Abstract
Cytosine residues in mammalian DNA occur in at least three forms, cytosine (C), 5-methylcytosine (M; 5mC) and 5-hydroxymethylcytosine (H; 5hmC). During semi-conservative DNA replication, hemi-methylated (M/C) and hemi-hydroxymethylated (H/C) CpG dinucleotides are transiently generated, where only the parental strand is modified and the daughter strand contains native cytosine. Here, we explore the role of DNA methyltransferases (DNMT) and ten eleven translocation (Tet) proteins in perpetuating these states after replication, and the molecular basis of their recognition by methyl-CpG-binding domain (MBD) proteins. Using recombinant proteins and modified double-stranded deoxyoligonucleotides, we show that DNMT1 prefers a hemi-methylated (M/C) substrate (by a factor of >60) over hemi-hydroxymethylated (H/C) and unmodified (C/C) sites, whereas both DNMT3A and DNMT3B have approximately equal activity on all three substrates (C/C, M/C and H/C). Binding of MBD proteins to methylated DNA inhibited Tet1 activity, suggesting that MBD binding may also play a role in regulating the levels of 5hmC. All five MBD proteins generally have reduced binding affinity for 5hmC relative to 5mC in the fully modified context (H/M versus M/M), though their relative abilities to distinguish the two varied considerably. We further show that the deamination product of 5hmC could be excised by thymine DNA glycosylase and MBD4 glycosylases regardless of context.
INTRODUCTION
5-hydroxymethylcytosine (5hmC) is a constituent of nuclear DNA, present in many tissues and cell types (1), but relatively enriched in embryonic stem cells (2) and Purkinje neurons (3). 5hmC in the mammalian genome depends on pre-existing 5-methylcytosine (5mC) (4). There are three mammalian ten eleven translocation (Tet) proteins that convert 5mC to 5hmC (2). In non-CpG context (CpA or CpT), 5mC is asymmetrical and 5hmC occurs largely DNA strand-specific (4). In the context of the palindromic CpG dinucleotide, 5hmC presumably exists as fully hydroxymethylated (H/H) form in cells and persists through cell division (5,6). However, the question remains how these H/H sites are maintained after semi-conservative DNA replication.
5hmC has also been proposed as a potential intermediate in active DNA demethylation via the base excision repair pathway (7,8). Methyl-CpG-binding domain 4 (MBD4) contains both an N-terminal MBD and a C-terminal thymine glycosylase domain that acts on G:T and G:U mismatches (9). Of particular interest is a recent report indicating that, in zebrafish, the activation-induced cytidine deaminase (AID) and MBD4 cooperate to demethylate DNA (10). Consistent with a role in DNA demethylation in mammals, AID is required to demethylate pluripotency genes during reprogramming of the somatic genome in embryonic stem cell fusions (11), and AID-deficient animals are less efficient in erasure of DNA methylation in primordial germ cells (12). It is noteworthy that AID promotes 5mC deamination, resulting in thymine (10,13), as well as 5hmC deamination, which would produce 5-hydroxymethyluracil (5hmU or hU) (8). Here, using in vitro biochemical methods, we investigated whether hydroxymethylation is maintained after DNA replication, whether methyl-DNA binding proteins inhibit Tet1 activity, and whether the deamination product of hydroxymetylcytosine could be excised by the glycosylase activities of MBD4 and thymine DNA glycosylase (TDG).
MATERIALS AND METHODS
Protein expression and purification
The expression constructs used to generate recombinant proteins in this study are listed in Supplementary Table S1 and detailed purification schemes are provided in Supplementary Methods. All constructs (except hDNMT1 full length) were expressed in Escherichia coli BL21 (DE3) Codon-plus RIL (Stratagene) harboring the RIL-Codon plus plasmid. 6×His-SUMO-tagged and non-cleavable 6×His-tagged proteins were expressed using modified pET28b vectors pETHisSumo (14) or pET6H (15), respectively. Glutathione S-transferase- (GST-) tagged proteins were expressed in pGEX-2T (GE healthcare) or a modified pET21d with GST inserted between BamHI and EcoRI restriction sites.
In general, cells were cultured in Luria–Bertani (LB) medium (supplemented with 1 mM MgCl2 and 1 mM ZnCl2, and either 50 mg ml−1 kanamycin or 100 mg ml−1 ampicillin) at 37°C until OD600 of ∼0.5–1.0 before shifting the temperature down to 14–24°C. After 2–3 h, 0.2–0.4 mM isopropyl β-d-thiogalactoside was supplied to induce protein expression and cells were cultured overnight (∼12–16 h). For MBD1, the induction time was 2 h. For UHRF1 (residues 124–628), expression cultures were grown overnight at 25°C in auto-induction medium (16). Cells were harvested and lysed as a 20% (v/v) suspension in 20 mM sodium phosphate, pH 7.4 (or 20 mM HEPES–NaOH, pH 7.0), 300–500 mM NaCl, 5% (v/v) glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) and 2 mM phenylmethylsulfonyl fluoride by two passes through an ice-cold French pressure cell press or by sonication (6 min total, 1 s on for 3 s off) for DNMT3 proteins. The lysate was clarified by centrifugation at 50 000g (or twice at 38 000g) for 50–60 min and filtered with cellulose nitrate membrane (Whatman).
The following chromatographic columns (all from GE Healthcare) were used for purification: nickel-charged HisTrap-HP, HiTrap-Q, HiTrap-SP, HiTrap-Heparin, Sephacryl-300 (16/60), Superdex-200 (16/60), Superdex-75 (16/60), GSTrap.
The 6×His-SUMO tag was cleaved by Ulp1 protease at 25 U ml−1 in room temperature for 2 h, whereas GST tag was cleaved by thrombin during dialysis in 20 mM Tris–HCl, pH 8.5, 150 mM NaCl, 5% glycerol and 0.5 mM TCEP for 6 h at 4°C or by PreScission protease (for GST-p66β).
Overexpression of human DNMT1 in Pichia pastoris
Human DNMT1 full-length (residues 1–1616) is overexpressed in P. pastoris using the Multicopy Pichia Expression pPIC3.5K (Invitrogen) modified to include a His-tag at the N-terminus of expressed proteins. An NdeI site was added so that the dnmt1 gene can be inserted into the vector. A 6L-scale fermentation resulted in ∼500 ml of the induced cell pellets, which were divided into 32 tubes. The amount of soluble full-length DNMT1 was estimated to be around 9 mg per 15 ml cell pellets.
Pichia expressed His6-tagged DNMT1 was purified with Ni-column, HiTrap-SP and Sephacryl-300 in the buffer of 20 mM HEPES–NaOH, pH 7.0, 300 mM NaCl, 5% glycerol and 1 mM dithiothreitol (DTT) and further purified from associated nucleic acid through Q column, eluting at ∼250 mM NaCl in the same buffer with 0.5 mM TCEP, instead of DTT.
Methyl transfer assays using oligonucleotides
Methyl transfer activity assays were performed in 50 mM Tris–HCl pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA) (for DNMT1) or in 50 mM Tris–HCl pH 7.5, 2.5% glycerol and 0.5 mM TCEP (for DNMT3A2/DNMT3L and DNMT3B2/DNMT3L). The reaction mixture (20 µl total volume) contained 5.5 µM [methyl-3H] AdoMet (10.0 Ci/mmol; Perkin Elmer) and 1.0 µM oligonucleotides (see below), and enzymes (20 nM DNMT1 N-terminal deletions Δ600, Δ644 and Δ728, or 200 nM DNMT1 FL and Δ350 or 300 nM DNMT3a2/3L and DNMT3B2/3L). All enzymes were pre-incubated with AdoMet for 10 min at 37°C before the addition of DNA.
The reaction times for DNMT1 were 0–15 min, and terminated by the addition of 1.5 mM unlabeled S-adenosyl-l-homocysteine (AdoHcy). The reactions for DNMT3A2/DNMT3L, DNMT3B2/DNMT3L and DNMT3A-C/DNMT3L-C complex were carried out at 37°C for 0–60 min, and terminated by the addition of 1% sodium dodecyl sulfate and 1 mg ml−1 of protease K and heated at 50°C for 15 min. The reaction mixtures were spotted on DE81 paper circles (Whatman), washed twice with 5 ml of cold 0.2 M NH4HCO3, twice with 5 ml of deionized water and once with 5 ml of ethanol. The dried circles were subjected to liquid-scintillation counting with Cytoscint scintillant. Each reaction was performed in duplicate.
To convert from counts-per-minute (cpm) to transferred methyl group concentration, we generated a linear calibration curve using 200 nM of HhaI methyltransferase to completely methylate 125, 250, 500 and 1000 nM of a 12-mer single CpG hemi-methylated DNA. After background subtraction, the resulting cpm's were plotted against the DNA concentration.
DNA binding assay
Fluorescence polarization measurements were carried out at 25°C on a Synergy 4 Microplate Reader (BioTek). A 10 nM of 6-carboxy-fluorescein (FAM)-labeled double strand DNA [FAM-5′-CCATGXGCTGAC-3′/5′-GTCAGYGCATGG-3′ where X and Y are C, 5mC (M), 5hmC (H), T, U or 5hmU] was incubated for 10 min with increasing amounts of proteins in binding buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5% glycerol and 0.5 mM TCEP). No change in fluorescence intensity was observed with the addition of protein. Curves were fit individually using GraphPad PRISM 5.0d software (GraphPad Software Inc.). Binding constants (KD) were calculated as [mP] = [maximum mP] × [C]/(KD + [C]) + [baseline mP], and saturated [mP] was calculated as saturation = ([mP] − [baseline mP])/([maximum mP]–[baseline mP]), where [mP] is milli-Polarization and [C] is protein concentration. Averaged KD and its standard error were reported.
DNA glycosylase activity assay
DNA glycosylase activity assay was performed similar to previously described except FAM labeled DNA oligonucleotides were used instead of radio-labeled DNAs (15). Purified MBD4 or TDG protein (0.5 µM) and 0.5 µM of double strand FAM labeled 32-mer annealed DNA (see below) were mixed in 20 µl nick buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.1% BSA) and incubated at 37°C for 1 h. Reactions were stopped by adding 2 µl of 1 N NaOH, and boiled for 10 min before 20 µl of loading buffer (98% formamide, 1 mM EDTA and 1 mg ml−1 of bromophenol blue and xylene cyanole) were added and boiled for another 10 min. Samples were immediately put into ice water to cool down and loaded on a 10 × 10 cm2 denaturing PAGE gel containing 15% acrylamide, 7 M urea and 24% formamide in 1 × TBE buffer. The gels were run at 200 V for 60 min. FAM-labeled single strand DNA was visualized by UV exposure. The following olignonucleotides were synthesized at the New England Biolabs:
(FAM)-5′-TCGGATGTTGTGGGTCAGXGCATGATAGTGTA-3′
3′-AGCCTACAACACCCAGTCGYGTACTATCACAT-5′
where X, Y = C, 5mC (M), 5hmC (H), U, T or 5hmU (hU).
RESULTS
DNMT3a and DNMT3b have comparable activities on C/C, M/C and H/C substrates
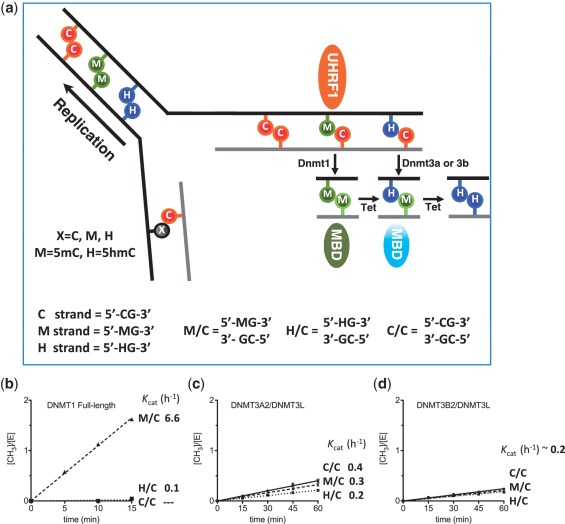
In mammals, DNA methyltransferases (DNMTs) include three members, in two families that are structurally and functionally distinct (17). The DNMT3A and DNMT3B (18,19), coupled with regulatory factor Dnmt3-Like (DNMT3L) protein (20,21), establish the initial methylation pattern de novo, while DNMT1 and its accessory protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) (22,23) maintain this pattern during chromosome replication. We first asked which Dnmts methylate the newly synthesized cytosine in the context of hemi-hydroxymethylated CpG site (H/C) (Figure 1). Using a 32-bp DNA oligonucleotide containing a single CpG site, either unmodified (C/C), hemi-methylated (M/C) or hemi-hydroxymethylated (H/C), we find that whereas the known maintenance methyltransferase DNMT1 (24) has high intrinsic activity for the M/C substrate (Kcat = 6.6 h−1; Figure 1b and Supplementary Figure S1), it has measurable but greatly reduced activity for the H/C substrate (Kcat ∼ 0.1 h−1) and no detectable activity on C/C (Figure 1b). Thus, unlike M/C, H/C is not a preferred substrate of DNMT1 and is unlikely to be methylated by DNMT1 after replication, in agreement with previous findings (25). Consistent with this notion, UHRF1, which is essential for DNMT1 function and selectivity for hemi-methylated CpG (M/C) sites in vivo (22,23), loses its intrinsic preference for hemi-methylated DNA when 5mC is replaced by 5hmC. UHRF1 (residues 124–628) shows a >10-fold reduced binding affinity for H/C DNA as compared to M/C DNA, that was similar in magnitude to its affinity for fully methylated (M/M), fully hydroxymethylated (H/H) and H/M DNA (Figure 2a and Supplementary Discussion). Therefore, neither DNMT1 nor UHRF1 are likely to be involved in post-replicative maintenance of H/C methylation, which, without the involvement of DNMT3 (see below), would lead to ‘passive demethylation’ of 5hmC.
Figure 1.
DNMT3A and DNMT3B can methylate the cytosine in the context of hemi-hydroxylmethylated CpG site (H/C). (a) Diagram showing the potential fate of single CpG sites that are either unmodified (C/C), fully methylated (M/M) or fully hydroxymethylated (H/H) at DNA replication. After strand synthesis, unmodified (C/C), hemi-methylated (M/C) or hemi-hydroxymethylated (H/C) sites are transiently generated. MBD indicates DNA methyl-binding domain proteins, while Tet refers to ten–eleven translocation proteins. (b–d) Enzymatic activity of recombinant DNMT1, DNMT3A2/DNMT3L and DNMT3B2/DNMT3L against a 32-bp DNA containing a single unmodified (C/C), hemi-methylated (M/C) or hemi-hydroxymethylated (H/C) CpG site. Note that DNMT1 (panel b) has robust preference for maintenance methylation at M/C sites over H/C and C/C sites in naked oligonucleotide DNA, whereas DNMT3A2/3L (c) and DNMT3B2/3L (d) have approximately similar activities on all three substrates.
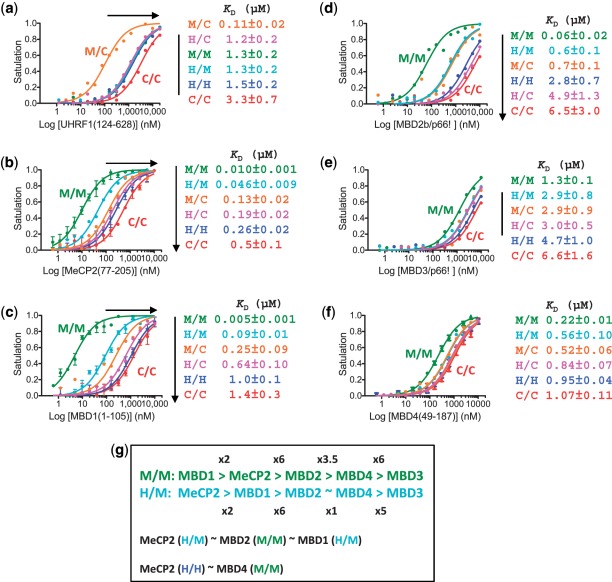
Figure 2.
Effect of hydroxymethylation on DNA methylation ‘readers’. Binding affinities of UHRF1 (residues 124–628) (a) and five MBD proteins (b–f) for a double stranded oligonucleotide containing a single CpG site with one of the six different modification states: unmodified (C/C), fully modified (M/M, H/H), hemi-(hydroxy)methylated (M/C, H/C) or hemi-methylated/hemi-hydroxymethylated (H/M) CpG site. Binding was assessed by fluorescence polarization. (a) UHRF1 has a strong preference for binding the hemi-methylated CpG (M/C) site. (b and c) MeCP2 and MBD1 have the strongest binding to fully methylated CpG (M/M). Although there is significantly lower affinity among the MBDs for H/M CpG dinucleotides (5-fold decreased affinity for MeCP2 and 18-fold for MBD1), this substrate is still preferred over hemi-methylated DNA (M/C), and is bound by MeCP2 and MBD1 with an affinity similar in magnitude to MBD2 (d), and greater affinity than MBD3 (e) and MBD4 (f) bind to fully methylated DNA (M/M). (g) Summary of relative binding affinities (by factor of x) of five MBD proteins for M/M and H/M substrates (top two lines). Note that the binding affinities of MeCP2 (and MBD1) for H/M and H/H are in the same order of magnitude as that of MBD2 and MBD4 to M/M substrate, respectively (lines 3 and 4).
De novo DNMT3 family includes two active enzymes, DNMT3A and DNMT3B (18,19), and one regulatory factor, DNMT3L protein (20,21). DNMT3L enhances methylation by both DNMT3A and DNMT3B (26–30) via direct interaction with the catalytic domain of DNMT3A and DNMT3B and stabilizing their binding of AdoMet (31). We purified DNMT3A2 [a shorter isoform of DNMT3A (32)] in complex with DNMT3L as well as the complex between DNMT3BΔ218 (deletion of N-terminal 218 residues, termed 3B2, approximately equivalent to DNMT3A2 in size) and DNMT3L (Supplementary Figure S2). Unlike DNMT1, DNMT3A2/3L and DNMT3B2/3L have approximately equal activities on all three (M/C, H/C, C/C) substrates, with Kcat values of 0.2–0.4 and ∼0.2 h−1, respectively (Figure 1c and d), suggesting that DNMT3A and DNMT3B do not distinguish various modifications in the hemi-modified context. The very slow catalytic turnover of DNMT3A and DNMT3B is probably due to the product binding of methylated DNA, considering that almost all of the cellular complement of DNMT3A and DNMT3B, but not DNMT1, is strongly anchored to nucleosomes containing methylated DNA (33,34).
Recognition of 5hmC by known MBDs
Tet proteins use 5mC as substrate to generate 5hmC (2). The amount of 5hmC, which inversely correlates with the amount of 5mC in Purkinje cell DNA and in granule cell DNA (3), could potentially be regulated by the combined cellular activities of Tet1-3 enzymes, DNMTs (through the regulation of the levels of 5mC substrate) and the methyl-specific DNA binding proteins (by binding and potentially masking 5mC), such as MeCP2, which is found in high concentrations in the brain (35). It was recently found that 5hmC levels in mouse cerebellum are negatively correlated with MeCP2 gene dosage (36). Consistent with this idea, we found that MeCP2 binding to methylated DNA inhibits Tet1 activity in vitro (Supplementary Figure S3).
To explore the effect of DNA hydroxylation on the function of MBD proteins, we measured the dissociation constants (KD) between five MBD proteins and double stranded oligonucleotides containing a single CpG dinucleotide with six different modification states (C/C, M/C, H/C, M/M, H/M and H/H as shown in Figure 1a) using fluorescence polarization analysis. MeCP2 (residues 77–205) binds fully methylated DNA (M/M) with a KD of 10 nM; changing a single methyl group to hydroxylmethyl on one strand (H/M) increased the KD to 46 nM (an ∼5-fold weaker binding) (Figure 2b). Full hydroxymethylation (H/H) weakened the binding by another factor of 5 with KD of 0.26 µM, a value similar to that of H/C (0.19 µM) and about half of C/C (0.5 µM) (Figure 2b), suggesting that the hydroxyl group of 5hmC disrupted the specific interaction between MeCP2 and 5mC, reducing the binding affinity to that of unmodified cytosine. In comparison, there was little apparent discrimination in the context of hemi-modification (0.19 µM for H/C versus 0.13 µM for M/C).
A similar trend was observed for the MBD domain of MBD1 (Figure 2c) and MBD2 (Figure 2d): each methyl group hydroxylation from M/M to H/M to H/H results in weaker binding (by a factor of 18 and 11 for MBD1 and 10 and 5 for MBD2, respectively). These results are in general agreement with the finding that MBD domains have a lower affinity toward sequences containing 5hmC (37,38). Nevertheless, it is noteworthy that the ability of the different MBDs to discriminate between these substrates (M/M versus H/M) varies considerably (from 2- to 18-fold), with MBD3 and MBD4 showing little discrimination (Figure 2e and f) (summarized in Figure 2g). Furthermore, although MeCP2 and MBD1 have significantly lower affinity for hemi-methylated and hemi-hydroxymethylated CpG dinucleotides (H/M) relative to M/M DNA (5- and 18-fold decreased affinity, respectively), this substrate is still preferred over hemi-methylated DNA (M/C) by a factor of ∼3, and is bound by MeCP2 and MBD1 with an affinity similar to that which MBD2 binds fully methylated (M/M) DNA, and a greater affinity than MBD3 and MBD4, bind to fully methylated DNA (M/M) (summarized in Figure 2g). These data suggest that in vivo there may be additional factors (e.g. relative local concentration, tissue-specific expression) that determine whether, and to what degree, 5hmC DNA is bound by MBDs.
Excision of 5hmU by MBD4 and TDG
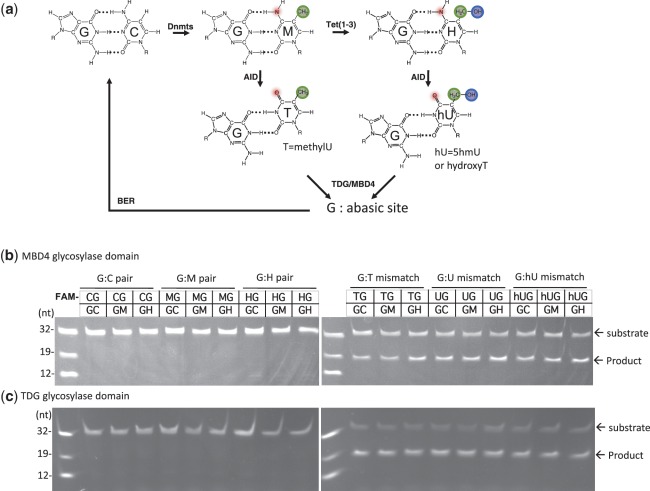
In the pathway shown in Figure 3a, Tet-mediated production of 5hmC becomes the substrate for AID, which converts 5hmC to 5hmU. We thus tested whether 5hmU can be excised by MBD4 and TDG glycosylase domains (15,39). We used the same 32-bp DNA duplexes containing a G:X mismatch within the CpG sequence context (where X = 5hmU, T or U) as the substrate. The X-containing strand was FAM labeled, and the excision of the mismatched base was monitored by denaturing gel electrophoresis following NaOH hydrolysis (Supplementary Figure S4). As expected, no glycosylase activity was observed on oligonucleotides bearing the ‘natural’ G:C and G:M base pairs, and there was efficient cleavage of substrates bearing G:T and G:U mismatches (Figure 3b and c). We extend these findings to show here that the TDG and MBD4 glycosylases are also inactive on G:H (which preserves Watson–Crick base-pair hydrogen bonds; see Figure 3a), but act on G:5hmU mismatched substrates, and further, that the modification status (methyl or hydroxymethyl) of the C in the opposite strand of neighboring G had no impact on the ability to remove G:T, G:U or G:5hmU mismatches (Figure 3b and c). Therefore, TDG and MBD4 are capable of acting on AID-generated 5hmU and completing the ‘demethylation’ of 5hmC.
Figure 3.
MBD4 and TDG are capable of excising 5-hydroxymethyluracil in the context of a double-stranded CpG dinucleotide. (a) A putative pathway of DNA demethylation involving DNA methylation by DNMTs, hydroxylation by Tet proteins, deamination by AID and glycosylation by MBD4 or TDG linked to base excision repair (BER). Double stranded 32-bp oligonucleotides bearing a single CpG dinucleotide and the indicated modification status (where M = 5mC and H = 5hmC) and labeled with FAM on the top strand were incubated with the glycosylase domain of MBD4 (b) or TDG (c) at 37°C for 1 h. The products of the reaction were separated on a denaturing polyacrylamide gel, and the FAM-labeled strand was excited by UV and photographed.
DISCUSSION
Taken together, we suggest that DNMT3A and DNMT3B are capable of acting on the hemi-hydroxymethylated (H/C) CpG sites generated during DNA replication, to yield hemi-hydroxylated and hemi-methylated (H/M) CpG sites. Recently, a maintenance methylation function has been proposed for DNMT3A and DNMT3B (40), which have been suggested to complete the methylation of sites missed by or resistant to DNMT1 activity during DNA replication (e.g. repetitive DNA). Assuming H/M sites are substrates for modification by Tet proteins (Supplementary Figure S5), there is the potential for regenerating a fully modified hydroxymethylated H/H site, recapitulating the parental DNA state after replication (Figure 1a). It is interesting to note that while Dnmt3a (but not Dnmt3b or Dnmt1) expressed in both paternal and maternal pronuclei of mouse zygotes, Tet3 (but not Tet1 or Tet2) expressed specifically in the male pronucleus (41). The expression patterns of Dnmt3a and Tet3 coincide with the loss of methylation and occurrence of 5hmC in the early mouse embryo (5,6).
We also showed that five MBD proteins have markedly varied abilities to discriminate M/M versus H/M substrates. MeCP2 and MBD1 bind H/M with an affinity similar to that which MBD2 binds fully methylated (M/M) DNA (Figure 2). These data suggest that MBD proteins (particularly MeCP2 and MBD1) may play a role in regulating the levels of 5hmC in vivo, by inhibiting Tet activities through their binding to M/M and H/M substrates. In mammals, all three Tet proteins, Tet1, Tet2 and Tet3 catalyze similar reactions, converting 5mC to 5hmC (42). One important future question will be to determine biochemically whether Tet proteins, like DNMTs, divide the labor among the family members, with one (or more) establishing the initial hydroxymethylation pattern de novo (i.e. using M/M as the substrate), and other(s) maintaining this pattern during chromosome replication (using H/M as the substrate). Finally, we demonstrated that MBD4 and TDG glycosylase domains are both active on AID-generated 5hmU and are thus capable completing the ‘active demethylation’ of 5hmC. An important future question will be to determine whether the two have largely complementary but context-dependent functions in 5hmC turnover. It would also be interesting to know whether Tet1-3 proteins interact with AID complex [including the damage response factor Gadd45 (10)] and TDG or MBD4 by some means.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–5, Supplementary Methods, Supplementary Discussion and Supplementary References [43–67].
FUNDING
U.S. National Institutes of Health (GM049245-18 to X.C.) and NIH (grants CA077337 and CA132065 to P.M.V.). X.C. is a Georgia Research Alliance Eminent Scholar and P.M.V. is a Georgia Cancer Coalition Distinguished Cancer Scholar. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank sincerely Brenda Baker of New England Biolabs for DNA oligo synthesis, Cris A. Lee of the Biochemistry, Cell and Developmental Biology program of Emory University for expression and purification of MeCP2 fragment, Dr Yi Zhang of University of North Carolina at Chapel Hill for providing initial Tet1 construct, Dr Robert M. Blumenthal of the University of Toledo College of Medicine for critical comments and editing the manuscript. H.H. performed purifications of DNMT1 N-terminal deletion fragments, purifications of MBD1 and MBD4, methylation assays, binding assays and glycosylase assays. Y.L. performed DNMT3 purifications and methylation assays. A.K.U. purified full-length DNMT1 and performed MBD inhibition of Tet1 activity. Y.C. purified MBD2 and MBD3; S.B.H. expressed full-length DNMT1 in P. pastoris; P.M.V. provided initial full-length human DNMT1 construct. X.Z. and X.C. organized and designed the scope of the study, and all were involved in analyzing data and helped in writing and revising the manuscript.
REFERENCES
- 1.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 5.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 7.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 10.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 14.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P, Qiu C, Sohail A, Zhang X, Bhagwat AS, Cheng X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 19.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 20.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 21.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 22.Bostick M, Kim JK, Estéve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 23.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 24.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 25.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 26.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 28.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 30.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 31.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 33.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell. Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 36.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 42.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 44.Mohan KN, Ding F, Chaillet JR. Distinct Roles of DMAP1 in Mouse Development. Mol. Cell Biol. 2011;31:1861–1869. doi: 10.1128/MCB.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee GE, Kim JH, Taylor M, Muller MT. DNA methyltransferase 1-associated protein (DMAP1) is a co-repressor that stimulates DNA methylation globally and locally at sites of double strand break repair. J. Biol. Chem. 2010;285:37630–37640. doi: 10.1074/jbc.M110.148536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estéve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat. Struct. Mol. Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 48.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 50.Syeda F, Fagan RL, Wean M, Avvakumov GV, Walker JR, Xue S, Dhe-Paganon S, Brenner C. The replication focus targeting sequence (RFTS) domain is a DNA-competitive Inhibitor of Dnmt1. J. Biol. Chem. 2011;286:15344–15351. doi: 10.1074/jbc.M110.209882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Achour M, Jacq X, Rondé P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 52.Pradhan M, Estéve PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 53.Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenöder S, Wang M, Qin W, Soding J, Spada F, et al. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS ONE. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 56.Lauster R, Trautner TA, Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol. 1989;206:305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- 57.Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A, Tajima S. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1) Proc. Natl Acad. Sci. USA. 2011;108:9055–9059. doi: 10.1073/pnas.1019629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 2002;277:40958–40966. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- 60.Feng Q, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Zhang Y. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell Biol. 2002;22:536–546. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gnanapragasam MN, Scarsdale JN, Amaya ML, Webb HD, Desai MA, Walavalkar NM, Wang SZ, Zu Zhu S, Ginder GD, Williams DC., Jr p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc. Natl Acad. Sci. USA. 2011;108:7487–7492. doi: 10.1073/pnas.1015341108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 64.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 65.Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 2008;283:34490–34494. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.