Abstract
The modified base 5-methylcytosine (m5C) is well studied in DNA, but investigations of its prevalence in cellular RNA have been largely confined to tRNA and rRNA. In animals, the two m5C methyltransferases NSUN2 and TRDMT1 are known to modify specific tRNAs and have roles in the control of cell growth and differentiation. To map modified cytosine sites across a human transcriptome, we coupled bisulfite conversion of cellular RNA with next-generation sequencing. We confirmed 21 of the 28 previously known m5C sites in human tRNAs and identified 234 novel tRNA candidate sites, mostly in anticipated structural positions. Surprisingly, we discovered 10 275 sites in mRNAs and other non-coding RNAs. We observed that distribution of modified cytosines between RNA types was not random; within mRNAs they were enriched in the untranslated regions and near Argonaute binding regions. We also identified five new sites modified by NSUN2, broadening its known substrate range to another tRNA, the RPPH1 subunit of RNase P and two mRNAs. Our data demonstrates the widespread presence of modified cytosines throughout coding and non-coding sequences in a transcriptome, suggesting a broader role of this modification in the post-transcriptional control of cellular RNA function.
INTRODUCTION
The presence of 5-methylcytosine (m5C) in DNA and its role as an epigenetic marker of genome activity is well established (1–3). This has been facilitated in large part by the ease of its detection using bisulfite sequencing, which involves chemical conversion of cytosine (but not m5C) to uracil (4–6). While DNA is relatively devoid of other modifications, 109 modifications have been identified in different classes of RNA across all three domains of life (7). tRNA is a particularly heavily modified RNA class, and m5C sites have been identified in numerous archaeal and eukaryotic tRNAs, commonly around the variable region and the anticodon loop. The modification has been shown to stabilize tRNA secondary structure, affect aminoacylation and codon recognition, and confer metabolic stability (8–13). m5C sites are also found in rRNA where they play roles in translational fidelity and tRNA recognition (14). Interestingly, work that led to the discovery of the mRNA cap structure also detected a low level of internal m5C in mammalian mRNA (15) and viral RNAs infecting mammalian cells (16–18), although specific m5C sites were not mapped and the methylation was not corroborated by all studies at the time (19–23). More recently, it was reported that the methyl-CpG binding protein 2 (MECP2) associates with RNA and can regulate mRNA splicing (24,25) and that reprogramming of cells to pluripotency can be achieved using m5C and pseudouridine-modified mRNAs encoding the four Yamanaka factors (26). These observations have reignited interest in the occurrence and function of m5C in mRNA and other non-coding RNA.
Two m5C methyltransferases (MTases) have been shown to catalyze the m5C modification of eukaryotic RNA. First, NCL1/TRM4 (Nuclear protein 1/tRNA–specific MTase 4) is responsible for all known m5C sites in yeast tRNA (27), however its human ortholog NOP2/Sun domain protein 2 [(NSUN2); also known as Misu (28,29)] may have a much narrower target range, selectively methylating the wobble position of tRNALeu(CAA) prior to intron splicing (30). Secondly, TRDMT1 (tRNA aspartic acid MTase 1, also known as DNMT2), has been shown to methylate position 38 on tRNAAsp(GUC) in eukaryotes (12,31,32), and tRNAVal(AAC) and tRNAGly(GCC) in Drosophila (12). TRDMT1 was previously thought to act as a DNA MTase; however, it is now primarily regarded as an RNA MTase (33). The range of RNA targets for these two enzymes in animals are largely unexplored (34). Importantly, NSUN2 is cell cycle-regulated (35), directly targeted by MYC (myelocytomatosis viral oncogene homolog) and implicated in cancer cell proliferation (28). NSUN2 knockout mice are small and have revealed a role of the enzyme in balancing stem cell self-renewal and differentiation (36). Loss of TRDMT1 enzymatic activity in zebrafish also leads to reduced body size and impaired differentiation of specific tissues (37). Of note, the anticancer drug 5-azacytidine was shown to prevent TRDMT1-dependent cytosine methylation of tRNAAsp(GUC) (38). These observations suggest that cytosine methylation in RNA is important to the control of cell growth and differentiation, thus motivating global screens for the occurrence of m5C in RNA.
Herein we have devised a method for transcriptome-wide detection of modified cytosine residues at single nucleotide resolution by combining RNA bisulfite conversion with next-generation sequencing. We report that RNA cytosine modification pervades the human transcriptome: we discovered over ten thousand novel candidate sites in mRNAs and various non-coding RNA types that are distributed in non-random patterns. Furthermore, we show that known and novel sites appear largely dependent on the m5C-specific MTase NSUN2. These data represent the first high-resolution view of cytosine modification across a transcriptome and provide a basis for exploration of its biological significance for mRNA and non-coding RNA function.
MATERIALS AND METHODS
Unless otherwise stated below, all kits and reagents were used according to the manufacturer’s instructions.
Cell culture and RNAi-mediated methyltransferase knockdown
HeLa cells were cultured in DMEM supplemented with 10% FBS (both GIBCO-Invitrogen), and incubated with 5% CO2 at 37°C. Total RNA was extracted from cells using TRIzol (Invitrogen). For RNAi-mediated knockdown of DNMT1, TRDMT1 and NSUN2, cells were transfected with siGenome SMART pool siRNAs or a non-targeting control pool (Dharmacon) using Lipofectamine RNAiMAX transfection reagent (Invitrogen). Cells were passaged and transfected again 72 h post-initial transfection. RNA was harvested from cells 6 days post-initial transfection. For verification of knockdown, 1 µg of total RNA was used for cDNA synthesis with the SuperScript III Reverse Transcriptase kit (Invitrogen). Real-time PCR was performed using SYBR Green I Master (Roche) on a LightCycler 480 (Roche) instrument and results were analysed using Relative Quantification Software (Roche). Oligonucleotides used for real-time PCR are listed in Supplementary Table S1.
RNA isolation, and bisulfite conversion of RNA
Resuspended RNA was treated with DNase (Ambion), and then phenol/chloroform extracted and re-precipitated with ethanol. The RNA was then enriched for mRNA by two iterations of oligo-dT-selection on magnetic beads using an mRNA isolation kit (New England Biolabs). Briefly, 100 μg of the total HeLa RNA was incubated with 200 pmol of biotin-(dT)18 conjugated to streptavidin magnetic beads (New England Biolabs). The sample was washed, eluted and then resubjected to the same process, followed by rRNA depletion using the RiboMinusTM kit (Invitrogen). About 4 μg of the mRNA-enriched sample was subjected to bisulfite treatment, after addition of 400 pg of in vitro transcribed Renilla luciferase (R-Luc) RNA (39) as a negative control and 4 ng of total HeLa cell RNA to ensure tRNA representation in the sample.
Bisulfite conversion of RNA was performed as previously described (40), with the following modifications. About 4 μg of RNA was mixed in 100 μl of 40% sodium bisulfite (Sigma), 600 μM hydroquinone (Sigma) solution (pH 5.1) and incubated at 75°C for 4 h. The reaction mixture was then desalted by two passages through Micro Bio-spin 6 chromatography columns (Bio-Rad). RNA was desulfonated by adding an equal volume of 1 M Tris (pH 9.0) to the reaction mixture and incubated for 1 h at 75°C, followed by ethanol precipitation.
Conventional bisulfite sequencing
A 220 ng aliquot of bisulfite-converted RNA was converted to cDNA using random hexamers and the Superscript III Reverse Transcriptase kit (Invitrogen). PCR conditions were optimized for each respective primer set (Supplementary Table S2) and the target products were isolated from primers and spurious products by agarose gel electrophoresis followed by band excision and purification using a Gel Extraction Kit (Qiagen). Amplicons were ligated into the pGEM-T Easy Vector system (Promega) and individual clones sequenced to determine bisulfite conversion efficiency of selected RNAs.
Novel m5C candidate sites were verified using new batches of HeLa cell total RNA. Where indicated, negative control transcripts corresponding to the local sequence around the candidate site were generated by in vitro transcription and spiked into the HeLa cell RNA to guard against non-conversion artefacts due to RNA sequence or structure. Briefly, multiple gene constructs were generated from unconverted HeLa cell cDNA to contain a stretch of sequence surrounding selected candidate m5C sites and flanked by unique priming sequences. Amplicons were cloned using the pGEM-T Easy Vector system (Promega) and used as templates for in vitro transcription using the MEGAscript high yield transcription kit (Ambion). Transcribed RNA was purified using MEGAclear columns (Ambion) and 400 pg of each in vitro transcript was spiked into HeLa cell total RNA (4 μg). The pool of RNA was then DNase treated, bisulfite-converted and conventionally sequenced as described above. Oligonucleotides used to generate in vitro transcription constructs and sequencing clones are listed in Supplementary Table S2.
Library preparation and SOLiD™ sequencing of bisulfite-converted RNA
To ensure decapping and phosphorylation of the 5′ end of the RNA, 500 ng of the bisulfite-converted sample was treated with five units of Tobacco Acid Pyrophosphatase (Epicentre Biotechnologies) at 37°C for 45 min followed by phenol/chloroform extraction. The sample was then subjected to T4 Polynucleotide Kinase (New England Biolabs) treatment and subsequent phenol/chloroform extraction. A next-generation sequencing library was prepared using the SOLiD™ Whole Transcriptome Analysis kit (Applied Biosystems). Since the sample was already sufficiently fragmented by the bisulfite-conversion reaction, the RNA fragmentation step prior to adaptor hybridization and ligation was not performed. cDNA with an approximate insert length of 50–120 nt were selected by polyacrylamide gel electrophoresis. Beads were prepared, deposited and sequenced on a SOLiD™ Version 3 instrument (Applied Biosystems). Sequenced read data was deposited in the National Center for Biotechnology Information sequence read archive (accession number SRA027832.1).
Sequenced read mapping and analysis
Sequenced reads were mapped against reference sequences consisting of all known human transcripts from the Ensembl v61 database (mRNA, rRNA, tRNA, mitochondrial RNA and all non-coding RNA) (41), predicted and known tRNA sequences obtained from the GtRNAdb (42) and tRNAdb (43), miRNA hairpin sequences from miRBase v16 (44), rRNA sequences obtained from the NCBI RefSeq database, and R-Luc spike-in negative control sequences (Supplementary Table S3). A ‘CCA’ sequence was appended to tRNAs lacking one since this non-templated addition is common to tRNA sequences. To reduce mapping ambiguity, identical sequences were collapsed and represented only once in the reference.
Sequenced reads were mapped using the SOCS-B program (45), an alignment tool designed to map bisulfite-converted SOLiD™ colour-space reads to nucleic acid reference sequences. This program disregards mismatches between ‘C’ in the reference and ‘T’ introduced in reads as a result of bisulfite conversion. SOCS-B mapping parameters were chosen such that low-quality reads (average quality across a read <18) and reads with ambiguous colour-call represented by ‘.’ in the colour-space sequence were discarded. During mapping, up to four colour-space mismatches were allowed between the reference and read sequences to accommodate sequencing errors and natural variation. Given the large proportion of ‘predicted’ tRNA sequences in the reference, reads multi-mapping to both known and predicted tRNA sequences were preferentially assigned to the known tRNA sequence. For all other reads, SOCS-B was set to assign multi-mapping reads to a single randomly selected mapped locus. Reads mapping to the anti-sense strand of the reference were discarded.
Identification of modified cytosine sites in RNA
Non-conversion of a cytosine in read sequences was taken to indicate the presence of m5C. To delimit m5C site prediction to high confidence candidates, threshold parameters were set to a read coverage ≥10 and conversion rate ≤80% per cytosine position in the reference (see ‘Results’ section). Read coverage was defined as the number of reads that overlap a given cytosine. The following equation describes the conversion rate calculations.
 |
Read coverage and cytosine conversion rate were calculated directly from mapped data for tRNA, rRNA and R-Luc spike-in negative control sequences. For all other RNA types, cytosine-mapping data was transferred to genomic coordinates before applying threshold criteria (Supplementary Figure S1). Ensembl Perl API v61 was used for coordinate transfers from transcript sequence to the human genome assembly version GRCh37 (hg19).
Analysis of modified cytosine location bias within the transcriptome
Enrichment of non-converted sites across genomic regions was computed by chi-squared and binomial tests, relative to the proportion of all potential sites (cytosines with read coverage ≥10) in each measured category. Enrichment of methylation sites for regulatory elements was estimated by overlap with the 3′-UTR structural RNA prediction set of (46) (binomial test relative to the proportion of structured to unstructured nucleotides in 3′UTR).
Enrichment of non-converted sites near binding regions for Argonaute (I–IV) and Pumilio 2 proteins was estimated by overlapping site coordinates with the PAR-CLIP dataset of (47) mapped as described in (48). Putative RNA binding protein (RBP) binding sites here defined as regions with more than one PAR-CLIP read. P-value of enrichment was computed by permutation test against a shuffled set of positions within each genomic region. A plot of RBP binding site density in the vicinity of the m5C sites was computed by averaging a 2000 bp window centred on all methylation sites.
RESULTS
A transcriptome-wide survey of m5C candidate sites in human RNA
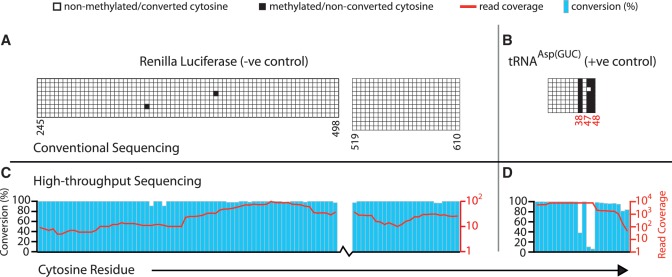
To develop a method for single nucleotide mapping of m5C sites in a cellular transcriptome we adapted an RNA bisulfite conversion protocol, originally devised for primer extension-based detection of m5C (40), for use with a sequencing-based readout (see ‘Materials and Methods’ section for details). As in DNA bisulfite sequencing, sites of m5C in RNA will be read as cytosine in cDNA sequence, while unmodified cytosines will appear as thymidine. Although some other types of modified cytosine can also be resistant to bisulfite treatment (see ‘Discussion’ section), for simplicity we refer in the following to sites of non-conversion as ‘m5C candidate sites’. We chose to analyse RNA preparations from HeLa cells (a human cervical cancer cell line) and used both positive and negative control RNAs to monitor success of the procedure. Our positive control was the endogenous tRNAAsp(GUC), which harbours three previously identified m5C sites at structural positions 38, 47 and 48 (7,31,32). Our negative control was in vitro transcribed Renilla luciferase (R-Luc) mRNA lacking m5C, which was spiked into the cellular RNA sample prior to bisulfite treatment. An aliquot of this RNA was used for conventional bisulfite sequencing reactions to examine the expected m5C patterns in our controls. The R-Luc negative control RNA exhibited virtually complete cytosine conversion (Figure 1A; 99.8% conversion overall), while all three known m5C sites in tRNAAsp(GUC) selectively displayed low levels of conversion (Figure 1B; position 38 and 48: 0%, position 47: 12.5% conversion). These results showed that our RNA conversion protocol was efficient and accurate detection of m5C sites is achieved.
Figure 1.
Single-nucleotide resolution mapping of m5C candidate sites in RNA. HeLa cell RNA preparations were spiked with a trace amount of in vitro transcribed R-Luc RNA and bisulfite-converted as detailed in the ‘Materials and Methods’ section. (A) Negative control R-Luc and (B) endogenous tRNA(AspGUC) as a positive control were first analyzed by conventional sequencing to establish the efficacy of bisulfite conversion (top panels; columns signify cytosine positions along the RNA sequence, rows represent individually sequenced alleles, open boxes indicate cytosine to uracil conversion read as thymidine in cDNA and filled boxes indicate a retained cytosine). Numbers below refer to cytosine positions in the primary RNA sequence. Nucleotide positions highlighted in red designate previously identified m5C sites in tRNA(AspGUC). Dual axis charts (bottom panels) display next-generation sequencing data mapped at 2 mm for the same control RNAs. Blue bars represent bisulfite-induced cytosine conversion, while red lines represent read coverage across individual residues. Top and bottom panels are aligned to each other by interrogated cytosine residues.
We then performed SOLiD™ next-generation sequencing of the converted RNA sample and obtained ∼96 million sequence reads (50 nt in length). Approximately 41 million reads could be mapped to a custom human transcriptome reference (described in the ‘Materials and Methods’ section) using the SOCS-B program (45) and allowing up to four colour-space mismatches without counting those resulting from a C to T sequence change (indicative of bisulfite conversion). We next analysed the coverage and extent of conversion at cytosine residues in our controls, setting successively more stringent cut-offs, from four to zero-mismatch (4–0 mm) mapping data. Our controls had sufficient coverage across cytosines and displayed the expected C to T conversion patterns (Figure 1C and D show results for 2 mm data; see also Table 1 and Supplementary Table S4). R-Luc RNA showed almost complete C to T conversion overall (e.g. 99.5% at 2 mm; Figure 1C), and no individual position showed less than 90% conversion at a coverage threshold of 10 irrespective of the number of mismatches allowed. The three known m5C sites in tRNAAsp(GUC) showed low levels of conversion; this was consistent at all stringencies of mapping (e.g. 17.9% at 2 mm; average across all three sites; Figure 1D). These data indicate that our combination of bisulfite conversion with next-generation sequencing can accurately detect m5C sites in RNA, and importantly, has a low false-positive rate.
Table 1.
Number of cytosine residues identified as m5C in different RNA categories in 2 mm data
| RNA type | # of Cytosine | # of Cytosine with ≥10 read coverage | # of m5C (≤80% conversion rate) | % m5C |
|---|---|---|---|---|
| R-Luc | 255 | 232 | 0 | 0.0 |
| tRNA | 14 584 | 2943 | 255 | 8.7 |
| mRNA | 16 785 229 | 2 247 702 | 8495 | 0.4 |
| Other non-coding RNA | 5 783 482 | 144 799 | 1780 | 1.2 |
Discovery of novel m5C candidate sites
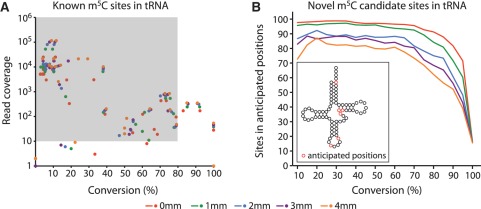
We next examined all reads mapping to cytoplasmic and mitochondrial tRNA sequences. There are 28 known m5C sites in human tRNAs (43) and 27 of these were detectable in our dataset with at least one read spanning the site (Supplementary Dataset S1, Supplementary Table S5). Confirming our approach, we called the large majority of these sites as modified when applying a C to T conversion threshold of ≤80% and read coverage of ≥10 (Figure 2A; 21 sites called as modified, with 24 sites having sufficient coverage when allowing ≥1 mm). We next extended our analysis to all cytosines in tRNAs. To reduce over- or under-calling of sites due to multi-mapping to highly similar tRNA sequences, reads were preferentially aligned to known tRNA sequences over predicted sequences (42,43). In animals, all known m5C sites are found at six discrete locations within the tRNA secondary structure (see inset in Figure 2B) (7,34,43). Thus, we posited that genuine novel m5C candidate sites in tRNAs should also be located primarily, if not exclusively, at these positions and used this notion to further refine the parameters for novel m5C site selection from our data. Up to a conversion threshold of 60% we found ∼80% or more of the novel candidate sites in these anticipated positions across all mismatch datasets, but for those with ≤2 mm this proportion remained at this level up to a conversion threshold of 80% (Figure 2B). Thus, taking into account our analyses of both known and novel tRNA sites, we defined ≤80% C to T conversion and ≥10 read coverage as suitable criteria to assign candidacy to novel m5C sites and applied them to reads mapped up to two colour-space mismatches.
Figure 2.
Defining parameters for m5C candidate site selection using tRNA data. (A) Plot of conversion against read coverage at 28 known m5C sites in human tRNAs (43). The majority of the previously identified tRNA m5C sites had a conversion of 80% or less and read coverage of at least 10 (shaded area). (B) Dependence of the proportion of novel tRNA candidate sites in anticipated tRNA structural positions (red circles in tRNA cloverleaf cartoon) on chosen conversion cut-off. Read coverage threshold was ≥10. Colour code for dots and lines refers to mapping at different colour-space mismatch limits.
In total, we detected 255 modified cytosine candidate sites in tRNA sequences using the above threshold criteria (Table 1 and Supplementary Dataset S2). Of these sites, 21 were previously known as m5C in human tRNAs, for 51 cases other human isodecoder tRNA variants were known to harbour m5C at those sites, 68 sites were reported as methylated in other animal tRNA orthologs, while 115 sites were entirely novel before this study.
Within rRNA sequences, we clearly detected two previously known m5C sites at positions 3782 and 4447 in human 28S rRNA (7,32). We independently verified these sites as well as a site within the decoding centre of 12S mitochondrial rRNA (position 841; Supplementary Figure S2) that was previously identified as m5C in hamster mitochondrial 12S rRNA (49,50). However, we also saw several prominent clusters of non-conversion (e.g. clusters of 10 or more sites in a 50-nt window, Supplementary Dataset S2), particularly in the cytoplasmic and mitochondrial large subunit rRNAs, which are among the longest, highly structured cellular RNAs. These are likely due to incomplete denaturation of some highly structured regions of RNAs, thus we did not consider further our mapping data for rRNAs. Only ∼7% of the m5C sites seen in other cellular RNAs occurred in such clusters, indicating a low proportion of false positive m5C predictions due to RNA structure in our data.
We then applied our selection criteria to identify novel m5C candidate sites throughout the transcriptome. To simplify the interpretation of results, we transferred the cytosine mapping information for the remaining RNA types to genomic co-ordinates and calculated cytosine conversion and coverage for each genomic locus. Approximately 2.4 million cytosine positions in the genome corresponding to known transcripts were covered by at least 10 reads, representing 10.6% of the total cytosines in known transcripts. Of these, 10 275 sites (0.43% of all assessed cytosines) were identified with ≤80% conversion rate; 1863 sites remained when applying the highly stringent requirement of ≤50% conversion rate. Genomic coordinates, extent of conversion and additional information on all identified sites are listed in Supplementary Dataset S2. About 1780 of these sites were present in a range of non-coding RNA types, including lincRNA, pseudogenes and processed pseudogenes, while 8495 sites were located in mRNA sequences (Table 1).
Validation of m5C candidate sites by conventional bisulfite sequencing
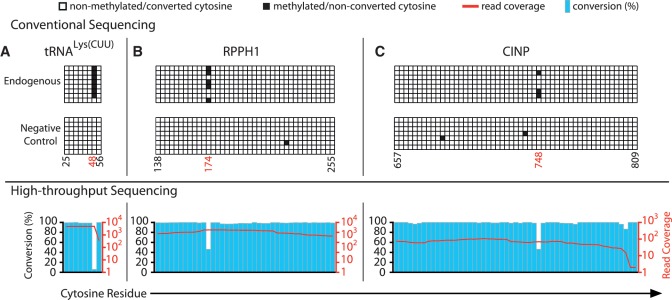
To validate our global site mapping data, we verified three candidate sites from mRNAs, eight from tRNAs, and two from other non-coding RNAs by conventional bisulfite sequencing (Figure 3 and Supplementary Figures S2 and S3). Among the selected sites there were six that had been previously identified as m5C in other animal homologs (tRNAGly(UCC), 3 sites; tRNALys(CUU), 1 site; mitochondrial tRNAGlu(UUC), 1 site; 12S mitochondrial rRNA, 1 site). The three sites in mitochondrial tRNASer(GCU) were entirely novel, as those were chosen in protein-coding mRNAs (cyclin-dependent kinase 2 interacting protein, CINP; nicotinate phosphoribosyltransferase domain containing 1, NAPRT1; NADH dehydrogenase 1 beta subcomplex 7 mRNA, NDUFB7; 1 site each) and a non-coding RNA (RPPH1, Ribonuclease P RNA component H1). To assess potential non-conversion due to local RNA structure, we designed and prepared in vitro transcribed negative control transcripts mimicking the sequence context of five of the sites to be verified. These negative controls were spiked into independently prepared HeLa cell RNA prior to bisulfite treatment. None of these negative controls showed any evidence of non-conversion at the test sites, whereas all sites except position 47 in tRNAGly(UCC were convincingly confirmed (Figure 3 and Supplementary Figure S3). This demonstrated that most of the novel m5C candidate sites predicted by our next-generation sequencing-based mapping are independently verifiable.
Figure 3.
Validation of novel m5C candidate sites. Conventional bisulfite sequencing data is shown for three novel sites, (A) residue 48 in tRNA(LysCUU), (B) residue 174 in the RNase P RNA component H1 (RPPH1), and (C) residue 748 in cyclin-dependent kinase 2 interacting protein mRNA (CINP). Top panels display results for endogenous transcripts. Data for spiked-in in vitro transcribed negative controls harboring the same sequence flanked by unique priming sites are also shown (middle panels) as are corresponding next-generation sequencing results (lower panels). Numbering of cytosine positions is as described in Figure 1, positions highlighted in red designate m5C sites identified by next- generation sequencing. See Supplementary Figure 3 for additional validation data.
Identification of NSUN2 and TRDMT1 dependent m5C sites
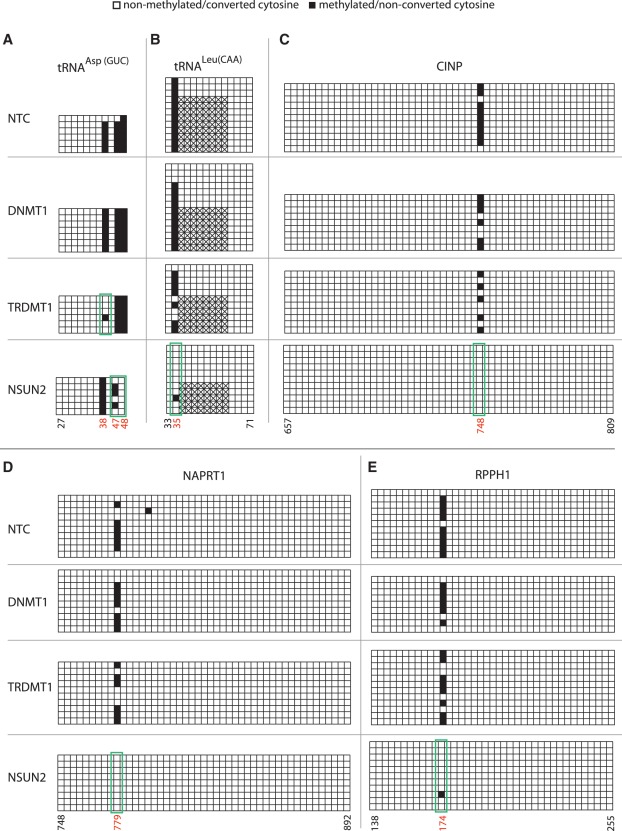
To gain insight into the enzymes mediating RNA methylation, we performed RNAi-mediated knockdown of the RNA methyltransferases TRDMT1 and NSUN2 by transient transfection of HeLa cells. Knockdown of the DNA methyltransferase DNMT1 was used as a negative control, with knockdown efficiency determined by quantitative RT–PCR (Supplementary Figure S4). Conventional bisulfite sequencing of target regions in tRNAAsp(GUC) and tRNALeu(CAA) revealed marked disappearance of known target m5C sites for TRDMT1 (residue 38 in tRNAAsp(GUC)) and NSUN2 (residue 34 in tRNALeu(CAA)) in the respective knockdown cells (Figure 4A and B). Furthermore, methylation of residues 47 and 48 in tRNAAsp(GUC) was also shown to disappear with NSUN2 knockdown. Novel sites in CINP and NAPRT1 mRNAs, and in the non-coding RPPH1, were all clearly NSUN2-dependent (Figure 4C–E). Two previously identified m5C sites in 28S rRNA, as well as two novel candidate sites in 12S mitochondrial rRNA and tRNALys(CUU) did not respond to either enzyme knockdown (Supplementary Figure S2). Modifications at these sites may be placed by other MTases, or they may correspond to other cytosine modifications that could protect against bisulfite conversion (see ‘Discussion’ section); most likely these sites reside in RNA species that are not turned over rapidly enough to be amenable to assay in short-term knockdown experiments. Of the 11 sites tested, we found that six were dependent on NSUN2 and one was dependent on TRDMT1, also corroborating the identity of the modification at these sites as m5C. This further implies a major role for NSUN2 in modifying the human transcriptome, increasing the number of its known target sites from one to six, and extending its substrate range to an additional tRNA as well as a non-coding RNA and two mRNAs.
Figure 4.
Analysis of methyltransferase target sites. HeLa cells were transfected with siRNAs targeting DNMT1, TRDMT1 and NSUN2 or a non-targeting control siRNA (NTC) as indicated on the left. Conventional bisulfite sequencing data was obtained as described in Figure 1 and is shown for (A) tRNAAsp(GUC) (residue 38 is a known TRDMT1 target) and (B) tRNALeu(CAA) (residue 34 is a known NSUN2 target) (C) CINP and (D) nicotinate phosphoribosyltransferase domain containing 1 (NAPRT1) mRNAs, and (E) RPPH1 non-coding RNA. Nucleotide positions highlighted in red below designate m5C sites identified by next-generation sequencing. Green boxes indicate sites selectively responding to MTase depletion. Hatched boxes indicate intronic sequence. See Supplementary Figure S4 for data on RNAi knock-down efficiency and Supplementary Figure S2 for analysis of additional target sites.
Analysis of m5C location bias within the transcriptome
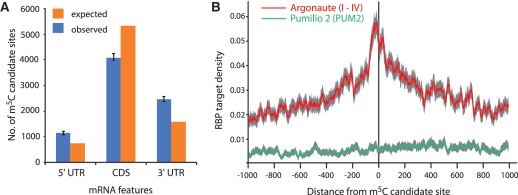
To discover the wider role of m5C in the human transcriptome, we searched our data for any bias in the location of m5C candidate sites. To this end, we considered a subset of 9177 sites residing in canonical transcripts (longest known cDNA coding sequence; 89% of all predicted sites). The distribution of m5C candidate sites varied across different RNA types (P-value <2.2 × 10−16, chi-squared test). Sites were significantly enriched in a variety of non-coding transcript types, including several pseudogene categories (P-value <2.2 × 10−16, binomial test), whereas they were significantly depleted in mRNA (P-value <2.2 × 10−16, binomial test) (Supplementary Table S6). Given that many non-coding RNAs are expected to be of low abundance, we deemed our dataset to be of insufficient coverage to further analyse the patterns of m5C distribution in these RNA types. By contrast, despite being relatively devoid of m5C, candidate sites in mRNAs constituted the great majority (∼83%) identified in our screen. We therefore, searched for any bias in site distribution within mRNA sequences. This revealed a significant enrichment within the untranslated regions (both 5′ and 3′ UTRs), and a relative depletion within coding regions (P-value <2.2 × 10−16, binomial test) (Figure 5A). Gene ontology enrichment analysis using the elim method (51) of genes harbouring m5C candidate sites in different transcript regions (3′UTR, 5′UTR and CDS) did not show any statistically significant enrichment at an FDR of 0.05 (data not shown).
Figure 5.
Analysis of bias in m5C candidate site location within mRNA. (A) Histogram showing relative enrichment of m5C sites in 3′ UTR and 5′ UTR compared to CDS (Error bars = 95% confidence interval; ‘Poisson’ distribution). (B) RNA-binding protein target density versus distance from m5C site in protein coding transcripts (CDS and UTR both). Confidence bands are shown in grey (SEM).
A complementary approach to look for functional links was to test for any spatial association between m5C candidate sites and cis-acting regulatory motifs within RNA. Thus, we overlaid our modification mapping data with a set of recently identified human regulatory RNA elements within 3′ UTRs (46). This showed a moderate but statistically significant enrichment of those elements in the vicinity of m5C sites (1.42-fold; P-value = 0.04, binomial test; data not shown). Furthermore, we overlaid our mapping data with publicly available transcriptome-wide maps of binding sites for major regulatory RNA-binding proteins in human cells generated by PAR-CLIP-Seq (47). This showed a substantial association of m5C candidate sites in mRNA with binding regions for the Argonaute I-IV proteins (Figure 5B), the central components of the miRNA/RISC complex (∼1.76-fold over 3′UTR; P-value <1 × 10−3, permutation test) but, interestingly, not for binding sites of another post-transcriptional regulator, the Pumilio 2 protein (1.15 fold; P-value = 0.13). We also inspected the local sequence context for candidate sites and saw an increased frequency of C and G as flanking bases and a depletion of U either side of the modified cytosine, but no strict requirement for a targeting context was evident (data not shown). While more parsimonious analyses of subclasses of m5C candidate sites may yield further insight, it appears that m5C context requirements differ from those for m6A where a clear sequence context was reported [e.g. Gm6A in yeast (52)]. The enrichment patterns detailed above persisted when the analyses were done with more stringently selected candidate sites (i.e. ≤50% conversion rate), and altogether they are highly suggestive of an involvement of cytosine modification in post-transcriptional gene regulation.
DISCUSSION
Herein we described the successful combination of bisulfite treatment with next-generation sequencing of a cellular RNA library. This allowed us to survey the modification status of over two million cytosine sites in the transcriptome of the human HeLa cell line, which led to the mapping of over ten thousand novel candidate m5C sites in diverse types of cellular RNA (Table 1; Supplementary Dataset S2). The accuracy of our approach and site selection criteria was supported by multiple tests, including verification of a subset of known and novel sites by conventional bisulfite sequencing and m5C MTase knockdown.
Some cytosine modifications other than m5C may also be resistant to bisulfite treatment, including 3-methylcytidine, N4-methylcytidine (m4C), N4,2’-O-dimethylcytidine (m4Cm), and N4-acetylated variants. Schaefer et al. (32) were able to detect m4Cm at position 1402 of bacterial 16S rRNA by locus-specific bisulfite sequencing. The equivalent region in mitochondrial 12S rRNA from the hamster is methylated in the motif ‘Gm4CCm5CG’ (49,50). We surveyed the orthologous region of the human counterpart by next generation and conventional sequencing and could only detect the m5C site (Supplementary Figure S2). This may suggest that human mitochondrial 12S rRNA does not carry the m4C modification or that m4C and m4Cm display differential sensitivity to bisulfite conversion. Perhaps, more likely, it reflects differences in treatment protocols as used by these colleagues and us. On balance, our evidence is consistent with m5C as the modification underlying the majority of novel candidates sites reported here, although definitive proof for any given site would require application of more elaborate direct detection methods. Transcriptome-wide bisulfite sequencing of material from cells depleted for specific MTases would be a further tractable option to confirm modification identity on a global scale. Our current method also has limits with regard to surveying extremely structured regions such as those found in rRNA, which are among the longest, highly structured RNAs, so we excluded suspect regions of rRNA from further analysis. In addition, we included unmodified spike-in sequences in our verification efforts to control for effects of local structure (Figure 3), and verified sites through other means, such as MTase knockdown (Figure 4). While this work was in progress, Schaefer and colleagues used their locus-specific bisulfite sequencing approach to detect known m5C sites in several tRNAs and rRNAs in different contexts (12,32,38), further underscoring the validity and scope of our transcriptome-wide method. Future efforts to improve the method will focus on optimization of RNA denaturation and conversion, while sufficiently preserving RNA integrity.
Our study detected 255 candidate sites for cytosine modification in tRNAs, most of them not previously mapped in humans. We found that most high-quality candidate sites were found at positions within tRNA secondary structure known to be occupied by m5C in animals (43) (Figure 2), broadly confirming existing expectations of the role of m5C in modulating tRNA function (8–13). Nevertheless, an emerging facet of tRNA biology is their processing into smaller regulatory RNAs species (53), and TRDMT1-mediated placement of m5C has been shown to protect several tRNAs against stress-mediated cleavage in Drosophila (12). The present work thus provides a wealth of mapped m5C sites that can now be assessed for a role in this phenomenon.
Human NSUN2 was thought to have a narrow substrate range, possibly confined to a single site within tRNALeu(CAA) (30). In contrast, our evidence demonstrates that its substrate range is likely to be much broader, as it is responsible for modification of further sites in tRNAAsp(GUC), the RNA subunit RPPH1 of RNase P (54), and even mRNAs (Figure 4). The unexpected relationship between a tRNA-modifying and a tRNA-processing enzyme is intriguing and experiments will now have to address whether RPPH1 modification affects the enzymatic properties of RNase P. In support of a broader role of NSUN2 in mRNA modification it was found to be among the most highly enriched proteins in an in vivo capture screen for HeLa cell proteins binding to polyadenylated RNA (A Castello & MW Hentze, personal communication). NSUN2 is frequently overexpressed in a number of different tumour types and varies in its expression and intracellular localization (i.e. nucleolar versus cytoplasmic) during the cell cycle, while its enzymatic activity is regulated by phosphorylation (28,35,55). Importantly, it furthermore functions in balancing stem cell self-renewal and differentiation (36). Given the broad role for NSUN2 in modifying different RNA types we uncovered here, it is thus plausible that aspects of the transcriptome-wide RNA methylation patterns we have observed are dynamically responding to, and/or supportive of, distinct states of cellular growth, differentiation and transformation. Comparative studies of differential RNA methylation, particularly in the cancer and stem cell contexts, and in cells depleted of the candidate RNA MTases are now warranted.
A key outcome of this study was the clear demonstration of a widespread presence of m5C within mRNAs. Pioneering work in the 1970’s had suggested the presence of m5C, as well as N6-methyladenosine (m6A) in mRNA, but their low abundance combined with a lack of suitable methods to map individual sites hampered investigation of their relevance. The identification of the methyltransferase responsible for m6A in mRNA subsequently allowed progress in its investigation (e.g. 52,56), but work on m5C in mRNA had all but seized since then. We detected candidate sites for cytosine modification in several thousand mRNAs, indicating that the presence of m5C is not limited to a highly specialized subset of mRNAs. Our analyses also indicate that m5C distribution within the transcriptome is not random, as we found the modification enriched within non-coding RNAs, whereas functional mRNAs were relatively devoid of it. Analyses of datasets with more complete mRNA coverage will be required to more precisely map the preferred locations of m5C within mRNAs, as they will likely hold clues to its molecular function(s). Our finding of an enrichment of m5C within the UTRs of mRNA and in the vicinity of Argonaute protein binding regions, already presents an intriguing pretext to further investigate a role of m5C in post-transcriptional gene regulation. Possibilities include, but are not limited to, a role in protecting mRNAs against innate antiviral defence mechanisms (26) or promoting their efficient translation, as has been suggested for m6A (52).
The potential of RNA editing and modification to further diversify functions of cellular transcriptomes has recently come to the fore (57). We provided here a first global map of the patterns of m5C modification in the human transcriptome, which link it to mechanisms of post-transcriptional gene regulation and control of cell growth and differentiation. Our approach to the transcriptome-wide mapping of m5C can now be utilized to survey distribution patterns of this modification in different organisms and tissues, as well as to assess its potential for dynamic change under different cellular conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–4, Supplementary Datasets 1–2.
FUNDING
The Victor Chang Cardiac Research Institute and The John Curtin School of Medical Research, as well as grants from the National Health & Medical Research Council (573726 and 514904 to T.P.). Funding for open access charge: The John Curtin School of Medical Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arthur Liu, Traude Beilharz and Cheryl Li for their contributions to early stages of this work, Jennifer Cropley and Paul D. Waters for helpful suggestions on the manuscript, and Gavin Huttley for allowing usage of computer resources. Author Contributions: J.E.S., M.N., C.M.S, and T.P. designed research; J.E.S. and T.S. performed research; J.E.S., H.P., and D.T.H. analysed data; B.J.P. identified and analyzed association of m5C with Ago-binding regions and structural motifs; J.E.S., C.M.S, H.P., B.J.P. and T.P. wrote the paper. All authors read and approved the final article.
REFERENCES
- 1.Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–238. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 4.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson JT, Wang XY. Nuclear RNA surveillance: no sign of substrates tailing off. Crit. Rev. Biochem. Mol. Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 10.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squires JE, Preiss T. Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics. 2010;2: 709–715. doi: 10.2217/epi.10.47. [DOI] [PubMed] [Google Scholar]
- 14.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubin DT, Stollar V. Methylation of Sindbis virus “26S” messenger RNA. Biochem. Biophys. Res. Commun. 1975;66:1373–1379. doi: 10.1016/0006-291x(75)90511-2. [DOI] [PubMed] [Google Scholar]
- 17.Dubin DT, Stollar V, Hsuchen CC, Timko K, Guild GM. Sindbis virus messenger RNA: the 5′-termini and methylated residues of 26 and 42 S RNA. Virology. 1977;77: 457–470. doi: 10.1016/0042-6822(77)90471-8. [DOI] [PubMed] [Google Scholar]
- 18.Sommer S, Salditt-Georgieff M, Bachenheimer S, Darnell JE, Furuichi Y, Morgan M, Shatkin AJ. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976;3:749–765. doi: 10.1093/nar/3.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255:28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 20.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl Acad. Sci. USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavi S, Shatkin AJ. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl Acad. Sci. USA. 1975;72:2012–2016. doi: 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976;7:227–237. doi: 10.1016/0092-8674(76)90022-2. [DOI] [PubMed] [Google Scholar]
- 24.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 25.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl Acad. Sci. USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 32.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 34.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakita-Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora-B regulates RNA methyltransferase NSUN2. Mol. Biol. Cell. 2007;18:1107–1117. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7:e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 39.Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell. Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ondov BD, Cochran C, Landers M, Meredith GD, Dudas M, Bergman NH. An alignment algorithm for bisulfite sequencing using the Applied Biosystems SOLiD System. Bioinformatics. 2010;26:1901–1902. doi: 10.1093/bioinformatics/btq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker BJ, Moltke I, Roth A, Washietl S, Wen J, Kellis M, Breaker R, Pedersen JS. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011;21:1929–1943. doi: 10.1101/gr.112516.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen J, Parker BJ, Jacobsen A, Krogh A. MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action. RNA. 2011;17:820–834. doi: 10.1261/rna.2387911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baer RJ, Dubin DT. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981;9:323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubin DT, HsuChen CC. The 3′-terminal region of mosquito mitochondrial small ribosomal subunit RNA: sequence and localization of methylated residues. Plasmid. 1983;9:307–320. doi: 10.1016/0147-619x(83)90008-2. [DOI] [PubMed] [Google Scholar]
- 51.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 52.Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Esakova O, Krasilnikov AS. Of proteins and RNA: The RNase P/MRP family. RNA. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, Ogawa I, Takata T, Shimamoto F, Tatsuka M. Frequent increased gene copy number and high protein expression of tRNA (Cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2011 doi: 10.1089/dna.2011.1446. [DOI] [PubMed] [Google Scholar]
- 56.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.