Abstract
During the last two decades, microRNAs (miRNAs) emerged as critical regulators of gene expression. By modulating the expression of numerous target mRNAs mainly at the post-transcriptional level, these small non-coding RNAs have been involved in most, if not all, biological processes as well as in the pathogenesis of a number of diseases. miR-132 and miR-212 are tandem miRNAs whose expression is necessary for the proper development, maturation and function of neurons and whose deregulation is associated with several neurological disorders, such as Alzheimer's disease and tauopathies (neurodegenerative diseases resulting from the pathological aggregation of tau protein in the human brain). Although their involvement in neuronal functions is the most described, evidences point towards a role of these miRNAs in many other biological processes, including inflammation and immune functions. Incidentally, miR-132 was recently classified as a ‘neurimmiR’, a class of miRNAs operating within and between the neural and immune compartments. In this review, we propose an outline of the current knowledge about miR-132 and miR-212 functions in neurons and immune cells, by describing the signalling pathways and transcription factors regulating their expression as well as their putative or demonstrated roles and validated mRNA targets.
INTRODUCTION
First discovered in 1993 by Lee et al. (1), microRNAs (miRNAs) form a class of small regulating RNA of ∼22 nt in length, able to post-transcriptionally regulate the expression of target mRNAs. In most cases, miRNAs interact with their target mRNAs via an imperfect matching occurring between the mRNA 3′-untranslated region (3′-UTR) and a region located between nucleotides 2 and 8 in the 5′ region of the miRNA, referred to as the miRNA ‘seed’ region. Depending on the complementarity degree of this interaction, miRNA can lead to the cleavage and degradation of their mRNA targets (when the matching is perfect or nearly perfect) or to their translational inhibition (when the interaction involves more mismatches) (2). Currently, over 1500 miRNA-encoding genes have been identified in the human genome (miRBase), and bioinformatical analyses based on the complementarity level between miRNA seed regions and mRNA 3′-UTR predict that each miRNA could regulate the expression of dozens to hundreds of mRNAs (2,3). In addition, a particular gene transcript could be the target of several miRNAs (4). However, the predicted impact of miRNAs on target mRNAs is probably underestimated since accumulating data indicate that miRNA regions located outside the seed region are involved in mRNA recognition and that miRNAs can bind other regions than mRNA 3′-UTR, including their 5′-UTR and coding sequence (2). Moreover, miRNAs could also positively regulate gene expression by enhancing mRNA translation and inducing gene expression via target gene promoter binding (5–8), thereby adding more complexity to their initially described mode of action. Altogether, miRNAs are believed to control the expression of one- to two-third of human genes, which explains their involvement in most, if not all, physiological processes as well as their association with numerous diseases when their expression is deregulated (2,3,9,10).
Almost unknown 5 years ago, miR-132 and miR-212, two miRNAs sharing close sequences highly conserved among vertebrates (Figure 1), have been described in an exponential number of publications during these last years, pointing out the pleiotropic feature of these two miRNAs. Most of what we currently know about their regulation and biological functions emerged from studies performed in the neuronal context. However, several studies have also highlighted their involvement in inflammation and other biological (dys)functions. In this review, we outline the main functions of miR-132 and miR-212 in the neural and immune compartments, through the description of their identified targets in different biological processes as well as the molecular pathways involved in the control of their expression.
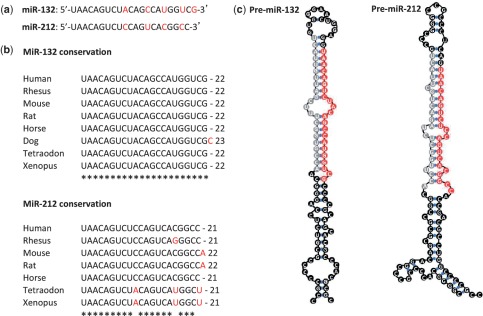
Figure 1.
Mature and precursor sequences of miR-132 and miR-212 in human. (a) miR-132 and miR-212 share similar mature sequences; in red are indicated the diverging nucleotides between the two human sequences. (b) miR-132 and miR-212 mature sequences are highly conserved among vertebrates; in red are indicated the nucleotides diverging from the human sequence. (c) Sequences and predicted stem–loop structures of human pre-miR-132 and pre-miR-212. The mature miRNA sequences are indicated in red, the miRNA* sequences are indicated in grey and predicted hydrogen bounds are indicated in blue. This figure was designed using miRBase (http://www.mirbase.org) for miRNAs and pre-miRNAs sequences and the Vienna RNAfold webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) for the prediction of the stem–loop structures.
THE GENOMIC FEATURES OF miR-212/132 CLUSTER
miRNA-encoding genes display different types of genomic organizations: intergenic miRNAs are located outside known transcription units whereas intragenic miRNAs are embedded in exons or introns, or even overlap exon–intron junctions of coding or non-coding genes (9,11). miR-132 and miR-212 were first reported to be transcribed from the first and stable intron of the non-coding transcript DQ223059, localized on chromosome 10 in rats, and from the first intron of the non-coding transcript AK006051, localized on chromosome 11 in mice (12,13). However, another transcript variant, likely resulting from an alternative splicing form of the AK006051 transcript was then identified in mice, with miR-132 and miR-212 sequences embedded in its second exon (Figure 2). Interestingly, these variants display tissue-specific patterns of expression, with the first variant being expressed in brain and testes and the second one in brain, testes, heart and mammary glands (14). Similarly to their rodent orthologues, hsa-miR-132 and hsa-miR-212 share the same primary transcript (A. Wanet and A. Tacheny, unpublished data). In addition, in human, they are found in an intergenic region located on chromosome 17p13.3. miR-132 and miR-212 exhibit similar mature sequences and share the same seed region (Figure 1); they may therefore target the same mRNAs. Nevertheless, this ‘double-targeting’ by both miR-132 and miR-212 was only demonstrated for few mRNAs so far and each of these miRNAs may also repress specific targets (Tables 1 and 2). Although much less studied than their guide-strand counterparts, it seems important to mention that the miR-212/132 locus maturation also generates two additional miRNAs, miR-212* and miR-132*, whose sequences are indicated in grey in Figure 1c. As the functions of these miRNAs are still unknown, the following sections will focus on miR-132 and miR-212. Nevertheless, one should keep in mind the existence of the star sequences of these miRNAs, which have been reported to be induced, like miR-132 and miR-212, in cortical neurons stimulated by neurotrophins (13), and that are expected to regulate different mRNA target subsets than miR-132 and miR-212 as they exhibit different sequences.
Figure 2.
Genomic features of the miR-212/132 locus in the mouse genome. The diagram indicates the genomic localization of miR-212 and miR-132 pre-miRNA sequences, the two isoform transcripts by which they are encoded in mice (thick lines indicate exons; thin lines indicate introns), as well as the mammal conservation (the sites predicted to be conserved are assigned positive scores, while sites predicted to be fast-evolving are assigned negative ones). These annotations are from the UCSC Genome Center and Ref. (14). Besides, the position of CREB and REST binding sites that were demonstrated to be involved in miR-212/132 transcription (12,13,16) are indicated. Interestingly, all of the indicated binding sites for transcription factors are conserved among the mouse, rat and human genomes.
Table 1.
miR-132 validated targets
| Target | References | |
|---|---|---|
| AChE | Acetylcholinesterase | (57) |
| AT1R | AngiotensinII type 1 receptor | (82) |
| BTG2 | B-cell translocation gene 2 | (35) |
| Cardiac l-type Ca channel β2 subunit protein | (83) | |
| HB-EGF | Heparin-binding EGF-like growth factor | (63) |
| JARID1A | Jumonji, AT-rich interactive domain 1A | (35) |
| MeCP2 | Methyl-CpG-binding protein | (24,35) |
| MMP-9 | Matrix metalloproteinase 9 | (14) |
| p120RasGAP | GTPase-activating protein | (75) |
| p250GAP | GTPase-activating protein | (12,21,84) |
| p300 | Transcriptional co-activator | (35,65) |
| PAIP2A | Polyadenylate-binding protein-interacting protein 2 | (35) |
| PTBP2 | Polypyrimidine tract binding protein 2 | (53) |
| Rb1 | Retinoblastoma tumour suppressor 1 | (77) |
| RXF4 | Regulatory factor X4 | (33) |
| SirT1 | Sirtuin | (85) |
| STAT4 | Signal transducer and activator of transcription 4 | (64) |
Table 2.
miR-212 validated targets
| Target | References | |
|---|---|---|
| HB-EGF | Heparin-binding EGF-like growth factor | (73) |
| MeCP2 | Methyl-CpG-binding protein | (41) |
| MMP-9 | Matrix metalloproteinase 9 | (14) |
| PED/PEA-15 | Antiapoptotic protein of the DED (death effector domain) family | (71) |
| Rb1 | Retinoblastoma tumour suppressor | (77) |
| SPRED1 | Sprouty-related, EVH1 domain containing 1 | (37) |
| STAT4 | Signal transducer and activator of transcription 4 | (64) |
| ZO-1 | Zonula occludens 1 | (86) |
Regarding its transcriptional regulation, the miR-212/132 locus was first identified in neuronal cells as a target of the cAMP-response element binding (CREB) protein transcription factor by a group using an approach called SACO (serial analysis of chromatin occupancy), enabling the identification of functional transcription factor binding sites at the genome scale (12,15). Soon after, another group using the same technique demonstrated that in non-neuronal cells the miR-212/132 locus is under the control of the transcriptional repressor Repressor Element 1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) (16). As illustrated in Figure 2, one REST and several CREB-binding sites are conserved among mammals in the vicinity of miR-212/132 locus, suggesting an evolutionary conserved involvement of CREB and REST in the control of miR-212/132 expression.
As far as the maturation of miR-132 and miR-212 from their primary transcript is concerned, relatively few data are available so far. Wibrand and co-workers analysed the pri-miR-212/132, pre-miR-212, pre-miR-132, miR-212 and miR-132 expressions during the long-term potentiation (LTP) of the rat adult dentate gyrus, and found parallel increases in the expression of pri-miR-212/132, pre-miR-212 and pre-miR-132, which suggests that the pri-miR-212/132 is rapidly transcribed and rapidly and efficiently processed in this context (the dentate gyrus is a part of the hippocampus, which is considered to contribute to memories formation; the LTP is thought to be one of the major cellular mechanisms that underlies learning and memory). The elevations in mature miRNAs were however not detected before 2 h later, suggesting a slower processing of the precursors into mature miRNAs. Besides, the elevation in mature miRNAs levels was found to be much smaller (<2-folds) than that of the primary and precursors forms (induced >50-folds), which may suggest that the precursor processing to mature miRNAs is limited, or may also reflect higher basal levels of mature miRNAs (17). If such patterns of maturation are observed in other contexts remains to be determined. Moreover, in a number of studies, one of the two miRNAs was found to be more induced than the other one. Whether this phenomenon results from a different stability of miR-132 and miR-212 and/or from a preferential processing of one of the two hairpin precursors, and if this is linked to the pre-miR or miR sequences also remains to be elucidated, in the different contexts in which these miRNAs have been studied.
miR-132 AND miR-212 IN THE NEURONAL CONTEXT
The miR-212/132 transcriptional regulation in neurons
As both CREB and REST are involved in processes related to neuronal development and function, it is not surprising that the majority of miR-132 and miR-212's functions were described in a neuronal context. Indeed, CREB is known to be involved in neuronal survival, maturation, differentiation and function, but also to control developmental plasticity, memory formation, adaptive behaviour, drug addiction and to regulate circadian rhythms (18). REST, on the other hand, is a transcriptional repressor known to actively repress neuronal gene expression in non-neuronal cells by acting as a scaffold protein enabling the recruitment, via the co-repressors interacting with its repressor domain, of histone deacetylases and other epigenetic factors themselves silencing the expression of REST target genes (19). Upon neuronal differentiation however, REST is conjugated to ubiquitin by the E3 ubiquitin ligase SCFβTRCP and targeted to proteolysis, enabling the expression of its target genes (20).
A functional binding site for REST has been located between the miR-212 and miR-132 sequences (Figure 2), and the involvement of REST in miR-132 expression has been demonstrated as the expression of a dominant negative form of REST in mouse embryonic fibroblasts results in an increased expression of miR-132 (16) (the report does not mention its effect on miR-212 expression, as that study focussed on brain-specific miRNAs and as miR-212 involvement in neuronal functions was still unknown at that time. Nevertheless, given that miR-212 and miR-132 are derived from the same primary transcript, one can deduce that the entire miR-212/132 locus is under the control of REST). Similarly, the involvement of CREB in the transcriptional control of miR-132 and miR-212 was first demonstrated in rat (12) and then in mouse (13) cortical neurons stimulated with neurotrophins. At least four CRE sites are involved in miR-212/132 transcription in mouse (Figure 2). Three of them were first identified in rat: one located between miR-212 and miR-132, near the REST binding site and the two others located immediately upstream of miR-212 sequence (12). In mouse, a fourth CRE binding site, highly conserved, was more recently identified in the promoter of the gene encoding AK006051 transcript, the three other CRE sites located in the first intron of this transcript corresponding to the sites previously identified in rat (13) (Figure 2).
Although only two transcription factors, REST and CREB, have been formally demonstrated to control the transcription of miR-212/132, other yet unidentified transcriptional regulators must be involved. Indeed, the deletion of the AK006051 transcript intron 1 nearly abolishes the expression of a luciferase reporter gene while the mutation of the three CRE sites localized in this particular intron only slightly reduces the induction of the luciferase following brain-derived neurotrophic factor (BDNF) stimulation (13). In addition, although BDNF-induced CREB-dependent miR-212/132 transcription has been shown to depend on extracellular signal-regulated kinases 1/2 (ERK1/2) activation and partly on mitogen- and stress-activated protein kinase 1/2 (MSK1/2) activation, an unidentified ERK1/2-dependent, MSK1/2- and CREB-independent mechanism may also contribute to miR-212/132 expression in BDNF-stimulated neurons (13) (Figure 3).
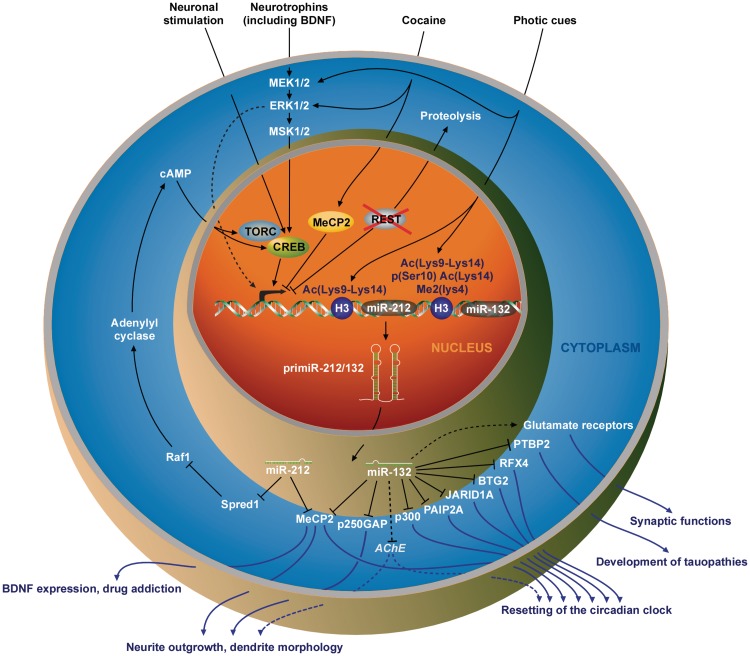
Figure 3.
Inducers and targets of the miR-212/132 locus in the neuronal compartment. In neurons, the transcriptional repressor REST is targeted to proteolysis, enabling the transcription of its target genes. Various stimuli (such as the exposition to neurotrophins or photic cues, or an extended access to cocaine) lead to the transcription of the miR-212/132 locus through CREB activation, although an unidentified ERK1/2-dependent, MSK1/2- and CREB-independent mechanism may also contribute to miR-212/132 expression in BDNF-stimulated neurons (dashed arrow). Histone 3 post-translational modifications are also involved in pri-miR-212/132 expression following light exposure. By repressing the expression of several mRNA targets (AChE is a probable but not yet demonstrated target of miR-132 in neurons), miR-212/132 are involved in neurite outgrowth and dendrite morphology as well as in the resetting of the circadian clock, and would participate to synaptic functions by up-regulating the expression of the glutamate receptors NR2A, NR2B and GluR1. miR-132 expression deregulation is also associated with the development of tauopathies through the targeting of PTBP2. Besides, miR-212 would be involved in regulating the vulnerability to cocaine addiction by targeting Spred1 mRNA.
miR-212/132 are required for neurons morphogenesis
miR-132 was shown to induce neurite outgrowth and to modulate the dendritic morphology of both immature cortical and hippocampal neurons by repressing p250GAP expression (12,21), a GTPase-activating protein involved in neuronal differentiation (22,23). Beside its interaction with p250GAP mRNA, miR-132 also modulates dendritic plasticity by controlling the expression of another target, methyl CpG-binding protein 2 (MeCP2) (24) (Figure 3). While a decrease in MeCP2 expression during the post-natal period postpones neuronal maturation and synapses formation (25), its over-expression triggers dendrite and axon arborization (26), which suggests the need of maintaining MeCP2 levels between narrow ranges to ensure a proper neuronal development (24). Interestingly, as MeCP2 was suggested to control BDNF expression, which itself leads to the induction of miR-212/132 expression, miR-132 would take part in a feedback mechanism involved in the homeostatic control of MeCP2 expression (24). miR-132 up-regulation resulting from the BDNF stimulation of cortical neurons is also involved, most likely indirectly, in the BDNF-induced expression of the glutamate receptors NR2A, NR2B and GluR1 (27) (Figure 3), adding new mechanisms about the involvement of miR-132 in synaptic functions.
Evidence emerging from in vivo studies demonstrated that miR-132 effects on neuron morphogenesis are not limited to in vitro cultured cells. Indeed, the deletion of miR-132/miR-212 locus is associated with a decrease in spine density and in dendrite length and arborization of newborn neurons in the mice adult hippocampus. As miR-132 is the main product of the miR-212/132 locus maturation in this context, miR-132 was proposed to be required for the dendritic growth and arborization of newborn neurons of the adult hippocampus (28). One may have noticed that the reported positive effect of miR-132 expression on dendritic growth and arborization of neurons appears in contradiction with the observation that MeCP2 over-expression also results in dendritic and axonal arborization (26), MeCP2 being a miR-132 target. The reason of this apparent contradiction may be related to the fact that miR-132 controls the expression of several targets involved both in stimulating (like MeCP2) or inhibiting neurite outgrowth and arborization (as p250GAP). In addition, one cannot exclude the participation of still unidentified miR-132 targets in the positive or negative control of neuron arborization.
Moreover, miR-212/132 primary transcript is up-regulated upon mice neuronal stimulation and it was suggested that miR-132 expression, that is specifically localized in active synapses, may play a role in long-term synapse activation and contribute to memorization processes (29). In addition, miR-132 participates to the integration of newborn neurons into the adult dentate gyrus, as it increases during neuron differentiation and maturation, while its knockdown results in decreased synapse formation and impairs the functional integration of newborn neurons (30).
Recent data also indicate that the miR-212/132 locus, that is induced in the visual cortex of light-exposed mice, would regulate the ocular dominance plasticity possibly through miR-132 action on dendritic spine morphology (ocular dominance is the tendency to prefer the visual input from one eye to the other; the ocular dominance plasticity refers to the possibility to modulate the ocular dominance through the ability of the brain to reorganize neuron connections in response to the visual experience, a phenomenon that could occur post-natally during a well-defined sensitive period) (31,32). Interestingly, the induction of pri-miR-212/132 expression in this model would depend on post-translational modifications of histone proteins located at CRE sequences close to miR-132 and miR-212. More precisely, it was shown that the light exposure increases the presence of histone marks known to be regulated by visual experience. Indeed, CRE loci located close to miR-132 sequence and upstream of miR-212 sequence exhibit enhanced lysine 9 and 14 acetylation on histone 3, and the CRE site located close to miR-132 moreover displays additional histone modifications associated with light exposure (enhanced phosphorylation of serine 10 and acetylation of lysine 14 on histone 3, and lysine 4 dimethylation on histone 3) (31).
miR-132 regulates circadian rhythms
Beside its induction by light exposure, a process dependent on histone modifications (31), miR-132 is regulated in a MEK1/2, CREB-dependent manner by photic cues in the suprachiasmatic nucleus and displays an expression profile associated with circadian rhythms, reaching a maximal expression during the subjective day (the subjective day is the part of the circadian cycle corresponding to the illuminated period during an entrainment by light–dark cycles) (33). From a functional point of view, miR-132 would be involved in a negative response to photic entrainment by attenuating the clock resetting triggered by light exposure, an effect that could be mediated by the repression of the RFX4 (regulatory factor X4) protein expression, a transcription factor highly expressed in suprachiasmatic nucleus and induced after light exposure during the night. miR-132 would also modulate the expression of period genes mPer1 and mPer2, which are involved in the normal resetting of the circadian clock (34), by repressing the expression of five targets, three of which being involved in chromatin remodelling, MeCP2, p300 and JARID1A (Jumonji, AT-rich interactive domain 1A) (also called ‘retinoblastoma binding protein 2′), and two being implicated in protein translation repression, B-cell translocation gene 2 (BTG2) and polyadenylate-binding protein-interacting protein 2 (PAIP2A) (35). By regulating the expression of these genes (Figure 3), miR-132 would act as a master factor for chromatin remodelling and protein translation thereby enabling the fine-tuned expression of genes involved in the circadian clock entrainment (35).
A CREB-miR-212-MeCP2 regulatory loop is involved in drug addiction
The best-known example of cross-regulation between transcription factors and miR-212/132 has recently been demonstrated in the field of drug addiction. It has been reported that miR-212, and in a smaller extent, miR-132, could themselves regulate the activity of both CREB and transducer of regulated CREB (TORC), one of its co-activators. Indeed, by repressing the expression of sprouty-related, EVH1 domain containing 1 (SPRED1), a protein known to inhibit Raf phosphorylation and activation (36), miR-212 is responsible for Raf1 activation (37). As Raf1 GTPases increase adenylyl cyclase activity (38), miR-212 over-expression leads to an increased cyclic AMP production, triggering PKA activation and CREB phosphorylation and activation. The CREB-dependent gene expression is further amplified by a positive effect of cAMP-dependent signalling on TORC, as cAMP can protect TORC against degradation by inducing its acetylation by p300 (37,39) and as PKA activity promotes TORC nuclear relocalization (40) (Figure 3). In this context, it was shown that by amplifying CREB activity, which is known to negatively modulate the reward response to cocaine, miR-212 expression can protect from cocaine addiction by decreasing the responsiveness to the motivational properties of the drug (37). Additional findings have shown that an interaction between miR-212 and MeCP2 [a target of both miR-132 (24) and miR-212 (41)], occurring in the dorsal striatum, is also involved in cocaine addiction. Indeed, striatal MeCP2, whose expression is increased in rats with extended access to cocaine, inhibits miR-212 expression that, in turn, represses MeCP2 expression. As MeCP2 levels are themselves correlated to BDNF expression, that controls cocaine intake, miR-212 would fine-tune the responses to drug abuse both by increasing CREB signalling and decreasing BDNF expression, resulting in a limited cocaine intake (42) [interested readers can refer to (43) for a figure illustrating this BDNF–CREB–MeCP2–miR-212 interplay]. The negative effect of MeCP2 on miR-212/132 expression identified in this study seems, a priori, in contradiction with the suggestion that the loss of MeCP2 would result in the down-regulation of miR-132 expression through decreased BDNF levels in rat cortical neurons (24). However, although the effect of MeCP2 on miR-212/132 transcription may depend on cell types, we cannot exclude that MeCP2 may exert an inhibitory effect on miR-212/132 transcription in cortical neurons as well, which would be hidden by the positive effect of BDNF on miR-212/132 expression.
miR-212/132 deregulation is associated with several brain-related diseases
Given their involvement in neuronal development and functions, it is not surprising that deregulated expression patterns for miR-132 and miR-212 have been associated with developmental defects as well as brain-related disorders. For example, a down-regulation of miR-212 has been identified in foetuses with anencephaly (44). miR-132 expression is down-regulated in two mouse models of Huntington's disease (45), as well as in the brain of human patients suffering from this pathology (46). Moreover, miR-132 and miR-212 are both deregulated in the prefrontal cortex of individuals affected by schizophrenia and bipolar disorders (47,48). Both miR-132 and miR-212 are down-regulated in the brain of α-synuclein (A30P)-transgenic mice, a model of Parkinson's disease (49). miR-212 was also found to be down-regulated in Alzheimer's disease patients, and its expression modulation correlates with the density of neurofibrillary tangles, a characteristic lesion of Alzheimer's disease (50). However, miR-132 down-regulation in Alzheimer's disease patients remains controversial, probably due to the differences in the populations studied, the tissues analysed and the protocols used by different groups (51,52). Recently, Smith and colleagues found that miR-132 is also down-regulated in patients suffering from progressive supranuclear palsy, a form of tauopathy linked to abnormal ratios of four repeats- and three repeats-tau isoforms (4R:3R tau ratios). The involvement of miR-132 in this disease would be explained by its ability to target the neuronal splicing factor PTBP2, whose over-expression would lead to abnormal 4R:3R tau ratios and the development of tauopathy (53). Interestingly, a recent report also mentioned that the in vivo microinjection of antagomirs targeting miR-132, which is induced in the mice hippocampus following epileptic seizures, diminished the seizure-elicited neuronal death (54).
Besides their numerous roles in neuronal development, functions and related diseases, increasing evidences point towards an important involvement of miR-132 and miR-212 in mediating inflammatory processes. Subsequently, miR-132 was recently designated as a ‘NeurimmiR’, a class of miRNAs regulating both neuronal and immune functions and was suggested to function as a cross-talk between both systems (55). This idea is strongly supported by the observation that, in vivo, miR-132 knockdown in newborn neurons impairs their integration in the adult dentate gyrus, while its knockdown in PC12 cells results in the up-regulation of pro-inflammatory gene expression (30). Indeed, Luikart et al. (30) suggested that the miR-132-regulation of pro-inflammatory gene expression may be part of the program necessary for the functional integration of newborn neurons in the adult central nervous system. In addition, the pre-incubation of cortical neurons with glucocorticoids prevents the BDNF-dependent miR-132 induction and the downstream up-regulation of the expression of glutamate receptors NR2A, NR2B and GluR1 above-mentioned (27), suggesting another possible link between neuronal and inflammatory processes. As described in the next section, miR-132 and miR-212 not only contribute to immune processes, but are also regulated by inflammatory signals.
miR-132 AND miR-212 REGULATE AND ARE REGULATED BY IMMUNE PROCESSES
These last 5 years, several groups performing miRNA expression profiling in diverse immune-related contexts came to highlight the induction of miR-132 (and miR-212) in several cell types including monocytes, macrophages, mast cells and lymphatic endothelial cells. miR-132 expression was first described in the inflammatory context by the Baltimore's group, who profiled the expression of 200 miRNAs in the LPS-stimulated human monocytic THP-1 cell line and showed that this miRNA, among others, is up-regulated by this pro-inflammatory signal (56). Intriguingly, the induction of pri-miR-212/132 is not detected in LPS-stimulated primary murine bone-marrow-derived macrophages, while it is in THP-1 cells (13). Whether these observations are due to the total absence of a miR-212 and miR-132 induction in those primary murine macrophages, or rather to a very transient induction of the primary precursor, or to modifications in the maturation of the primary and precursor forms leading to increased miR-212/132 levels despite an absence of transcriptional activation, remains to be determined.
miR-132 is also induced in LPS-stimulated primary human macrophages, as well as in the bone marrow and splenocytes of LPS-treated mice, where it represses the acetylcholinesterase (AChE) expression (57). By repressing the expression of AChE, an enzyme hydrolysing the acetylcholine, considered as an important inhibitor of peripheral inflammation, miR-132 would play a role in the brain-to-body resolution of inflammation. Although the regulation of AChE by miR-132 has been demonstrated in splenocytes and macrophages, it probably also occurs in the brain as suggested by the authors, and in this manner plays a role in circadian rhythms (57). Indeed, whereas AChE expression is maximal during sleeping hours and minimal during activity periods (58), miR-132 expression demonstrates an opposite expression profile (33). Incidentally, mice overexpressing AChE exhibit circadian irregularities when subjected to a reversal of the light/dark cycles (59). Besides, the miR-132-AChE interaction may also regulate neuron morphogenesis, as AChE is involved in neurite outgrowth and extension (60,61). A regulation of AChE by miR-132 in neuronal cells would also confer an indirect role for this miRNA in the regulation of cholinergic anti-inflammatory effects attributed to AChE (62). It would therefore be of great interest to determine if the AChE/miR-132 interaction occurs in neuronal cells, given that recognized effect of the miR-132/212 locus expression in neurons may be mediated partly through their interaction with AChE. These observations strongly suggest that the consequences of miR-212/132 deregulated expression on circadian rhythms and neuron morphogenesis are the results of changes in the expression of several targets, and not only in those that were identified as targets in these particular contexts.
In mouse and human mast cells activated by immunoglobulin E (IgE), the induction of miR-132 has been shown to repress heparin-binding epidermal-like growth factor (HB-EGF) expression. HB-EGF, which is involved in cell proliferation, cell migration and wound healing among other functions, also exhibits an increased expression in IgE-induced mast cells. Therefore, by targeting HB-EGF mRNA, miR-132 might be involved in a negative response to mast cells activation, limiting the remodelling and stimulation of the tissue environment upon chronic allergen exposure (63). Similarly, the induction of miR-132 and miR-212 in IL-12-stimulated primary human natural killer cells negatively regulates the IL-12 signalling pathway through the repression of signal transducer and activator of transcription 4 (STAT4) expression (64).
miR-132 and in a lesser extent, miR-212, are also induced during primary human lymphatic endothelial cells (LEC) and human foreskin fibroblasts infection by the Kaposi's sarcoma-associated herpes virus (KSHV), as well as during the infection of THP-1 by KSHV, herpes simplex virus-1 (HSV-1) and human cytomegalovirus (HCMV) (65). The induction of miR-212/132 locus in KSHV-infected LEC depends on the activation of ERK and p38 MAPK but also on the activation of the cAMP/PKA pathway, all of these signalling pathways subsequently leading to the phosphorylation and activation of CREB. miR-132 induction upon viral infection might also be part of the viral defensive strategy to evade immunity. Indeed, miR-132 targets the transcriptional co-activator p300 (65), described to interact with central actors of the inflammatory response (66,67) and to play a role in the initiation of the antiviral response (68). Therefore, the induction of miR-132 upon viral infection would facilitate viruses replication by inhibiting the expression of genes known to be induced by the interferon response (65). Incidentally, the targeting of p300 is a mechanism developed by several oncogenic DNA viruses enabling them to evade the innate immunity (69). It is worth noting that the induction of miR-132 upon viral infection is transient, what could be, at least partly, explained by the fact that the transcriptional co-activator p300 binds CREB to allow gene transcription activation (65,66) and thus controls miR-132 expression. Therefore, the repression of p300 expression by miR-132 would be part of a negative feed-back loop leading to decreased miR-132 expression (65).
From a clinical point of view, miR-132 is also, among other miRNAs, up-regulated in peripheral blood mononuclear cells (PBMC) of patients suffering from rheumatoid arthritis (70). Both the monocyte and macrophage population of PBMC cells display a higher expression of these miRNAs when compared with the lymphocyte fraction, suggesting that monocytes and macrophages most likely predominantly contribute to the general up-regulation of these miRNAs—and particularly of miR-132—in PBMC (70). This is consistent with data obtained in our group, indicating that miR-132 and miR-212 are also induced in THP-1 monocytes and macrophages stimulated by various inflammatory cytokines (A. Tacheny, unpublished data). This observation, in consideration with the reported induction of miR-132 in primary human macrophages treated with CpG oligonucleotides, a TLR9 ligand (57), suggests that miR-132 expression in human monocytes and macrophages would be induced by bacterial and viral ligands and more generally by pro-inflammatory molecules. In addition, as several validated targets of miR-132 and miR-212 participate in the inflammatory response (see Tables 1 and 2), like STAT4, AChE, HB-EGF, p300, MeCP2 and SirT1, one might consider these miRNAs as part of mechanisms allowing the resolution of inflammation.
miR-212 AND miR-132: BESIDES THEIR NEURIMMIR FUNCTIONS
Besides their involvement in neuronal processes and inflammation, miR-132 and miR-212 have also been described in a number of other fields, in studies enabling the identification of numerous other targets (Tables 1 and 2). Although not developed in this review, it is worth mentioning that a number of reports suggested that miR-212/132 expression could be regulated by hormones, nutrition and metabolism, unveiling emerging additional contexts in which miR-212/132 would exert their regulatory roles.
The most documented emerging field in which miR-212/132 have been involved is probably cell transformation and tumourigenesis—although this may likely result from the increasing interest devoted to miRNA deregulation in cancer. As shown in Table 3, both miR-132 and miR-212 were shown to be up-regulated or down-regulated in different cancer types. Moreover, both tumour-promoting and tumour-suppressing functions (depending on cancer types) were attributed to miR-212/132, unveiling the complexity of their involvement in tumourigenesis. For example, miR-212 down-regulation has been associated with the resistance/bad response to several anti-cancer treatments (71–73), while a reduced miR-132 expression was observed in good responders to ifosfamide, an alkylating agent used in the treatment of cancer (74). Other reports suggest that miR-212 would negatively affect gastric cancer cells proliferation (41) while miR-132 would contribute, in human tumours and hemangiomas, to the angiogenic switch by stimulating endothelial cell proliferation as well as their tube-forming capacity through the targeting of p120RasGAP (75). Regarding the involvement of miR-212/132 in pancreatic cancer, some authors reported the down-regulation of miR-132 in carcinoma tissues compared with their non-cancerous counterparts (76) whereas others found an elevated miR-212/132 expression in pancreatic cancer patients (77), with opposite effects on pancreatic cancer cell proliferation. Although one could speculate that the divergent data obtained in these studies may partly result from different genetic backgrounds in the populations studied, further studies are needed to clarify the effect of miR-212/132 expression on the proliferation of pancreatic cancer cells. Nevertheless, these reports suggest the possibility that a same miRNA may both positively and negatively regulate certain cell functions.
Table 3.
miR-212 and miR-132 expression are deregulated in several cancer types
| Cancer type | miRNA | Induced (↑) or repressed (↓) in cancer versus normal tissues/ cells | References |
|---|---|---|---|
| Chronic lymphoblastic leukaemia | miR-132 | ↑ | (87) |
| Haemangioma and endothelium of human tumours | miR-132 | ↑ | (75) |
| Squamous cell carcinoma of the tongue | miR-132 | ↑ | (88) |
| Pancreatic cancer | miR-132 | ↓ | (76) |
| miR-132, miR-212 | ↑ | (77) | |
| Gastric cancer | miR-212 | ↓ | (41) |
| Non-small cell lung cancer | miR-212 | ↓ | (71) |
| Oral squamous cell carcinoma | miR-212 | ↑ | (89) |
CONCLUSION AND FUTURE DIRECTIONS
As outlined in this review, numerous studies have highlighted the important role of the miR-212/132 locus in neuronal morphogenesis and function. However, the activities of miR-212/132 are not limited to the neuronal compartment and an increasing number of publications point towards a participation of these miRNAs in inflammation and immune processes and importantly, in the cross-talk between neural and immune functions. The identification of a number of targets enabled to propose mechanisms by which miR-132 and/or miR-212 are involved in these processes. However, as miRNAs are predicted to target numerous mRNAs, one should keep in mind that the resulting effects of a deregulated expression of these miRNAs might potentially results from modifications in the expression of other targets as well, as herein exemplified with the miR-132/AChE interaction. From a transcriptional point of view, although REST and histone modifications were both demonstrated to regulate miR-212/132 transcription, the transcription factor that transcends all the context-specific inductions of miR-212/132 is undoubtedly CREB, which appears to control their transcription in a wide variety of situations and biological processes.
Although miR-212 and miR-132 are simultaneously transcribed, several studies have highlighted that one or the other of these miRNAs was more strongly induced and/or more expressed than the other one in different contexts, raising the currently obscure question of the regulation of miRNAs maturation and turnover. Several proteins have been involved in the enhanced processing or stability of particular subsets of miRNAs or even of specific miRNAs [interested readers can refer to the following reviews (78,79)], but if such actors are involved in the preferential maturation and/or processing of miR-132 or miR-212 remains to be determined. Remenyi et al. (13) suggested that the loop sequences of these miRNAs, which are highly conserved among vertebrates, may play a role in their processing, but the putative proteins binding these loops are still unknown. On the other hand, Lagos et al. (65) suggested that miR-212 would be less stable rather than differentially processed compared to miR-132 in lymphatic endothelial cells, but the maturation and stability of miR-212 and miR-132 may also depend on cell types and physiological contexts. Further studies are therefore clearly required to get deeper insights into the post-transcriptional regulation of miR-212/132.
Another incompletely addressed question so far relates to the relative redundancy of miR-132 and miR-212 functions. The similarity of miR-212 and miR-132 mature sequences and more importantly their identical seed sequences suggest that these miRNAs may result from a gene duplication event in the evolution course. It is therefore not surprising that these miRNAs share several target mRNAs. However, as most studies do not systematically investigate miR-212 and miR-132, it is currently not well defined if these miRNAs really have specific targets, or if they mostly exert numerous and overlapping functions. The currently prevalent model of miRNA target recognition defines the miRNA seed region as the critical determinant of the specific targets recognition. According to this model, miR-212 and miR-132 would be functionally equivalent. However, increasing evidence also point towards a role of miRNA non-seed nucleotides, including in their 3′ region, in the target recognition (2). Indeed, base-pairing between the mRNA target and the miRNA 3′ region can either supplement the traditional seed pairing, or compensate for single-nucleotide bulge or mismatch in the seed region (80,81). The diverging nucleotides between miR-132 and miR-212 sequences (see Figure 1a) may therefore be involved in specific target recognition. Moreover, another likely important parameter of the interaction between a miRNA and a target mRNA is the expression level of the two partners. This is illustrated in the work of Lagos and co-workers, who demonstrated that while miR-132 inhibition in KSHV-infected lymphatic endothelial cells (that overexpress miR-132 compared with non-infected cells) reverses its effect on p300 expression, its inhibition in non-infected cells does not affect p300 levels. These results suggest that a critical level of miR-132 is necessary for its ability to repress p300 expression (65). Therefore, although both miR-132 and miR-212 may target a same mRNA when expressed at sufficiently high levels, their physiological expression levels in different cell types and conditions likely influence their overall ability to repress the expression of a specific mRNA target. A differential processing of miR-132 and miR-212 may therefore account for their putative different abilities to repress a target expression. A particular care should therefore be taken when analysing the potential inhibitory role of a miRNA on a target mRNA expression, as experiments involving the over-expression of miRNAs may potentially identify interactions between miRNAs and mRNA targets that cannot occur at physiological concentrations. Further studies are therefore necessary in order to determine the extent to which miR-132 and miR-212 functions are overlapping. A possible scenario would be that miR-212 and miR-132 exert both redundant and specific functions, by being kept under the same transcriptional control and regulated differently at the post-transcriptional level, and/or by targeting both common and specific mRNA targets.
FUNDING
Fonds National pour la Recherche Scientifique (FNRS, Belgium) (doctoral fellowship to A.W.). Funding for open access charge: University of Namur (FUNDP), Belgium.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
A. Wanet is recipient of the doctoral fellowship from the Fonds National pour la Recherche Scientifique (FNRS, Belgium). The authors thank Michel Savels for his contribution to the figure layout.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int. J. Biochem. Cell Biol. 2009;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27:27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 5.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 6.Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L. Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 2009;53:245–257. doi: 10.1387/ijdb.082691av. [DOI] [PubMed] [Google Scholar]
- 7.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 11.Olena AF, Patton JG. Genomic organization of microRNAs. J. Cell Physiol. 2010;222:540–545. doi: 10.1002/jcp.21993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl Acad. Sci. USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem. J. 2010;428:281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- 14.Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PA, Thum T, Groner B, Chowdhury K. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat. Genet. 2010;42:1101–1108. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- 15.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl Acad. Sci. USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 2010;31:636–645. doi: 10.1111/j.1460-9568.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 19.Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system–what's the REST of the story? Neurosci. Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl Acad. Sci. USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakazawa T, Watabe AM, Tezuka T, Yoshida Y, Yokoyama K, Umemori H, Inoue A, Okabe S, Manabe T, Yamamoto T. p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol. Biol. Cell. 2003;14:2921–2934. doi: 10.1091/mbc.E02-09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Komiya M, Sone K, Hirose E, Gotoh N, Morii H, Ohta Y, Mori N. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 2002;22:8721–8734. doi: 10.1128/MCB.22.24.8721-8734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J. Neuropathol. Exp. Neurol. 2005;64:537–544. doi: 10.1093/jnen/64.6.537. [DOI] [PubMed] [Google Scholar]
- 26.Jugloff DG, Jung BP, Purushotham D, Logan R, Eubanks JH. Increased dendritic complexity and axonal length in cultured mouse cortical neurons overexpressing methyl-CpG-binding protein MeCP2. Neurobiol. Dis. 2005;19:18–27. doi: 10.1016/j.nbd.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165:1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 28.Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl Acad. Sci. USA. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat. Neurosci. 2011;14:1237–1239. doi: 10.1038/nn.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 2011;14:1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mper2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 2010;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 37.Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Q, Gros R, Gray ID, Taussig R, Ferguson SS, Feldman RD. Raf kinase activation of adenylyl cyclases: isoform-selective regulation. Mol. Pharmacol. 2004;66:921–928. [PubMed] [Google Scholar]
- 39.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Wada R, Akiyama Y, Hashimoto Y, Fukamachi H, Yuasa Y. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int. J. Cancer. 2009;127:1106–1114. doi: 10.1002/ijc.25126. [DOI] [PubMed] [Google Scholar]
- 42.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J, Nestler EJ. MeCP2 and drug addiction. Nat. Neurosci. 2010;13:1039–1041. doi: 10.1038/nn0910-1039. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z, Chang H, Li Y, Zhang T, Zou J, Zheng X, Wu J. MicroRNAs: potential regulators involved in human anencephaly. Int. J. Biochem. Cell Biol. 2010;42:367–374. doi: 10.1016/j.biocel.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, et al. Altered microRNA regulation in Huntington's disease models. Exp. Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 47.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr. Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillardon F, Mack M, Rist W, Schnack C, Lenter M, Hildebrandt T, Hengerer B. MicroRNA and proteome expression profiling in early-symptomatic alpha-synuclein(A30P)-transgenic mice. Proteomics Clin. Appl. 2008;2:697–705. doi: 10.1002/prca.200780025. [DOI] [PubMed] [Google Scholar]
- 50.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 52.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 53.Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buée L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 54.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, et al. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soreq H, Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol. Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 58.Schiebeler H, von Mayersbach H. Circadian variations of acetylcholine esterase (E.C.3.1.1.7) in rat brains. Int. J. Chronobiol. 1974;2:281–289. [PubMed] [Google Scholar]
- 59.Cohen O, Erb C, Ginzberg D, Pollak Y, Seidman S, Shoham S, Yirmiya R, Soreq H. Neuronal overexpression of “readthrough” acetylcholinesterase is associated with antisense-suppressible behavioral impairments. Mol. Psychiatry. 2002;7:874–885. doi: 10.1038/sj.mp.4001103. [DOI] [PubMed] [Google Scholar]
- 60.Grisaru D, Sternfeld M, Eldor A, Glick D, Soreq H. Structural roles of acetylcholinesterase variants in biology and pathology. Eur. J. Biochem. 1999;264:672–686. doi: 10.1046/j.1432-1327.1999.00693.x. [DOI] [PubMed] [Google Scholar]
- 61.Sklan EH, Berson A, Birikh KR, Gutnick A, Shahar O, Shoham S, Soreq H. Acetylcholinesterase modulates stress-induced motor responses through catalytic and noncatalytic properties. Biol. Psychiatry. 2006;60:741–751. doi: 10.1016/j.biopsych.2006.03.080. [DOI] [PubMed] [Google Scholar]
- 62.Rosas-Ballina M, Olofsson PS, Ochani M, Valdés-Ferrer SI, Levine YA, Reardon C, Tusche MW, Pavlov VA, Andersson U, Chavan S, et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science. 2011;334:98–101. doi: 10.1126/science.1209985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molnár V, Ersek B, Wiener Z, Tömböl Z, Szabó PM, Igaz P, Falus A. MicroRNA-132 targets HB-EGF upon IgE-mediated activation in murine and human mast cells. Cell. Mol. Life Sci. 2011 doi: 10.1007/s00018-011-0786-3. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Y, Lei Y, Zhang H, Hou L, Zhang M, Dayton AI. MicroRNA regulation of STAT4 protein expression: rapid and sensitive modulation of IL-12 signaling in human natural killer cells. Blood. 2011;118:6793–6802. doi: 10.1182/blood-2011-05-356162. [DOI] [PubMed] [Google Scholar]
- 65.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 66.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 67.Matt T. Transcriptional control of the inflammatory response: a role for the CREB-binding protein (CBP) Acta Med. Austriaca. 2002;29:77–79. doi: 10.1046/j.1563-2571.2002.02010.x. [DOI] [PubMed] [Google Scholar]
- 68.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 69.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 70.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res. Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Incoronato M, Garofalo M, Urso L, Romano G, Quintavalle C, Zanca C, Iaboni M, Nuovo G, Croce CM, Condorelli G. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70:3638–3646. doi: 10.1158/0008-5472.CAN-09-3341. [DOI] [PubMed] [Google Scholar]
- 72.Rui W, Bing F, Hai-Zhu S, Wei D, Long-Bang C. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1) J. Cell. Mol. Med. 2010;14:206–214. doi: 10.1111/j.1582-4934.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough WG, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PLoS One. 2010;5:e12702. doi: 10.1371/journal.pone.0012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gougelet A, Pissaloux D, Besse A, Perez J, Duc A, Dutour A, Blay JY, Alberti L. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int. J. Cancer. 2011;129:680–690. doi: 10.1002/ijc.25715. [DOI] [PubMed] [Google Scholar]
- 75.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 77.Park JK, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, Postier RG, Brackett DJ, Schmittgen TD. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem. Biophys. Res. Commun. 2011;406:518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 79.Davis BN, Hata A. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Commun. Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 82.Elton TS, Kuhn DE, Malana GE, Martin MM, Nuovo GJ, Pleister AP, Feldman DS. MiR-132 regulates angiotensin II type 1 receptor expression through a protein coding region binding site. Circulation. 2008;18:S513. [Google Scholar]
- 83.Carrillo ED, Escobar Y, González G, Hernández A, Galindo JM, García MC, Sanchez JA. Posttranscriptional regulation of the β2-subunit of cardiac L-type Ca2+ channels by microRNAs during long-term exposure to isoproterenol in rats. J. Cardiovasc. Pharmacol. 2011;58:470–478. doi: 10.1097/FJC.0b013e31822a789b. [DOI] [PubMed] [Google Scholar]
- 84.Godoy J, Nishimura M, Webster NJ. Gonadotropin-releasing hormone induces miR-132 and miR-212 to regulate cellular morphology and migration in immortalized LbetaT2 pituitary gonadotrope cells. Mol. Endocrinol. 2011;25:810–820. doi: 10.1210/me.2010-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol. Clin. Exp. Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 87.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 89.Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int. J. Immunopathol. Pharmacol. 2010;23:1229–1234. doi: 10.1177/039463201002300427. [DOI] [PubMed] [Google Scholar]