist RNA gene might have been originated gradually, allowing for a period of time where non-coding and coding isoforms of the gene coexisted (76). The coding and non-coding isoforms of SRA also originate from the same gene, suggesting that this gene system might be a rare and unique capture of this stage of evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Laboratory Directed Research and Development program. Funding for open access charge: Laboratory Directed Research and Development program.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: neutral evolution of ‘non-coding' complementary DNAs. Nature. 2004;431 1 p following 757; discussion following 757. [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb. Perspect. Biol. 2011;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterlee JS, Barbee S, Jin P, Krichevsky A, Salama S, Schratt G, Wu DY. Noncoding RNAs in the brain. J. Neurosci. 2007;27:11856–11859. doi: 10.1523/JNEUROSCI.3624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian D, Sun S, Lee JT. The long noncoding RNA, jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 14.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 16.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 18.Hussein-Fikret S, Fuller PJ. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol. Cell. Endocrinol. 2005;229:149–160. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Lanz RB, Chua SS, Barron N, Söder BM, DeMayo F, O'Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003;23:7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–4193. [PubMed] [Google Scholar]
- 21.Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 2000;60:6266–6271. [PubMed] [Google Scholar]
- 22.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emberley E, Huang GJ, Hamedani MK, Czosnek A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC, Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem. Biophys. Res. Commun. 2003;301:509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 24.Coleman KM, Lam V, Jaber BM, Lanz RB, Smith CL. SRA coactivation of estrogen receptor-alpha is phosphorylation-independent, and enhances 4-hydroxytamoxifen agonist activity. Biochem. Biophys. Res. Commun. 2004;323:332–338. doi: 10.1016/j.bbrc.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 25.Cavarretta IT, Mukopadhyay R, Lonard DM, Cowsert LM, Bennett CF, O'Malley BW, Smith CL. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation. Mol. Endocrinol. 2002;16:253–270. doi: 10.1210/mend.16.2.0770. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deblois G, Giguere V. Ligand-independent coactivation of ERalpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J. Steroid Biochem. Mol. Biol. 2003;85:123–131. doi: 10.1016/s0960-0760(03)00225-5. [DOI] [PubMed] [Google Scholar]
- 28.Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, Redfern AD, Miles LE, Barker A, MacDonald LM, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 29.Kurisu T, Tanaka T, Ishii J, Matsumura K, Sugimura K, Nakatani T, Kawashima H. Expression and function of human steroid receptor RNA activator in prostate cancer cells: role of endogenous hSRA protein in androgen receptor-mediated transcription. Prostate Cancer Prostatic Dis. 2006;9:173–178. doi: 10.1038/sj.pcan.4500867. [DOI] [PubMed] [Google Scholar]
- 30.Lanz RB, Razani B, Goldberg AD, O'Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc. Natl Acad. Sci. USA. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 32.Xu B, Koenig RJ. An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J. Biol. Chem. 2004;279:33051–33056. doi: 10.1074/jbc.M404930200. [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Yang WH, Gerin I, Hu CD, Hammer GD, Koenig RJ. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol. Cell. Biol. 2009;29:1719–1734. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Caretti G, Lei EP, Sartorelli V. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle. 2007;6:1172–1176. doi: 10.4161/cc.6.10.4228. [DOI] [PubMed] [Google Scholar]
- 36.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 39.Chooniedass-Kothari S, Hamedani MK, Troup S, Hubé F, Leygue E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int. J. Cancer. 2006;118:1054–1059. doi: 10.1002/ijc.21425. [DOI] [PubMed] [Google Scholar]
- 40.Hubé F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leygue E. Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucleic Recept. Signal. 2007;5:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper C, Vincett D, Yan Y, Hamedani MK, Myal Y, Leygue E. Steroid receptor RNA activator bi-faceted genetic system: heads or tails? Biochimie. 2011;93:1973–1980. doi: 10.1016/j.biochi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AA, Wang X, Deng G, Myal Y, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 44.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, et al. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa M, Christian EL, Michel F. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA. 1998;4:1055–1068. doi: 10.1017/s1355838298980670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgstaller P, Kochoyan M, Famulok M. Structural probing and damage selection of citrulline- and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–4776. doi: 10.1093/nar/23.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beniaminov A, Westhof E, Krol A. Distinctive structures between chimpanzee and human in a brain noncoding RNA. RNA. 2008;14:1270–1275. doi: 10.1261/rna.1054608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novikova IV, Hassan BH, Mirzoyan MG, Leontis NB. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res. 2011;39:2903–2917. doi: 10.1093/nar/gkq1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 53.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 54.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, Karolchik D, Barber GP, Harte RA, Hillman-Jackson J, et al. The ENCODE project at UC santa cruz. Nucleic Acids Res. 2007;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 57.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 60.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lockard RE, Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981;9:5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziehler WA, Engelke DR. Probing RNA structure with chemical reagents and enzymes. Curr. Protoc. Nucleic Acid Chem. 2000;Chapter 6:Unit 6.1. doi: 10.1002/0471142700.nc0601s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols NM, Yue D. Ribonucleases. Curr. Protoc. Mol. Biol. 2008;Chapter 3:Unit 3.13. doi: 10.1002/0471142727.mb0313s84. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011 doi: 10.1126/science.1212642. Epub. [DOI] [PubMed] [Google Scholar]
- 66.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 67.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 68.Regad L, Martin J, Nuel G, Camproux AC. Mining protein loops using a structural alphabet and statistical exceptionality. BMC Bioinformatics. 2010;11:75. doi: 10.1186/1471-2105-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cruz JA, Westhof E. Sequence-based identification of 3D structural modules in RNA with RMDetect. Nat. Methods. 2011;8:513–521. doi: 10.1038/nmeth.1603. [DOI] [PubMed] [Google Scholar]
- 70.Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 71.Pace NR, Thomas BC, Woese CR. The RNA World. 2nd edn. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1999. Probing RNA structure, function and history by comparative analysis; p. 113. [Google Scholar]
- 72.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 73.Chooniedass-Kothari S, Vincett D, Yan Y, Cooper C, Hamedani MK, Myal Y, Leygue E. The protein encoded by the functional steroid receptor RNA activator is a new modulator of ER alpha transcriptional activity. FEBS Lett. 2010;584:1174–1180. doi: 10.1016/j.febslet.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 74.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 75.Colley SM, Leedman PJ. Steroid receptor RNA activator - a nuclear receptor coregulator with multiple partners: insights and challenges. Biochimie. 2011;93:1966–1972. doi: 10.1016/j.biochi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 77.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 78.Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, Brockdorff N, Zakian SM. A dual origin of the xist gene from a protein-coding gene and a set of transposable elements. PLoS One. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.