Abstract
The microRNA (miRNA)-mediated repression of protein synthesis in mammalian cells is a reversible process. Target mRNAs with regulatory AU-rich elements (AREs) in their 3′-untranslated regions (3′-UTR) can be relieved of miRNA repression under cellular stress in a process involving the embryonic lethal and altered vision family ARE-binding protein HuR. The HuR-mediated derepression occurred even when AREs were positioned at a considerable distance from the miRNA sites raising questions about the mechanism of HuR action. Here, we show that the relief of miRNA-mediated repression involving HuR can be recapitulated in different in vitro systems in the absence of stress, indicating that HuR alone is sufficient to relieve the miRNA repression upon binding to RNA ARE. Using in vitro assays with purified miRISC and recombinant HuR and its mutants, we show that HuR, likely by its property to oligomerize along RNA, leads to the dissociation of miRISC from target RNA even when miRISC and HuR binding sites are positioned at a distance. Further, we demonstrate that HuR association with AREs can also inhibit miRNA-mediated deadenylation of mRNA in the Krebs-2 ascites extract, in a manner likewise depending on the potential of HuR to oligomerize.
INTRODUCTION
MicroRNAs (miRNAs) are ∼21-nt-long non-coding RNAs acting as post-transcriptional regulators of gene expression in eukaryotes. In mammals, hundreds of different miRNAs are expressed and they are predicted to control the activity of ∼50% of all genes. Thus, miRNAs regulate most of the investigated developmental and cellular processes and their altered expression is observed in many human pathologies, including cancer (1–5).
In metazoa, miRNAs regulate gene expression by base pairing to target messenger RNAs (mRNAs), bringing about their translational repression and/or deadenylation, which leads to mRNA degradation. Generally, miRNAs base pair imperfectly with sequences in mRNA 3′-untranslated regions (3′-UTRs), where positions 2–8 of the miRNA, referred to as a seed sequence, are most important for this association. However, some miRNAs base pair with perfect or nearly perfect complementarity and induce endonucleolytic cleavage of mRNA by an RNA interference (RNAi) mechanism, similar to that mediated by short interfering RNAs (siRNAs) (1,6–9).
miRNAs function in the form of ribonucleoprotein particles, miRNPs or miRISCs (miRNA-induced silencing complexes). Argonaute (Ago) proteins are the best-characterized essential components of miRISC and four Ago proteins, Ago1 through Ago4, all directly associate with miRNAs in mammals and function in the repression. Of the four mammalian Ago proteins, only Ago2 is catalytically competent to endonucleolytically cleave the target RNA. GW182 proteins are additional important components of miRISC. They associate with Ago proteins and function as effectors in the repression (8,9).
Until recently, miRNAs were identified primarily as negative regulators of cellular mRNAs and it was not known whether the inhibition of a particular mRNA can be effectively reversed. The ability to disengage miRNAs from the repressed mRNA, or render them inactive, would result in miRNA regulation being more dynamic and more responsive to specific cellular events. Indeed, it was found recently that miRNA repression of different mRNAs can be strongly modulated or even reversed by factors as diverse as cellular stress, developmental cues or neuronal stimulation (10–27). Moreover, miRNAs in cell cycle-arrested cells were found to activate rather than repress translation of selected mRNA targets (28–30). Many of these reports document a crucial role for RNA-binding proteins (RBPs) in modulating miRNA function. Hundreds of different RBPs are expressed in metazoan cells and dozens of them are known to interact, like miRNAs, with mRNA 3′-UTRs and regulate mRNA expression and stability (31). Since 3′-UTRs of mammalian mRNAs can be as long as 10 or more kilobases and can associate with many different miRNAs and RBPs, these findings indicated a potentially very complex interplay between the two classes of regulators interacting with the 3′-UTR (23,24).
Previously, we demonstrated that CAT-1 mRNA that encodes the high-affinity cationic amino acid transporter and which is translationally repressed by miR-122 in human Huh7 hepatoma cells, can be relieved of inhibition by subjecting cells to various stress conditions. We also found that the stress-induced alleviation of miR-122 repression requires the binding to the CAT-1 mRNA 3′-UTR of HuR, an RBP known to interact with AU-rich elements (AREs) (11). Reporter RNAs bearing sites targeted by let-7 miRNA showed regulation similar to that of CAT-1 mRNA and reporters bearing the CAT1 3′-UTR (11). HuR is an ubiquitously expressed member of the embryonic lethal and altered vision (ELAV) family of proteins, which also includes three neuron-specific proteins, HuB, HuC and HuD (32–34). HuR consists of three RNA-binding RRM domains, with RRM1 and RRM2 together being responsible for binding to the ARE. HuR and other members of the ELAV family have the potential to oligomerize along RNA and a hinge region separating RRM2 and RRM3, and RRM3 of ELAV proteins were shown to be important for this process (35–40). Although HuR is predominantly a nuclear protein, it shuttles between the nucleus and the cytoplasm and the hinge region is essential for shuttling and nuclear accumulation of the protein (41). In response to different types of cellular stress, HuR translocates from the nucleus to the cytoplasm, where it modulates the translation and/or stability of many mRNAs (34,42,43).
Although the role of HuR in the reversal of miRNA repression of CAT-1 mRNA as well as its function in modulating miRNA-mediated repression of other mRNAs, is well documented (11,12,17,21,22), the mechanism of these HuR effects has not been established. Likewise, it is not known whether factors other than HuR, possibly induced in cells subjected to stress, participate in the process. In the present study, we have used recombinant HuR and its mutants, and purified miRISC to study the derepression process in vitro.
MATERIALS AND METHODS
Expression plasmids
Plasmids expressing FLAG-HA-Ago1, FLAG-HA-Ago2 and the FLAG-HA tagging vector were obtained from G. Meister (44). Myc-Ago2, Ago2D669A and Ago2H634A mutant clones were generously provided by G. Hannon (45). Full-length cDNAs-encoding mutant Ago2 were amplified from Myc-Ago2 plasmids and subcloned into FLAG-HA tagging vector between NotI and EcoRI sites. pLET7a encoding pri-let-7a was kindly provided by J. Belasco (46). For expression of HuR and mutant proteins [HuRΔH: hinge region (H) (amino acids 186–242) deleted; HuRΔ3: RRM3 (amino acids 243–326) deleted; and HuRΔH3: both hinge and RRM3 (amino acids 186–326) deleted] in Escherichia coli, appropriate regions were PCR amplified from plasmids kindly provided by A. B. Shyu (47) and the inserts were cloned in pET42a(+) (Novagen) between NdeI and XhoI sites for His-tag purification. Plasmids HA-Ago3 (48) and HA-TNRC6B (49) were described before. Primers for amplification are presented in Supplementary Table S1. Correctness of all plasmids was confirmed by sequencing.
Purification of His6-HuR fusion proteins
Overnight cultures of E. coli BL21, transformed with plasmids expressing HuR or its mutants were diluted at 1:200 with the LB medium. At A600 of 0.3, cultures were induced with IPTG (0.1 mM) and grown overnight at 18°C. Cells were spun down and lysed in buffer containing 20 mM Tris–HCl, pH 7.5, 300 mM KCl, 2 mM MgCl2, 5 mM β-mercaptoethanol (β-me), 50 mM imidazole, 0.5% Triton X-100, 5% glycerol, 0.5 mg/ml lysozyme, 1 × EDTA-free Protease Inhibitor Cocktail (Roche). The lysate was centrifuged at 16 000g for 30 min at 4°C. The supernatant was incubated with Ni-NTA Agarose beads (Qiagen) for 2 h at 4°C. After washing the beads with buffer A (20 mM Tris–HCl, pH 7.5, 150 mM KCl, 2 mM MgCl2, 5 mM β-me, 100 mM imidazole, 0.5% Triton X-100, 2.5% glycerol) and buffer B (as buffer A but containing 250 mM imidazole), each for 15 min at 4°C, protein was eluted with buffer C (as buffer A but containing 500 mM imidazole, 5% glycerol and 1 × EDTA-free Protease Inhibitor Cocktail (Roche). The eluted protein was dialyzed against storage buffer (20 mM Tris–HCl, pH 7.5, 200 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 5% glycerol and EDTA-free Protease Inhibitor Cocktail) and stored at −80°C. To avoid precipitation, concentration of recombinant proteins was kept below 3 µM.
Purification of miRISC
MiRISC complexes, preferentially loaded with let-7a miRNA, were purified from HEK293T cells (grown in 10 cm dish) doubly transfected with plasmids (5 μg each) expressing pri-let-7a and indicated FLAG-HA-tagged Ago proteins or their mutants. Forty-eight hours after transfection, cells were lyzed in 1 ml of 50 mM Tris–HCl, pH 7.4 containing 150 mM NaCl, 1 mM EDTA and 0.5% Triton X-100. After centrifugation at 14 000g, the supernatant was used for IP purification with 40 μl anti-FLAG M2 Affinity Gel (Sigma). The miRISC complex was eluted with 3xFLAG peptide (Sigma), following the manufacturer's protocol.
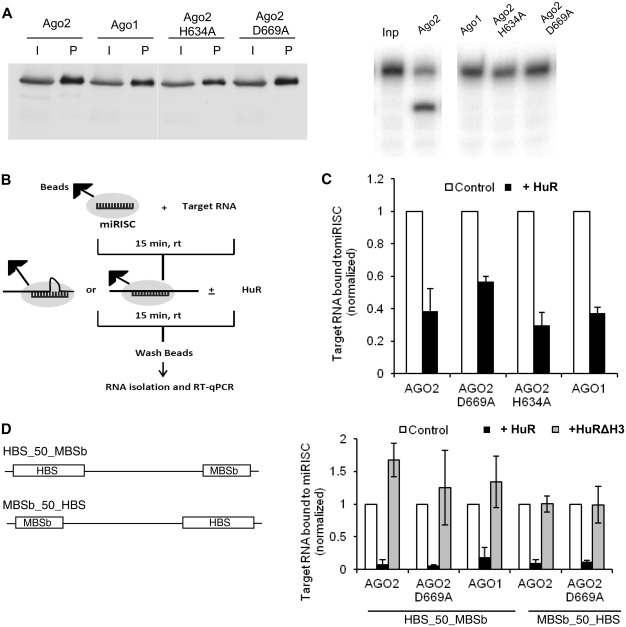
Concentration of active Ago2-miRISC was determined as described in ref. (50). In brief, the cleavage of labeled target RNA present in excess was quantified in a time course, the amount of product (y-axis) was plotted against time (x-axis), and slower steady-state cleavage rate line was extrapolated back to y-axis. The y-intercept at the zero time point denoted the amount of active miRISC. Since concentration of Ago2-mutant- and also Ago1-containing purified miRISCs could not be measured by the cleavage assay, calculation of their amounts was based on western analysis and comparison with the Ago2 miRISC (Figure 5A).
Figure 5.
HuR promotes dissociation of miRISC from target RNAs bearing either perfectly complementary or bulged let-7 sites. (A) Characterization of purified miRISC complexes containing Ago1, Ago2 or indicated Ago2 mutants. (Left panel) Western blot performed with anti-HA antibody, showing purification of miRISCs containing Ago1, Ago2 or indicated Ago2 mutants. ‘I’ denotes input cell extracts and ‘P’ denotes miRISCs purified on anti-FLAG M2 Affinity beads. (Right panel) miRISC cleavage assay demonstrating that only miRISC containing wt Ago2 is catalytically active. ‘Inp’ denotes input 5′-32P-labeled RNA substrate HBS_50_MBSp. (B) Schematic overview of experiments to determine the effect of HuR on association of miRISC with target RNA. MiRISC-bound beads were incubated with target RNA for 15 min at 23°C followed by additional 15 min incubation at 23°C in the absence (control) or presence of 150 nM HuR. The RNA remaining bound to beads was subjected to RT-qPCR (for details see ‘Materials and Methods’ section). (C) Quantification, by RT-qPCR, of the effect of HuR on association of indicated different forms (Ago2, Ago1, Ago2D669A and Ago2H634A) of miRISC with HBS_50_MBSp RNA. Values for RNA remaining bound with different miRISC forms upon addition of 150 nM HuR are normalized to values measured in the absence of HuR which are set to 1 (means ± SD; n ≥ 3) (D) (Right panel) Quantification, by qRT-PCR, of the effect of HuR or HuRΔH3 mutant on association of indicated forms (Ago2, Ago1 and Ago2D669A) of miRISC with RNA targets bearing bulged let-7 site positioned either downstream (HBS_50_MBSb) or upstream (MBSb_50_HBS) of the HuR binding site. Values for RNA remaining bound with different miRISC forms upon addition of 150 nM HuR or HuRΔH3 are normalized to values measured in the absence of HuR (means ± SD; n ≥ 3). (Left panel) Schemes of target RNAs used for the assay. ‘b’ denotes the bulged site.
Generation of templates for in vitro transcription
Templates for generation of HBS_20_MBSp, ΔHBS_20_MBSp and HBSMut_20_MBSp target RNAs were prepared by annealing synthetic antisense DNA oligonucleotides (Microsynth; for sequences, see Supplementary Table S2) to the T7 promoter sense oligonucleotode (T7s; Supplementary Table S2). Mixture of oligonucleotides, each 50 µM, was heated at 95°C for 3 min in 1 × annealing buffer (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 0.1 mM EDTA) and then allowed to slowly cool down to room temperature.
For synthesis of HBS_50_MBSp, HBS_50_MBSb and MBSb_50_HBS target RNAs, double-stranded DNA templates were used. The templates were prepared by annealing equimolar amounts of two oligonucleotides (T7s and 5′-phosphorylated sense; Supplementary Table S2) forming a sense strand with two oligonucleotides (reverse and 5′-phosphorylated antisense; Supplementary Table S2) forming an antisense (template) strand, followed by ligation by T4 DNA ligase (NEB). The resulting double-stranded products were used as templates for PCR amplification with T7s and reverse oligonucleotides as primers, gel purified and used as templates for in vitro transcription.
In vitro transcription, RNA labelling, and DNA and LNA oligonucleotide annealing
MBSp and MutMBSp substrate RNAs were purchased from Dharmacon RNA Technologies. For other target RNAs, transcription reactions (50 µl), containing 1 × transcription buffer (Promega), 500 µM each ATP, CTP and UTP, 200 µM GTP, 3.5 mM guanosine, 1.25 mM MgCl2, 2.5 mM DTT, 40 U of RNase Out (Invitrogen) and 300 U of T7 RNA polymerase (Epicenter Biotechnologies), were performed at 37°C for 3 h with 200 nM of template. Inclusion of excess of guanosine, compared to GTP, resulted in its incorporation at the RNA 5′-end, thus making the 5′-end dephosphorylation step unnecessary (51). Sequences of RNA transcripts are shown in Supplementary Table S3. The transcripts were analyzed by 10% polyacrylamide/8 M urea PAGE, excised, eluted from the gel with 0.3 M sodium acetate (pH 5.2), 0.5 mM EDTA and 0.1% SDS, precipitated, dissolved in water and stored at −80°C. When indicated, the transcripts were 5′-end-labeled, using T4 polynucleotide kinase (NEB) and [γ-32P] ATP (5000 Ci/mmol; Hartmann Analytic) according to manufacturer's protocol. Following labelling, transcripts were subjected to column purification (Micro Bio-Spin 30 Columns, RNase-free; Bio-Rad Laboratories), precipitated and stored at −20°C in water before use.
For annealing of HBS_50_MBSp RNA to a 50-mer DNA (complementary to the entire spacer) or 27-mer LNA oligonucleotide (complementary to the region starting from 4th nt of the spacer), they were mixed at the RNA to oligonucleotide ratio of 1:3, heated at 95°C for 3 min in 1 × annealing buffer (10 mM Tris–HCl, pH 8.0, 100 mM NaCl), followed by slow cooling to room temperature. Annealing of HBS_20_MBSp to 20-mer LNA oligonucleotide sequence (complementary to the entire spacer) was done in a similar way. The sequences of oligonucleotides are given in Supplementary Table S4.
Target RNA cleavage assay
The reactions (10 µl), containing 1 × cleavage buffer (20 mM HEPES-KOH, pH 7.4, 150 mM KCl, 2 mM MgCl2, 0.01% Triton X-100, 5% glycerol, 1 mM DTT), 0.1 µg yeast tRNA, purified miRISC (∼0.2 nM), 5′-end labeled target RNA (0.1 nM) and increasing amounts of indicated HuR proteins, were incubated for 15 min at 30°C. Reactions were stopped by adding urea stop dye (8 M urea, 50 mM EDTA, 0.04% bromophenol blue, 0.04% xylene cyanol), followed by heating at 95°C for 2 min. The products were analyzed by 8 M urea PAGE. Reaction products were quantified using a PhosphorImager (Molecular Dynamics, Inc). For analysis of the effect of HuR on cleavage of target RNA containing pre-bound miRISC, the 1× cleavage buffer initially contained 5 mM EDTA in place of MgCl2. At the indicated time point, MgCl2 was added to the final 5 mM concentration.
For quantification of cleavage reactions, the radioactivity present in bands corresponding to cleaved (RC) and uncleaved (RU) RNA in each lane was measured using ImageQuant v5.2 (Molecular Dynamics, Inc). The degree of cleavage was always calculated as a fraction of radioactivity present in RC band compared to the total measured radioactivity [RC/ (RC + RU)]. The values for reactions containing increasing concentrations of HuR or its mutants were normalized to values obtained in the absence of HuR which were set to 1.
Electrophoretic mobility shift assays
Interactions of HuR and its mutants with target RNAs were analyzed by electrophoretic mobility shift assays (EMSA), using non-denaturing gels. The reactions (10 µl) contained 1× cleavage buffer, 1 µg of yeast tRNA, 2 fmol of 5′-32P-labeled RNA, and indicated increasing concentrations of purified proteins. Following 10 min incubation on ice, the reactions were immediately processed for a non-denaturing 6% PAGE performed at 4°C. The gel was dried and the resolved RNA:protein complexes were visualized by phosphorimaging.
Analysis of miRISC association with target RNA
For measurements of miRISC association with target RNA, the purified miRISC was immobilized on α-FLAG beads and then first incubated with target RNA, followed by addition of HuR (see scheme in Figure 5B). A 30 μl α-FLAG M2 Affinity Gel beads (Sigma) were pre-incubated with 4 μl of purified miRISC in 1 ml of binding buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 × EDTA-free Protease Inhibitor Cocktail (Roche)]. The beads were spun down and resuspended in 100 µl of the binding buffer additionally containing 50 fmol of target RNA, 1 μg tRNA, 0.5 mM DTT and 5 mM EDTA (in a case of the Ago2 miRISC) or 2 mM MgCl2 (in a case of other catalytically inactive miRISCs). Samples were incubated for 15 min at 23°C, followed by additional 15 min incubation at 23°C in the presence or absence of 150 nM of HuR. The beads were spun down by centrifugation at 5000g for 1 min and washed twice with binding buffer. The RNA bound to beads was extracted with TRIzol reagent (Invitrogen), reverse transcribed using a reverse primer (GCTCATCAGATGTTGAGTCC; complementary to the 3′-end of the spacer region) and 25 U of Superscript III RT (Invitrogen). One-fourth of the RT reaction was then used for quantitative real-time PCR (RT-qPCR), using the ABI 7500 Fast Sequence Detection System (Applied Biosystems) and Platinum SYBR Green qPCR Super Mix (Invitrogen). Sequence of the forward PCR primer for HBS_50_MBSp and HBS_50_MBSb are GGTATTTATTTATTTATTTGTTTG and that for MBSb_50_HBS is GGTCCAAAGAGTGACTGCACAGCC. The reverse primer was the same as that used for RT.
RNA deadenylation assay
Plasmids used in the synthesis of RNAs tested in deadenylation assays were prepared by inserting the AU-rich downstream (ARD) region from the 3′-UTR of human CAT-1 mRNA (11) into the NotI site downstream of the 6xB and 6xBMut sequences of RL-6xB-pA and RL-6xBMut-pA (52) to generate RL-6xB-ARD-pA and RL-6xBMut-ARD-pA, respectively. The sequences of primers are given in Supplementary Table S1. The preparation of all internally labeled RNAs by in vitro transcription and the assay were performed as previously described (53). Quantification of deadenylation reactions was performed similarly to the quantification of cleavage reactions described above. The percentage of deadenylation was always calculated as a fraction of radioactivity present in the band corresponding to poly(A)− RNA compared to the amount of radioactivity corresponding to the sum of poly(A)− and poly(A)+ RNAs [pA−/(pA+ + pA−)] present in each lane.
RESULTS
Recombinant HuR inhibits cleavage of RNA by miRISC in vitro
To investigate whether it is possible to recapitulate the HuR-mediated alleviation of miRNA repression in vitro, using purified components, we first used the miRISC-mediated cleavage assay. miRISC bearing Ago2 as an effector protein, endonucleolytically cleaves target RNAs, which contain sites perfectly complementary to miRNA (44,45). The let-7-miRNA-enriched miRISC was prepared by the co-expression of human FLAG-HA-tagged Ago2 and let-7a primary precursor (pri-miRNA) in HEK293T cells, followed by affinity purification on anti-FLAG antibody beads. In HEK293T cells, let-7 RNAs are expressed only at a relatively low level (54) that allows for enrichment of the FLAG-tagged miRISC in this specific miRNA. The expression of tagged Ago2 was verified by western blotting (Supplementary Figure S1A) and functionality of the purified miRISC was ascertained by incubating it with the 5′-32P-labeled model RNA substrates bearing one perfectly complementary let-7 site (MBSp) or its mutated version (MutMBSp) (Supplementary Figure S1B). Only miRISC enriched in the co-expressed let-7 miRNA catalyzed target RNA cleavage and the cleavage was dependent on the presence of a fully complementary site (Supplementary Figures S1C and S1D).
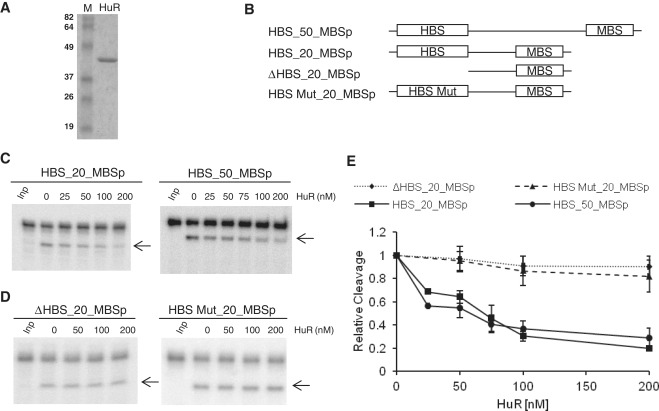
We tested whether addition of purified recombinant HuR, bacterially expressed as a His-tag fusion (Figure 1A), affects miRISC-mediated cleavage of RNA containing the let-7 MBSp and the HuR-binding site [HBS; a high-affinity AU-rich element (ARE) originating from IL-1β mRNA; (55)] positioned upstream (Figure 1B). As shown in Figure 1C and E, addition of HuR resulted in the concentration-dependent repression of cleavage of substrates containing HBS and MBS elements. Since a similar effect was seen with RNAs bearing the elements either 20 nt (HBS_20_MBSp) or 50 nt (HBS_50_MBSp) apart, it is unlikely that binding of HuR to HBS caused a direct steric hindrance of miRISC association. HuR had no effect on cleavage of RNAs bearing mutated HBS or having no HBS (Figure 1D and E). The latter two substrates also did not bind HuR, as assessed by a native gel EMSA (data not shown). Unless indicated otherwise, all target RNA cleavage assays were incubated for 15 min to stay within a linear range of the reaction (Supplementary Figure S2). This was particularly important when effects of HuR and its mutants on the miRISC-mediated cleavage were compared (see below).
Figure 1.
Purified recombinant HuR specifically inhibits RNA cleavage mediated by the let-7 miRISC. (A) Coomassie blue-stained SDS–polyacrylamide gel showing purity of the recombinant full-length HuR. Lane M, protein size markers (in kDa). (B) Schemes of target RNAs used for the cleavage assay. HBS, HuR Binding Site; MBS, miRNA Binding Site; ΔHBS and HBS Mut, HBS deleted or mutated. ‘p’ denotes perfect let-7 complementarity of the MBS site. (C and D) Representative in vitro cleavage reactions performed with different 5′-32P-labeled target RNAs and increasing indicated concentrations of HuR. ‘Inp’ denotes input 5′-32P-labeled RNA substrate. The cleavage product is indicated by an arrow. (E) PhosphorImaging quantification of cleavage reactions similar to those shown in panels C and D. Values (means ± SD; n ≥ 3) were normalized to the reactions in the absence of HuR.
HuR oligomerization mutants are defective in attenuating miRISC cleavage
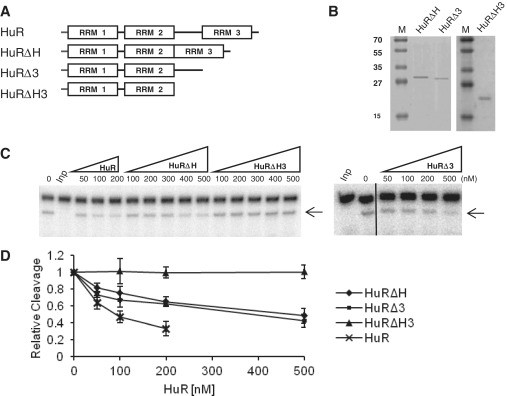
HuR and related proteins, such as HuB, HuD and Drosophila ELAV, are known to oligomerize along RNA substrates (35–40). Furthermore, for HuR and Drosophila ELAV, the hinge separating RRM2 and RRM3, and the RRM3 were identified as domains contributing to the formation of cooperative HuR–RNA complexes (36,40). We investigated whether HuR oligomerization might play a role in miRISC interference of RNA cleavage. To this end, we have purified three different HuR mutants: HuRΔH (devoid of a hinge region separating RRM2 and RRM3), HuRΔ3 (RRM3 deleted) and HuRΔH3 (missing both the hinge region and RRM3) (Figure 2A and B). Consistent with previous findings (36,40), a full-length HuR, and HuRΔH and HuRΔ3 mutants formed complexes with HBS_50_MBSp RNA, gradually increasing in size in a concentration-dependent manner. In contrast, the potential of HuRΔH3 to oligomerize was clearly diminished as seen in Supplementary Figure S3 (36). We then compared activity of different HuR mutants to interfere with the miRISC function. Interestingly, the two mutants able to oligomerize, HuRΔH and HuRΔ3, inhibited miRISC-mediated cleavage of HBS_50_MBSp RNA, although at a concentration higher than the full-length HuR. In contrast, the cleavage of target RNA remained unaffected in the presence of the mutant HuRΔH3, which is defective in oligomerization (Figure 2C and D).
Figure 2.
Activity of different HuR mutants in alleviating miRISC cleavage. (A) Schemes of wt HuR and its deletion mutants. (B) Coomassie blue-stained SDS–polyacrylamide gel showing purification of different HuR mutant HuR. Lane M, protein size markers (in kDa). (C) Representative in vitro cleavage reactions of HBS_50_MBSp RNA in the presence of increasing concentrations of indicated HuR mutants. (D) Quantification of the effect of different HuR mutants on HBS_50_MBSp RNA cleavage. Values represent means ± SD; n ≥ 3.
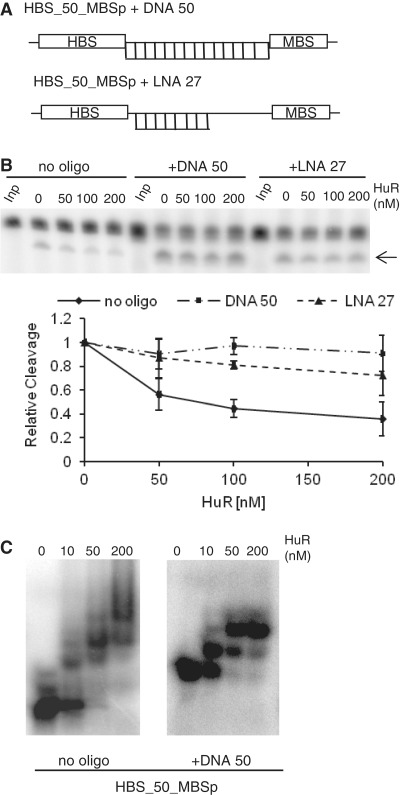
To obtain additional evidence that HuR oligomerization may be important for interference with the miRISC-mediated target cleavage, we tested the effect of pre-hybridizing the oligonucleotides complementary to the spacer separating HBS and MBSp sequences in the target RNA. Hybridization of a 50-nt-long oligodeoxynucleotide complementary to the entire spacer of the HBS_50_MBSp RNA or a 27-mer oligonucleotide bearing several LNA nucleotides (Figure 3A) and forming a duplex of stability comparable to that formed with the 50-mer DNA oligonucleotide, almost entirely eliminated the inhibitory effect of HuR on RNA cleavage by miRISC (Figure 3B). Similar result was obtained with HBS_20_MBSp RNA, having the 20-nt-long spacer separating HBS and MBSp sites. Annealing of the 20-mer LNA oligonucleotide complementary to the HBS_20_MBSp spacer completely eliminated the effect of HuR (Supplementary Figure S4). We have verified that hybridization of complementary 50-mer DNA oligonucleotide to the HBS_50_MBSp RNA strongly reduced a potential of the duplex to form multimeric complexes with HuR, as analyzed by EMSA (Figure 3C). It was also noted that the IL-1β HBS element is 33-nt long and probably able to accommodate two HuR molecules, either monomers or dimers (36,56). Incubation of HuR with duplexes of HBS_50_MBSp RNA and LNA oligonucleotide resulted in formation of complexes that did not enter the non-denaturing gel, thus preventing analysis of the LNA effect on HuR oligomerization by EMSA (data not shown).
Figure 3.
Oligonucleotides complementary to the spacer separating HBS and MBS sites prevent the HuR effect on miRISC cleavage in vitro. (A) Schemes of the HBS_50_MBSp target RNA hybridized to complementary 50-mer DNA and 27-mer LNA oligonucleotides. (B) Prehybridization of DNA and LNA oligonucleotides interferes with the alleviating effect of HuR on miRISC cleavage. (Upper panels) Effect of increasing concentrations of HuR on cleavage of control HBS_50_MBSp RNA or its duplexes with either DNA or LNA oligonucleotides. (Lower panel) Quantification (means ± SD; n = 3) of experiments similar to those shown in upper panels. (C) Prehybridization of 50-nt DNA oligonucleotide complementary to the spacer region of HBS_50_MBSp RNA interferes with HuR oligomerization along RNA as visualized by EMSA. Although comparison of the left (no oligonucleotide) and right (complementary oligonucleotide added) panels reveals the decrease in a number of HuR-associated bands in response to hybridizing the oligonucletide, the precise number and nature of HuR molecules associated with each EMSA band is difficult to assign. The IL-1β HBS element itself is 33-nt long and is likely to accommodate two HuR molecules; in addition, HuR may associate with RNA as either monomer or homodimer (36,56).
Taken together, these results indicate that the oligomerization potential of HuR along target RNA may be important for the protein interference with miRISC activity.
HuR can displace miRISC pre-bound to the target RNA
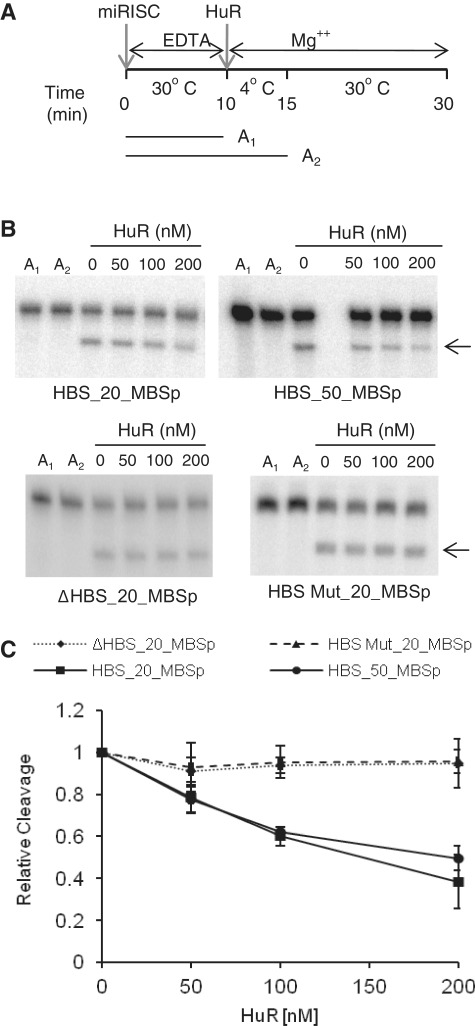
In the experiments described so far, miRISC and HuR were always mixed together prior to addition of target mRNA. We investigated whether the addition of HuR can inhibit miRISC or dissociate it from the target even when it has been pre-bound to the RNA. First, we tested the effect of HuR addition on the cleavage activity of miRISC pre-incubated with target RNAs. Pre-incubation of miRISC with either HBS_20_MBSp or HBS_50_MBSp was performed in the presence of EDTA to inhibit Mg2+-dependent cleavage of RNAs containing perfectly complementary let-7 sites. Following a 10-min incubation at 30°C, Mg2+ and HuR were added and the incubation continued initially at 4°C (to inhibit miRISC cleavage but to allow HuR binding) and then at 30°C (Figure 4A). We have verified that inclusion of EDTA or incubation at 4°C effectively prevents substrate RNA cleavage by miRISC (Figure 4A and B, conditions A1 and A2). Importantly, as shown in Figure 4B and C, inclusion of HuR led to concentration-dependent inhibition of target cleavage, indicating that HuR can interfere with the activity of miRISC pre-bound to RNA. The observed effect was dependent on the presence of functional HuR-binding site since addition of HuR had no effect on activity of miRISC pre-bound to RNA bearing either mutated HBS or having no HBS (Figure 4B and C).
Figure 4.
HuR inhibits cleavage by the miRISC pre-bound to RNA. (A) Overview of the experimental set up for cleavage assay with miRISC pre-bound to target RNA. RNA was pre-bound to miRISC for 10 min in the presence of EDTA to ensure blocking of the cleavage (time-point A1), followed by incubation on ice for 5 min (time-point A2) after simultaneous addition of HuR and Mg2+. The cleavage reaction was incubated for 15 min at 30°C. (B) Representative in vitro cleavage reactions of HBS_20_MBSp and HBS_50_MBSp RNAs (upper panels) and ΔHBS_20_MBSp and MutHBS_20_MBSp RNAs (lower panels), as a function of increasing concentration of HuR. Reactions followed the experimental set up presented in A. The RNA remains uncleaved when incubated with miRISC in the presence of EDTA at 30°C (lane A1) or upon addition of Mg2+ when the incubation is performed on ice (lane A2). (C) Quantification of reactions similar to the one shown in B (mean ± SD; n = 3).
To determine directly whether HuR is able to displace miRISC bound to the target, we measured the association of the HBS_50_MBSp RNA with miRISC following the HuR addition. In addition to the miRISC containing a wild-type (wt) Ago2, we also used miRISCs in which let-7 is loaded onto FLAG-HA-Ago1 or FLAG-HA-tagged Ago2 mutants Ago2D669A and Ago2H634A (Figure 5A, left panel), which like Ago1 are catalytically inactive [(45), Figure 5A, right panel)]. Pre-binding of the wt-Ago2 miRISC was carried out in the presence of EDTA but pre-binding of the three remaining catalytically incompetent miRISCs was performed in the presence of Mg2+. Following incubation with HuR, the amount of HBS_50_MBSp RNA remaining associated with the immobilized miRISC was quantified by real-time PCR (RT-qPCR) (Figure 5B). We found that addition of HuR decreased by 2- to 3-fold the association of HBS_50_MBSp RNA with all tested miRISC variants, consistent with HuR-mediated dislocation of miRISC from the RNA target (Figure 5C).
HuR can effectively displace miRISC from target RNAs bearing a bulged miRNA site
We also tested the effect of HuR addition on the association of the Ago2-miRISC with targets containing the let-7 site, which is not complementary to let-7 RNA in its central region [‘bulged’ let-7 site, identical to that used in RL-3xB reporters responding to let-7 miRNA. Reporters bearing these sites and also HuR-binding AREs were shown previously to be relieved from the let-7-mediated repression in cells subjected to stress (11)]. The two RNA targets analyzed by us, HBS_50_MBSb and MBSb_50_HBS, differ in the relative orientation of HBS and MBSb sites in RNA (Figure 5D, left panel). Inclusion of HuR decreased the association of targets with both wt (Ago2 and Ago1) and mutant (Ago2D669A) miRISCs by ∼10-fold. This effect was independent of the orientation of MBSb and HBS sites and, most significantly, was not observed when the non-oligomerizing HuR mutant HuRΔH3 was used instead of the wt protein (Figure 5D, right panel). Notably, the efficiency of miRISC displacement from HBS_50_MBSb was much higher than that seen with HBS_50_MBSp containing a perfectly complementary let-7 site. This difference was also apparent when the effect of HuR on miRISC association with both substrates was compared in the same experiment (data not shown).
Taken together, these results indicate that HuR promotes dissociation of miRISC from substrates containing both perfectly complementary and bulged miRNA sites and that the potential of HuR to oligomerize along RNA is important for this effect.
HuR does not interact with miRISC proteins
The requirement for oligomerization described above suggested that HuR has to get into proximity of miRISC and possibly interact with its components to exert its derepressive function. To address this possibility, we tested whether HuR interacts with miRISC components, such as Ago or GW182 proteins. We performed immunoprecipitation (IP) experiments of HuR and Ago proteins from cell extracts prepared from either stressed or non-stressed cells. Use of antibodies against endogenous proteins revealed that HuR did not co-immunoprecipitate Ago2 and that Ago2 did not co-immunoprecipitate HuR (Supplementary Figure S5A) in either type of cells. The IP experiments with ectopically expressed epitope-tagged proteins similarly provided no evidence of HuR interacting with Ago2, Ago3 or protein TNRC6B (Supplementary Figures S5B and S5C). As expected, the IP experiments revealed interaction of Ago2 and TNRC6B (Supplementary Figure S5B). We conclude that HuR does not stably associate with miRISC protein components (see ‘Discussion’ section).
Addition of HuR to Krebs ascites cell extract alleviated deadenylation of an miRNA reporter
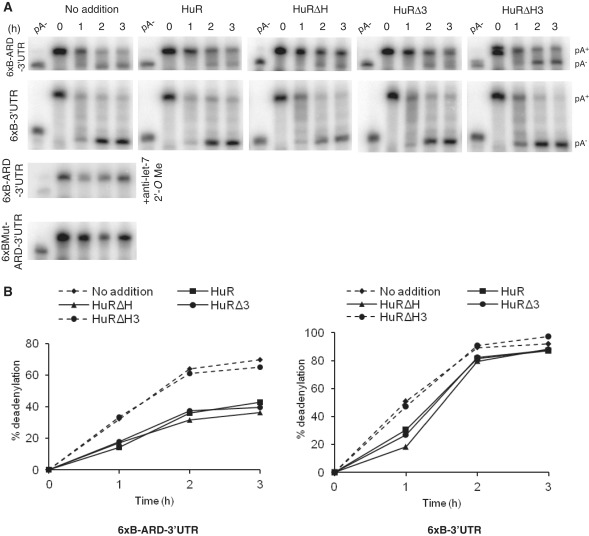
We previously described an in vitro extract derived from Krebs-2 ascites cells that faithfully recapitulates both RNA deadenylation and translational repression induced by miRNAs (52,53). We tested whether addition of recombinant HuR or its mutants interferes with the miRNA-induced deadenylation of RNA bearing let-7 miRNA sites and the HuR binding region ARD, derived from the CAT-1 3′-UTR [6xB-ARD-3′-UTR; for detailed description of ARD, see ref. (11)]. We found that addition of wt HuR or its mutants HuRΔH and HuRΔ3, which have the potential to relieve repression in other assays (see above), markedly inhibited deadenylation of 6xB-ARD-3′-UTR RNA in vitro. In contrast, addition of the HuR mutant HuRΔH3 that was inactive in the derepression had no effect (Figures 6A, upper row and 6B, left panel). In control experiments, we verified that addition of any of the four forms of HuR had no major effect on deadenylation of RNA containing no ARD (6xB-3′-UTR; Figures 6A, second row and 6B, right panel). As expected, addition of anti-let-7 2′–O-methyl oligonucleotide blocked deadenylation of 6xB-ARD-3′-UTR and RNA bearing mutated let-7 sites (6xBMut-ARD-3′-UTR) did not undergo deadenylation (Figure 6A, two bottom panels).
Figure 6.
Effect of HuR and its mutants on miRNA-mediated deadenylation of target RNAs in Krebs-2 ascites extract. (A) Effect of HuR and its mutants on deadenylation of 6xB-ARD-3′UTR (upper row) and 6xB-3′-UTR (second row) RNAs as analyzed by PAGE. RNAs were incubated in the presence or absence of 250 nM of HuR or its mutants for indicated time. Positions of pA+ and pA− RNAs are marked on the right. (Bottom panels) Control assays performed in the absence of HuR with 6xBMut-ARD-3′-UTR and 6xB-ARD-3′-UTR RNAs, the latter assay containing anti-let-7 2′-O-methyl oligonucleotide. (B) Quantification of the deadenylation reactions of 6xB-ARD-3′-UTR (left panel) and 6xB-3′-UTR (right panel) shown in A.
In conclusion, HuR can interfere with the inhibitory function of let-7 in the in vitro system recapitulating miRNA-mediated deadenylation. Similarly, as in the case of assays involving purified miRISC (Figure 2C and D), the presence of either the HuR hinge region or the RRM3 was required for the protein to counter the let-7 induced mRNA deadenylation, consistent with the possibility that the HuR effect involves its oligomerization.
DISCUSSION
We showed previously that both CAT-1 mRNA and reporter mRNAs bearing AREs in their 3′-UTR can be relieved of miRNA-mediated repression in mammalian cells subjected to various stress conditions. The derepression required the binding of the ELAV family protein HuR to the 3′-UTR AREs, which occurred even when AREs were positioned at a considerable distance from the miRNA sites (11). These observations raised questions about the mechanism of HuR action and that of other possibly stress-induced factors that may participate in the process. In this present report, we show that the relief of miRNA-mediated repression involving HuR can be recapitulated in different in vitro systems in the absence of stress, indicating that HuR alone is sufficient to execute derepression upon binding to the RNA ARE. Nevertheless, it is possible that additional factors can assist HuR in its activity in a cellular context. Using in vitro assays with purified miRISC and recombinant HuR and its mutants, we show that HuR leads to the dissociation of miRISC from target RNA and that this function of HuR correlates with its property to oligomerize along RNA. Further, we demonstrate that HuR association with AREs can also inhibit miRNA-mediated deadenylation of mRNA in the Krebs-2 ascites extract, in a similar manner depending on the potential of HuR to oligomerize.
Our findings that addition of HuR could interfere with miRISC activity irrespective of whether the miRISC site was located 20- or 50-nt away from the HuR-binding region suggested that the HuR effect is unlikely to be caused by direct sterical occlusion of the miRISC site by the protein (Figure 1). Since HuR and related proteins, such as HuD and Drosophila ELAV are known to oligomerize along RNA substrates (35–40), we investigated whether this property of HuR plays a role in the suppressive effect of HuR on miRISC activity. Three types of experiments provided strong support for the multimerization of HuR being important: (i) only full-length HuR and its mutants HuRΔH and HuRΔ3, all able to multimerize [Supplementary Figure S3; and (36)], inhibited the miRISC-mediated cleavage of RNA bearing HBS and MBS sequences. In contrast, an HuR mutant (HuRΔH3) that is devoid of a hinge and RRM3 and that binds target RNA but is impaired in multimerization [Supplementary Figure S3; and ref. (36)] did not interfere with miRISC activity (Figure 2C and D). (ii) Pre-hybridization of target RNA with oligonucleotides, either DNA or LNA, complementary to the spacer separating HBS and MBS sequences eliminated its inhibitory effect on RNA cleavage by miRISC (Figure 3B). (3) Finally, the full-length HuR and its mutants able to oligomerize but not the mutant HuRΔH3, which lacks a hinge and RRM3, interfered with the let-7-induced mRNA deadenylation in vitro (Figure 6), consistent with the possibility that this HuR effect requires its oligomerization.
The apparent role of oligomerization in the derepressive function of HuR suggested that the protein has to get into proximity of miRISC to interfere with its activity. Consistently, our results indicate that HuR may exert its function by displacing the miRISC from RNA. This conclusion is supported by in vitro experiments performed with purified HuR and let-7-enriched miRISC and RNAs bearing both HuR and miRNA sites. These experiments revealed that addition of HuR, either simultaneously with miRISC or after miRISC has been pre-bound to RNA, results in a concentration-dependent dislocation of miRISC from the target RNA (Figures 1, 4 and 5). This effect occurred irrespective of whether miRISC was base paired with target RNA by perfect or imperfect complementarity, although the HuR effect was much more pronounced for substrates associating with miRISC through an imperfect, central bulge-forming interaction (Figure 5).
The less effective displacement of miRISC from a target having perfect complementarity, and hence engaged in a more stable interaction with let-7 miRNA, suggested that one possible mode of HuR function is through the destabilization of the miRNA–mRNA base pairing. This notion is supported by our observation that, in transfected cells, HuR does not alleviate repression induced by direct tethering of Ago2 to mRNA, a process not involving miRNA base pairing with the target RNA (Bhattacharyya, S.N., Kundu, P. and Fillipowicz, W.; unpublished results). The possibility that HuR functions by displacing miRISC from the target, and not by interfering with the repressive function of the miRISC protein components, is also supported by the results of immunoprecipitation experiments. We found no evidence that Ago or GW182 proteins interact with endogenous HuR (Supplementary Figure S5). We note, however, that some authors have reported that HuR, either endogenous (17) or overexpressed (57), interacts in an RNA-dependent manner with Ago2. In contrast, others did not detect HuR–Ago2 interaction even in the absence of RNase treatment (58), or did not find HuR among proteins identified by mass spectrometry as associated with tagged Ago1, Ago2 or three human GW182 proteins, but found it in complexes with Ago3 and Ago4 (59).
The HuR effect described in this study, leading to the relief of miRNA repression from a distance in a process likely involving HuR oligomerization, represents a novel mechanism of miRNA regulation, differing from other examples of the cross-talk between miRNAs and RBPs at the 3′-UTR of metazoan mRNAs. Dead end (Dnd1), an RBP expressed in germ cells of zebrafish and mammals, prevents miRNA repression by binding to sequence elements in the 3′-UTR that overlap with miRNA sites, thus effectively blocking miRNA access to target mRNA (15). Similarly, hnRNP protein L competes with miRNA for binding to the same CA-rich elements present in the 3′-UTR of VEGFA mRNA (14), and the CRD-BP RBP competes with miR-340 for binding to the MITF mRNA 3′-UTR (60). In contrast, Pumilio 1 (PUM1), an ubiquitously expressed RBP, was found to facilitate miRNA repression. Binding of PUM1 to the p27 mRNA 3′-UTR induces a local change in RNA structure that favors association with specific miRNAs (16).
Regulation of miRNA repression by RBPs interacting with the 3′-UTR is probably a widespread phenomenon (8,23,24). In addition to the examples discussed above, further cases of cross-talk between RBPs and miRNAs have been reported (13,18,61), although their mechanisms are unknown. Furthermore, a comparative study of mRNAs interacting with Pumilio (PUF) proteins showed a considerable enrichment of PUF-binding sites in the vicinity of predicted miRNA recognition sequences in human mRNAs (62), suggesting that PUF-miRNA cross-talk is a common event. Importantly, several of the mechanistically unsolved cases also involve HuR. Gorospe and collaborators found that HuR antagonizes the miR-548c-3p-mediated repression of topoisomerase IIα (TOP2A) mRNA (21) and miR-494-mediated repression of nucleolin mRNA (22) in HeLa cells. Similarly, as in the case of CAT-1 mRNA (11), interaction sites of HuR and co-regulated miRNAs in both mRNAs were positioned at a considerable distance, suggesting that the mechanism of HuR effect might be related to that reported in our study. Interestingly, in two other examples involving HuR, the protein appeared to have a positive rather than alleviating effect on miRNA-mediated repression. Specifically, it was found that HuR enhances c-Myc mRNA repression by let-7 (17) and RhoB mRNA repression by miR-19 (12). Possibly, HuR can enhance miRNA repression by locally modifying mRNA structure and increasing the accessibility of miRNA-binding sites, as was shown for PUM and p27 mRNA (16).
The HuR-miRNA cross-talk most probably extends far beyond the examples described above. This is supported by the results of recent transcriptome-wide mapping of HuR binding sites in mammalian cells (63–65). These studies revealed that HuR sites are enriched near predicted miRNA sites in mRNAs and frequently overlap with them. Mukherjee et al. (2011) demonstrated that functional depletion of miRNAs in HeLa cells results in a significantly lower upregulation of mRNAs containing overlapping miRNA and HuR sites when compared to the mRNAs bearing no overlapping sites, suggesting that the competition between HuR and miRNAs to access the target sites can prevent miRNA repression. However, these findings do not explain the ’long-distance’ alleviating effects of HuR observed in the case of CAT-1, TOP2A and nucleolin mRNAs (11,21,22) and the reporters used in this study.
It will be important to investigate which factors determine the outcome, either negative or positive, of the HuR effect on miRNA repression. The activity of HuR is known to be controlled by various post-translational modifications and proteolytic cleavages, and also by interaction with protein ligands (34). Moreover, HuR has recently been reported to have an RNA 3′-terminal adenosyl transferase activity, residing in RRM3 (66). Hence, it is perhaps not surprising to find that the protein contributes to miRNA regulation in different ways.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary tables 1–4, Supplementary figures 1–5, Supplementary methods and a Supplementary reference [67].
FUNDING
Indo-Swiss Joint Research Project of Swiss National Science Foundation and Dept. of Science and Technology, Govt. of India, and HFSPO Short Term Fellowship and Young Researcher Award Fund from Lady Tata Memorial Trust, India (to S.N.B.); Novartis Research Foundation (to Friedrich Miescher Institute); and Canadian Institute of Health Research (MOP-93607 to N.S. and M.F.). Funding for open access charge: Novartis Research Foundation (to Friedrich Miescher Institute).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ann-Bin Shyu, Joel Belasco, Gunter Meister, Greg Hannon and Zissimoss Mourelatos for kindly providing different reagents, Stefan Ameres for his input at initial stage of this work, and Nicole Meisner-Kober for valuable discussions.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J. Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 9.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 10.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Glorian V, Maillot G, Polés S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;12:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 14.Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 17.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 20.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, Kulshrestha R, Panchakshari R, Kim HH, Yang X, et al. Translational Control of TOP2A Influences Doxorubicin Efficacy. Mol. Cell Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of Nucleolin expression by HuR and miR-494. Mol. Cell Biol. 2011;30:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 24.Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- 25.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schratt G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 27.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 28.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 30.Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs) Proc. Natl Acad. Sci. USA. 2011;108:8281–8286. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 32.Pascale A, Govoni S. The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell Mol. Life Sci. 2011;69:501–517. doi: 10.1007/s00018-011-0810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisner NC, Filipowicz W. Properties of the regulatory rna-binding protein HuR and its role in controlling miRNA repression. Adv. Exp. Med. Biol. 2011;700:106–123. doi: 10.1007/978-1-4419-7823-3_10. [DOI] [PubMed] [Google Scholar]
- 35.Devaux A, Colegrove-Otero LJ, Standart N. Xenopus ElrB, but not ElrA, binds RNA as an oligomer: possible role of the linker. FEBS Lett. 2006;580:4947–4952. doi: 10.1016/j.febslet.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J. Biol. Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J. Cell Sci. 1996;109(Pt 3):579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 38.Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soller M, White K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol. Cell Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toba G, White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res. 2008;36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 44.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 49.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 51.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 2004;279:42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 52.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 53.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meisner NC, Hackermüller J, Uhl V, Aszódi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 56.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 57.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 59.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J. Biol. Chem. 2010;285:20532–20540. doi: 10.1074/jbc.M110.109298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 64.Uren PJ, Burns SC, Ruan J, Singh KK, Smith AD, Penalva LO. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J. Biol. Chem. 2011;286:37063–37066. doi: 10.1074/jbc.C111.266882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the rna-binding protein HuR couples Pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meisner NC, Hintersteiner M, Seifert JM, Bauer R, Benoit RM, Widmer A, Schindler T, Uhl V, Lang M, Gstach H, et al. Terminal adenosyl transferase activity of posttranscriptional regulator HuR revealed by confocal on-bead screening. J. Mol. Biol. 2009;386:435–450. doi: 10.1016/j.jmb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 67.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O'Doherty U, Mourelatos Z. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.