Abstract
The dimer initiation site/dimer linkage sequence (DIS/DLS) region of the human immunodeficiency virus type 1 (HIV-1) RNA genome is suggested to play essential roles at various stages of the viral life cycle. Through a novel assay we had recently developed, we reported on the necessary and sufficient region for RNA dimerization in the HIV-1 virion. Using this system, we performed further detailed mapping of the functional base pairs necessary for HIV-1 DLS structure. Interestingly, the study revealed a previously unnoticed stem formation between two distantly positioned regions. Based on this and other findings on functional base pairing in vivo, we propose new 3D models of the HIV-1 DLS which contain a unique pseudoknot-like conformation. Since this pseudoknot-like conformation appears to be thermodynamically stable, forms a foundational skeleton for the DLS and sterically restricts the spontaneous diversification of DLS conformations, its unique shape may contribute to the viral life cycle and potentially serve as a novel target for anti-HIV-1 therapies.
INTRODUCTION
The genome of a retrovirus such as human immunodeficiency virus type 1 (HIV-1) is a single-stranded, positive-sense RNA. The viral genome always exists as a dimer in virions, and they interact non-covalently, as heating easily dissociates purified dimeric genomes into monomers. It has been suggested that the genome dimerization plays an important role at various stages of the viral life cycle, including genome packaging and reverse transcription, as well as genome recombination processes involved in viral diversification (1–3). Electron microscopy studies have suggested that the 5′ region of both viral RNAs contains a primary contact point and is referred to as the dimer linkage structures (DLS) (4–6). Thus far, there have been a certain number of reports regarding in vivo HIV-1 RNA dimerization (7–11) and we are gradually accumulating further knowledge and insight. The presumptive primary DLS of HIV-1 has been mapped to a region near the major splice donor and biochemical analysis indicates that the DLS probably consists of multiple stem–loop structures (12). Within the DLS, stem–loop 1 (SL1) has been regarded as the most important region, which forms a stem–loop structure with a hairpin loop containing a 6-nt palindromic sequence (13). The dimer formation would occur through a kissing hairpin mechanism by which the two RNAs would form an initial loop–loop contact based on complementary anti-parallel base pairing at the SL1 loops (14). So far, partial structures within DLS, such as SL1, 2, 3 and 4, have been solved and characterized to some extent (15–24). Furthermore, an in vitro NMR structural model of the entire HIV-1 DLS was released recently (25), stimulating increasing demand for the further detailed structural information about the in vivo structure of the HIV-1 DLS.
We previously developed a novel system to assess the DLS operation within the HIV-1 virion and identified the region that is necessary and sufficient for HIV-1 genome dimerization in the virion (26). Using this system coupled with loss- and gain-of-function studies using site-directed mutagenesis, we performed a more detailed mapping of the functional base pairs required for HIV-1 genome dimerization and replication. Interestingly, the study revealed a previously unnoticed base-pairing interaction between two distantly positioned 5 nt in the DLS (GCGUC-GACGC duplex). Based on this and other findings, we propose a new secondary structure model of a minimal DLS element that contains a unique pseudoknot-like conformation, as well as two 3D structure models of the DLS element, which are consistent with current and previous experimental findings.
MATERIALS AND METHODS
Constructs
The replication-competent HIV-1 pro-viral clone pNL4-3 (subtype B) (27), its Env mutant pNLNh (1), or its derivative DDNLp4Δ2 (renamed as DDNd2) (26) were used as progenitors for the mutant constructs described below. Mutant plasmids were constructed with standard methods. To make the construction of recombination clones more convenient, several base-substitution mutations were introduced into the N-terminal region of the matrix (MA) gene of pNLNh to create the SgrAI recognition site (pNLAINh) (2). Although the one amino acid substitution (Ser to Thr) at the ninth position of MA in pNLAINh was inevitably introduced as a result of these base substitutions, similar levels of virion production were observed in both cells transfected with pNLAINh or pNLNh, suggesting that the effect of the mutations on particle formation was negligible. Precise details of the plasmid construction are found in the Supplementary Methods and Supplementary Table S1.
DNA transfection
In total, 293 T cells (28) (∼3 × 106) were seeded on dishes (diameter, 100 mm) the day before transfection with plasmid DNA (5 µg) by means of the calcium phosphate precipitation method (29). The day after transfection, the supernatant was replaced with fresh medium.
Virus infection
At 48–72 h post-transfection, the media was centrifuged and the supernatant was used for infection into MT-4 cells. Equal amounts of p24 were inoculated into the cells. The supernatants of the MT-4 were harvested every 3 or 4 days for multiple replication assays. About 10 µl of each cell supernatant was subjected to exogenous RT assay as described previously (30).
Isolation of RNA from virions
At 48–72 h post-transfection, virus particles were collected from the media as described elsewhere (31). The physical virus titer was determined with an ELISA assay kit for quantitation of CA-p24 (ZeptoMetrix, Inc., Buffalo, NY, USA). To isolate RNA from particles, virions were disrupted by the addition of 1% sodium dodecyl sulfate (SDS) and treated with proteinase K (300 µg/ml) at room temperature for 60 min, followed by TE-saturated phenol/chloroform extraction, chloroform extraction and ethanol precipitation.
Northern blotting analysis
Pelleted RNA was resuspended in T-buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1% SDS, 100 mM NaCl and 10% formamide) and the thermostability of the dimeric viral RNA was determined by incubating RNA aliquots for 10 min at the prescribed temperatures (32). RNA folding under native conditions is regarded to be achieved by incubation at 25 or 35°C, since viral RNA was always treated at or below room temperature during purification. RNA electrophoresis on native agarose gel and northern hybridization analysis were performed as described elsewhere (1). Plasmid T7pol (32) was used to synthesize a complementary RNA probe for northern hybridization. In experiments designed to assess the conversion of dimers to monomers, relative amounts of both RNA species were quantitated by phosphorimager analysis (Fujifilm Co., Tokyo, Japan) to determine ratios of dimers and monomers. Since the ratio of monomer content to total RNA of virion varied from experiment to experiment, we calculated the ‘D–value’, which is the index of the dimerization efficiency, for the fair validation (Figure 1). The calculation of D-value was described elsewhere (1). In short, we made an Equation (1) to give an index value of dimer formation efficiency (D) for each of the mutants, to map the functional DLS area fairly and squarely.
Where W, B and M represent ratios of monomer content to total RNA of virions produced from wild-type, DDNd2 and each DDNd2 derivative mutant construct at room temperature, respectively. The D-value of the wild-type is 0 and that of DDNd2 is 1. When the combination of mutants severely affects dimer formation, the monomeric genome content of the virion is reduced and D becomes close to 0. Thus, the D-value was expected to represent the magnitude of the effect of the DLS mutation in virions.
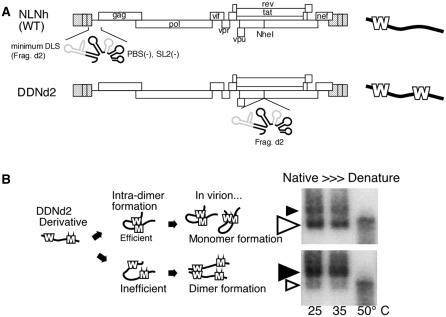
Figure 1.
System utilized to map the functional sequence of HIV-1 DLS. (A) Schematic representation of the constructs utilized. On the right, bold lines represent viral genomes. Ws represent DLS at native and ectopic positions. Ectopically inserted DLS (d2 fragment) expands from nucleotide 557 to nucleotide 806 (NL4-3 DNA sequence, Genbank Accession no. M19921.2) with deletions of partial PBS stem–loop and SL2. (B) Schematic of the genome dimerization assay. Bold lines represent viral genomes. Ms represent d2 fragments with mutations. Native northern blots of viral RNA of DDNd2 derivatives are shown on the right. Black and white arrowheads indicate the dimeric and monomeric viral genome, respectively. Where the size of the arrowheads indicate the relative strength of the signal. The temperatures of RNA incubation are shown.
Selective 2′-hydroxyl acylation analyzed by primer extension analysis
RNA selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) analysis was performed as described elsewhere (33,34) with minor modifications. Ex virion HIV-1 RNA was applied for the analysis and RNA reactivity to the chemical modification reagent at specific sequence positions were calculated. Experimental details are found in Supplementary Methods.
Construction of 3D structural model of the DLS dimer
We predicted the 3D structure of dimer of the DLS by the following procedures. First, we predicted secondary structure of the DLS monomer by using bioinformatics algorithms without using information on the base pairing predicted from present mutation study. We explored most likely base pairs in the DLS monomer by CentroidFold (http://www.ncrna.org/centroidfold) (35,36) without restraints of any base pairs (Supplementary Figure S4, red lines). We subsequently explored other likely base pairs by MC-Fold (http://www.major.iric.ca/MC-Pipeline/) (37), with constraints of the most likely base pairs as predicted by CentroidFold (Supplementary Figure S4, green lines). The explored base pairs included all of the base pairs that were predicted from data with our RNA dimerization assay. Next, using the secondary structure information of the DLS monomer, 3D structural models of the DLS monomer were predicted with MC-Sym (37). We only considered authentic base pairs, A-U, G-C or G-U pairs, at the ends of each stem to minimize instability of the structure. With these constraints, we could obtain only two monomer models, since pseudo-knot like conformation in the DLS restrained conformational flexibility of the monomer. Then, dimer structures were constructed by superposing the 3D structure of SL1 in the predicted monomer with that in the reported structure of SL1 dimer [PDB code: 2D19 (24)]. For the superposition, we ignored intra-molecular base pairs at the dimerization site in SL1 to preserve inter-molecular base pairs critical for the RNA dimerization. Last, energy minimization was achieved for the constructed dimer structures, by 10 000 steps of the steepest descent method and the subsequent 10 000 steps of the conjugated gradient method. At the minimization, energy calculation was performed with parmbsc0 force field (38) and generalized born solvent energy function (39) by sander module in AMBER9 (40).
RESULTS
In vivo evaluation of the branched multiple hairpin formation of HIV-1 DLS
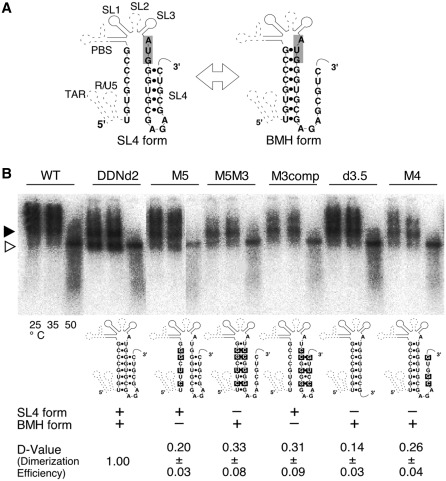
In our previous report, we identified the region that is necessary and sufficient for HIV-1 genome dimerization in the virion (d2 fragment) utilizing a unique analysis system that we developed (Figure 1A and B) (26). The mapping of the DLS has been performed based on the predicted secondary structural model which includes the PBS stem–loop and SL1-4 (12). So far, several reports suggested that the HIV-1 genome leader region could form a possible structure called branched multiple hairpin (BMH) (41,42). In this model, the 5′ junction sequence formed a stem structure with a gag AUG sequence that has been suggested to form the base of SL4. As shown in Figure 2A, gag AUG region could interact with either the immediate downstream region to form SL4, or with the 5′ junction to make the BMH form. The BMH model has been previously validated by in vitro experiments (41,42), and our in vivo DLS studies included all sequences required for proper BMH formation. To test whether the BMH or SL4 formation occurs in virus particles, several mutants derived from DDNd2 (renamed from DDNLp4Δ2) (26) were constructed. A series of mutants were constructed by introducing several base substitutions in the inserted d2 fragment of DDNd2 (Figure 2A). The mutant M5 was constructed to disrupt an interaction between AUG and the 5′ junction by introducing a mutation in the 5′ junction. M5M3 was constructed to restore the AUG–5′ interaction and to disrupt SL4 formation by an additional mutation in the AUG region. M3comp was constructed to force SL4 formation through mutations in both stems of SL4. The mutants d3.5 and M4 contained an 8-base deletion and a 3-base substitution at the 3′-end of DLS, respectively, to eliminate SL4 formation. The mutants d3.5 and M4 did not make any notable contributions to BMH structure formation (Figure 2B). The viral RNAs from these mutants were analyzed by native northern blotting. As shown in Figure 2B, all of the genomic RNA from the mutants formed monomeric genomes inefficiently in comparison with that of the wild-type (WT). These data indicated that neither the BMH nor SL4 forms sufficiently represented the functional structure of HIV-1 DLS. This observation strongly suggested that the actual functional active form of DLS in vivo might be an entirely novel secondary structure.
Figure 2.
Evaluation of previously proposed HIV-1 DLS models. (A) Schematic representations of the alternative forms of DLS (12,41,42). The SL4 form contained a stem–loop at 3′-end, whereas the BMH form contained a stem–loop at 5′-end. (B) Virion RNA profiles detected by northern blotting in a native agarose gel. Viruses were prepared by transfection of 293 T cells with the wild-type (WT; pNLNh) virus or its derivative mutants. Aliquots of RNA extracted from virions were resuspended in T-buffer and incubated for 10 min in parallel reactions. The temperatures in which aliquots were incubated are indicated. Below the blot, schematic figures of the mutants and their dimerization efficiency (D-Value) are shown. Results are the average of three independent experiments with standard error of mean (SEM).
Discovery of a novel interaction within the DLS for HIV-1 RNA dimerization and replication
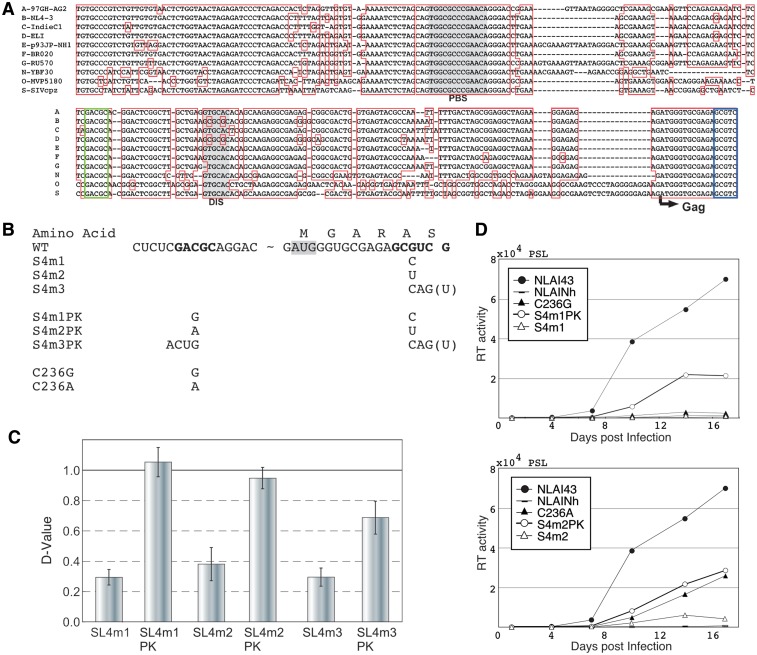
We focused our analysis on the functional significance of the RNA sequence at the 3′-end of the DLS on the HIV-1 life cycle, since a complete deletion or subtle base substitutions of the 3′ half of SL4 severely crippled the dimerization ability of DLS (Figure 2, M4 and d3.5). When comparing the DLS sequences from nine HIV-1 subtypes and SIVcpz, we found highly-conserved complementary sequences immediately upstream of the SL1 stem (GACGC) and at the 3′-end of the DLS (GCGUC) (Figure 3A, green and blue boxes). To assess whether the two complementary sequences can form a functional duplex for RNA dimerization and support HIV-1 replication, mutational analysis was performed. As the GCGUC sequence is in Gag frame, several silent mutations at GCGUC and their respective restorative substitutions at GACGC were introduced into the d2 fragment of DDNd2 to observe the roles of RNA sequence on genome dimerization (Figure 3B). Notably, the dimerization abilities of the silent mutants were markedly reduced and the subsequent compensatory substitutions restored its ability, to a level comparable with that of WT (Figure 3C and Supplementary Figure S2). The same mutations were introduced into the backbone of the HIV-1 pro-viral construct (pNLAI43) and similar defects and subsequent restorations were also observed in the viral replication process (Figure 3D). These data strongly suggest that this duplex formation (GACGC-GCGUC duplex) occurs within the virus and grossly affects its activity in vivo.
Figure 3.
Demonstration of a novel long-range interaction within the DLS. (A) DLS sequence alignment of nine HIV-1 subtypes and SIVcpz. PBS and DIS are presented as shaded. The evolutionarily-conserved complementary sequences are in blue boxes (GCGUC) and green boxes (GACGC). (B) The sequence of base substitution mutants. The bases for mutation are shown in bold. The G to U substitutions at the 3′-end are an inevitable change for silent mutation for S4m3, although the G is not included in the DLS in this report. Thus, we introduced a 4-base substitution in the compensatory mutant (S4m3PK). (C) Dimerization ability of the mutants. Results are the average of at least three independent experiments with SEM. (D) Replication of DLS mutants. Growth curves of two sets of 1-base substitution and compensatory mutants are shown. Data are representative of three independent experiments.
Recently, in vivo HIV-1 RNA secondary structures have been described in several reports, based on the results of chemical or enzymatic probing, such as SHAPE analysis (10,33,34). In these presented structural models, there were no traces of the GACGC-GCGUC duplex. To get further information about the duplex by chemical probing, we have performed SHAPE analysis with ex virion RNA. The reactivity of the duplex mutants (NLAIS4m1, NLAIS4m1PK and NLAIC236G) along with the wild-type virus was calculated for each base in the duplex and adjacent U5-AUG duplex (Supplementary Figure S3A and B). Consistent with the previous SHAPE reports, the overall GACGC-GCGUC duplex reactivity was very low and not significantly affected by the mutations to destabilize (C236G and S4m1) and to restore the duplex (S4m1PK). Although it was very faint, there were some changes in reactivity, notably at GCGUC sequence exclusively in the destabilizing mutant. In addition, coinciding with the results from dimerization assay above (Figure 4B) and previous SHAPE assays, the 5′-end of DLS (U105-U107) was relatively reactive, suggesting the absence of base paring. At the same time, similar to the GACGC-GCGUC duplex, the overall U5-AUG duplex reactivity was not significantly affected by the mutations.
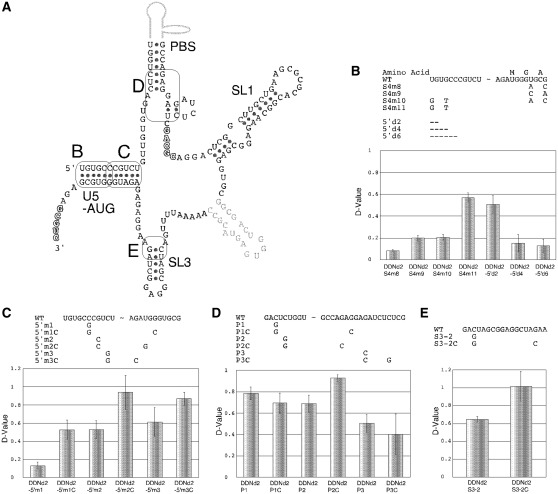
Figure 4.
Confirmation of the stem formations within the DLS. (A) Schematic representations of the stems in HIV-1 DLS studied in this report. The specific positions examined are enclosed with rectangles and tagged with capitals. (B–E) The dimerization assay of the mutants. Upper tables describe the mutations introduced. Hyphens represent base deletions. Lower graphs represent dimerization ability of the mutants. Results are the average of at least three independent experiments with SEM.
Confirmation of reported stem formations within DLS in vivo
We next closely examined the roles of several stem formations within the DLS that have been predicted by computational or in vitro biochemical analysis, for viral RNA dimerization using our in-virion assay system (Figure 4 and Supplementary Figure S2) (26). Figure 4A illustrates a schematic diagram of the DLS and stems analyzed in this report. The importance of the long stem, which establishes the BMH form (U5-AUG stem), was examined by multiple experiments (Figure 4B and C). The 2-base substitution at the 3′-end of the U5-AUG stem resulted in a reduction in dimerization abilities (Figure 4B, S4m8 and m9) and compensatory mutations did not rescue the ability (Figure 4B, S4m10). On the other hand, a 2-base substitution in 5′-end of the stem triggered a mild reduction in dimerization (Figure 4B, S4m11). To better understand the role of the 5′-end of the DLS, 2-, 4- and 6-base deletions were introduced. The 2-base deletion showed a mild effect, whereas the 4- and 6-base deletions nearly abolished dimerization activity completely (Figure 4B, 5′d2, 5′d4 and 5′d6). We next examined the importance of duplex formation at the top of the U5-AUG stem (Figure 4C). A single base substitution of G to C, or C to G, reduced dimerization activity, and respective compensatory substitutions fully recovered dimerization activity. Taken together, these results strongly suggest that the top stem of the U5-AUG structure was functionally important for the DLS structure. The functional importance of the PBS and SL3 stem base pairings were also explored. Of the three potential base-pairings analyzed around the PBS stem, only one of the base-pairings (Figure 4D: P2-P2C) confirmed to be important for DLS structure, as evidenced by the loss- and gain-of-function mutagenesis studies. Similarly, within the SL3, base pairing of the stem was proved to be functionally required (Figure 4E).
The computer-aided modeling of the DLS 3D structure
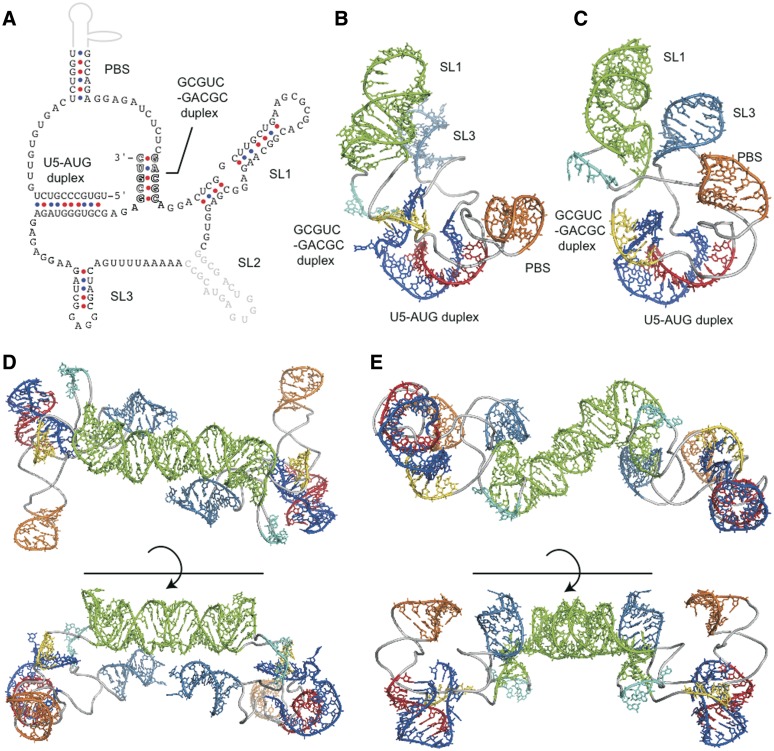
Upon the accumulated results, we draw the two-dimensional (2D) structural schema of DLS (Figure 5A). The 2D schema shows that the DLS is characterized by a unique pseudoknot-like conformation and three stem–loops. Such a pseudoknot-like conformation was predicted to form upon performing secondary structure prediction analysis using MC-Fold, without using additional experimental information. We further attempted the 3D structural modeling of the DLS by using RNA structure prediction computer programs that have shown marked improvements as of recent. We then confirmed whether the DLS could indeed have conformations similar to our modeling, without unfavorable interactions such as steric clash. The modeling predicted only two plausible 3D structures of the DLS monomer having the pseudoknot-like conformation (Figure 5B and C). In each of the DLS structures, the SL1 loop neighbored the SL3 loop and the pseudoknot-like conformation was located near the bases of these loops. In contrast, the two structures had distinct PBS stem orientations. Furthermore, we constructed dimer predictions based on these monomer structures (Figure 5D and E). In the dimer structures, palindromic sequences within the SL1 had interactions between monomers, whereas the pseudoknot-like conformations were furthest apart from each other. We also predicted 3D structural models of the DLS without a connection region between SL3 and the 3′-end part of the pseudoknot-like conformation. The modeling continually generated more structures, suggesting the pseudoknot-like conformation in the DLS would restrict its conformation. In the two models, an AUG codon at the translation initiation site of Gag p17 ORF formed a ‘U5-AUG duplex’, which was embedded as unexposed, translation-incompetent structures.
Figure 5.
New models of 2D and 3D structure of the DLS. The 2D and 3D structures of the DLS. (A) The 2D DLS schematic model that contains a previously unknown GCGUC-GACGC duplex. Other duplexes that were proposed previously and suggested to play roles in HIV-1 RNA dimerization within the virion in this and other studies (12,41) are also indicated. (B, C) Two 3D structural models of the DLS monomer. The 3D structural models of the DLS monomer were predicted with the MC-Sym software (37) by incorporating information on the duplexes within the DLS described in Figure 5A. Only two monomer models that were thermodynamically stable were obtained, due to the pseudoknot-like conformation that was formed with the two duplexes (GCGUC-GACGC and U5-AUG) restraining the conformational flexibility of the monomer. (D, E) Two 3D structural models of the DLS dimer. The 3D structural models of the DLS dimer were constructed by superposing 3D structure of SL1 with SL1 in a reported dimer structure [PDB code: 2D19 (24)], followed by energy minimization as described in ‘Materials and Methods' section.
DISCUSSION
The retroviral RNA genome dimerization is an essential event for the viral lifecycle, especially for genome packaging, reverse transcription and recombination. Thus, the genome dimerization signal (DLS) of HIV-1 could be a probable target for anti-retroviral therapy. Although the DLS on the RNA strand is known to be located at the 5′-end of the genome, its precise sequence and its functional structure has never been determined. We previously developed a unique analysis system to map the functional region of the DLS working within the HIV-1 virion (1) (Figure 1A and B). This assay is very sensitive and even subtle mutations within the DLS are often detectable, as mutations dramatically alter RNA dimerization abilities. Through this system, we determined entire functional sequences that are required for mediating the RNA–RNA interactions within the HIV-1 virion (26). We built upon our previous observations and attempted to determine the DLS functional structure through loss- and gain-of-function studies using site-directed mutagenesis. We initially tried to choose the right structural model from two previously proposed models (BMH and SL4), but were unsuccessful in fully restoring the WT dimerization phenotype (Figure 2). All base substitution mutations on either 5′- or 3′-end of the DLS, as well as the deletion of the 3′-end crippled dimerization abilities, and we were unable to confirm a definitive stem formation. The data suggested that the 3′-end of the DLS is responsible for viral RNA dimerization in vivo. On the other hand, mutations in the 5′-end of DLS also notably affected dimerization abilities. As both the 3′-end region in the BMH model and the 5′-end region in the SL4 form do not contribute to maintain the overall DLS structure, these existing models may not fully represent the actual DLS structure. Thus, it was necessary to explore a new structural model to better compensate for the conflicts apparent in the previous models. In the northern blots of Figure 2, there are several lower migrating bands than viral RNA dimers. We believe they represent heterologous form(s) of viral genome dimer. In our recent report, we noted that the viral RNA profiles transform variously during maturation (3), thus under certain conditions, it is plausible that viral RNAs could form heterologous profiles.
We next tried to get further insight into the importance of the 3′-end of the DLS by searching for nucleotide sequence conservation among variants of HIV-1 (Figure 3A). As expected, complete conservation of the SL4 region (downstream of AUG) was found with nine HIV-1 subtypes and SIVcpz. In addition, we discovered a completely conserved 5-base sequence upstream of SL1 (GACGC), which formed a perfect complementary base paring with the DLS 3′-end sequence (GCGUC). It is conceivable that these two areas form a previously unknown stem structure (GCGUC-GACGC duplex). Therefore, we performed mutational analysis to assess the functionality of this base-pairing using genome dimerization and viral viability assays (Figure 3B–D). All mutations that disrupted this base pairing severely reduced DLS activities and viral replication abilities, whereas subsequent corrections of these mutations fully restored DLS activity and viral replication. These results strongly suggest that the GCGUC-GACGC duplex plays very important roles for DLS function.
Our consecutive exploration to determine DLS structure resulted in the accumulation of informative data (Figure 4). The data suggested the existence of important structures in the upper portion of the U5-AUG stem, a part of the PBS stem and the SL3 stem. Additionally, the data also suggested the importance of the DLS 5′-end (Figuure 4B, DDNd2-5′d2, d4 and d6) and the base just downstream of the PBS stem (Figure 4D, DDNd2P3), although the structural status of those bases remains unknown.
From these data, we constructed a 2D DLS schematic model which contained a pseudoknot-like conformation and three stem–loops (Figure 5A). We further attempted to construct a 3D structural model through in silico analysis. The modeling predicted only two plausible monomer models and two corresponding dimer models, due to limitations imposed by steric restriction from the pseudoknot-like conformation. In those two models, conformations and orientations of the SL1 and SL3 pseudoknot-like regions were similar to each other. This suggests that the pseudoknot-like conformation would be solid, and formed a foundational skeleton for the DLS to maintain each angle of stem–loops, including SL1.
Several studies were published reporting on the HIV-1 packaging signal (psi) and DLS (psi/DLS) structural models (10,14,25,33,34,43,44). However, none of the works predicted the stem formation of the GCGUC-GACGC duplex reported here, and we believe there are several reasons to explain this discrepancy. First, some models were originated by in vitro experiments (14,25,43,44). As HIV-1 RNA dimerization and packaging occurs during virion production, Gag– and/or GagPol–RNA binding is believed to be one of the essential steps for RNA dimerization and DLS formation (3,45–47). Thus, even in the presence of exogenous viral nucleocapsid (NC) or Gag proteins in the reaction, the conditions for RNA folding in vitro would not be the same as in vivo. These subtle differences may mask, or cause oversight, details of these interactions that were unexpected. Second, other reports were based on the results of chemical or enzymatic probing of viral RNA in vivo (10,33,34). These RNA probing analyses basically provide us with the information of single- or double-stranded RNA formation tendencies within the RNA strand, yet no information on specific stem formation. It should be noted that SHAPE analysis-based structural modeling still depends on empirical and predictive computer calculations, which are susceptible to the aforementioned discrepancies. In fact, most of the bases in GACGC-GCGUC were unresponsive to probing in some of the previous reports (33,34), suggesting the stem formation could occur between these strands. The SHAPE analysis we performed in which we targeted the duplex with ex virion RNA (Supplementary Figure S3A) reconfirmed previous reports that the overall SHAPE reactivity of duplex forming bases was very low. It was not surprising that the duplex reactivity of the mutants was not significantly affected, as the mutations only disrupt one out of five possible base pairings. In addition, the viral RNA dimerization status of the destabilizing mutants looked unchanged in the viral particle (Supplementary Figure S3C). Slight elevations in reactivity that were observed at the counterpart of each mutated stem in the destabilizing mutant (GACGC of S4m1 and GCGUC of C236G) might reflect the reduction in mutant viability (Figure 3D). To pursue and confirm this interaction, as well as the proposed 3D model, additional in vitro experiments might be beneficial such as cross-linking [as in (44)], NMR [as used in (25)] or/and fluorescence resonance energy transfer (FRET) experiments providing distance constraints [for a review, (48)].
We do not rule out the possibility of many RNA–RNA interactions presented hitherto. As we mentioned in our previous reports (1,26), we assume that RNA dimerization is one of the multiple steps involved in RNA packaging, and that psi/DLS RNA must be recognized as an intact packaging signal by viral protein only after a secondary or higher structure is composed by two psi/DLS molecules. Within the cytoplasm of virus producing cell, non-specific Gag–viral RNA binding would occur just after Gag translation. If that were indeed the case, transient RNA secondary structure formation and/or Gag–RNA interaction would be continually required to form the accurate DLS through a trial-and-error process. During this process, various stem formations would be possible, and the suggested stems in previous reports, such as SL4 (12) or GACGC and the lower part of SL1 (33,34), might exist transiently.
According to our previous data, SL2 is dispensable for RNA dimerization in virion (26). This does not mean that SL2 is unnecessary for RNA packaging, as we repeatedly mentioned that genome packaging process would require additional steps other than genome dimerization for completion. Indeed, certain RNA regions outside DLS such as SL2 were reported to be required for genome encapsidation (17,18,31), suggesting SL2 could be required for formation of psi, which is larger than and wholly involves DLS. SL2 region showed high conservation and GC-rich profiles (Figure 3A), which may suggest it as a target for a stem formation other than SL2 during psi construction. Moreover, SL2 is definitely essential for viral RNA splicing, thus its sequence must be highly conserved for solid structure formation of SL2.
We mainly discussed a potential of intra-strand long-range RNA–RNA interaction in the 5′-UTR of HIV-1. However, based on presented results, this interaction could also involve inter-strand RNA interactions (within a dimer of viral RNA). The dimerization assay we performed (Figures 2, 3 and 4) only validated intra-strand interactions since loss-and-gain mutations were introduced in one DLS of the constructs. On the other hand, the results of viral replication assay and SHAPE analysis could reflect both intra- and inter-strand interactions. During virion maturation, dimerized DLS is suggested to change its form from kissing-loop to extended-duplex by the action of the NC protein (49). Elongation of extended-duplex form might result in the inter-strand encounter between long-range interaction sites. Further analysis focusing on inter-strand base pairing may give us unforeseen possibility of DLS structure.
There are several reports suggesting viral RNA pseudoknot formations [for a review, (50)] and their importance in viral replication have been well discussed. However, most of the reported studies focus on internal ribosomal entry site (IRES) formulations or ribosomal frameshifts. Although there was a report describing long range pseudoknot in the 5′-untranslated and MA coding regions of HIV-1 genomic RNA (51), it was in vitro study with partial RNA fragments. As far as we know, this is the first report suggesting that the RNA pseudoknot-like conformation is essential to maintain viral genome dimerization in virion and subsequent viral replication.
In conclusion, we discovered a long-range interaction within the HIV-1 DLS which has been previously unnoticed and successfully constructed novel 3D models of the DLS through in silico modeling. The structure around this long-range interaction was very conserved and formed a solid pseudoknot-like conformation; therefore its unique shape could be an attractive target for novel anti-HIV therapies. So far, the development of anti-HIV drugs that target RNA dimerization has been dominated by the DIS site (52–55), therefore the characteristic viral RNA structure presented here could be a good target candidate for multi-drug therapy strategies. Further investigation of both the function and structure of the DLS would be required for effective drug development, as well as a better understanding of the viral life cycle.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4, Supplementary Methods and Supplementary References [54–58].
FUNDING
Funding for the open access charge: The Ministry of Education, Culture, Sports, Science and Technology; the Ministry of Health, Labour and Welfare; the Health Science Foundation, Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Seiga Ohmine of the Mayo Clinic, for critically reading the manuscript and Dr Shota Nakamura of the RIMD, Osaka University for helpful discussions.
REFERENCES
- 1.Sakuragi J, Ueda S, Iwamoto A, Shioda T. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 2003;77:4060–4069. doi: 10.1128/JVI.77.7.4060-4069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakuragi J, Sakuragi S, Ohishi M, Shioda T. Direct correlation between genome dimerization and recombination efficiency of HIV-1. Microbes Infect. 2010;12:1002–1011. doi: 10.1016/j.micinf.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Ohishi M, Nakano T, Sakuragi S, Shioda T, Sano K, Sakuragi JI. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 2011;39:3404–3417. doi: 10.1093/nar/gkq1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kung HJ, Bailey JM, Davidson N, Vogt PK, Nicolson MO, McAllister RM. Electron microscope studies of tumor virus RNA. Cold Spring Harb. Symp. Quant. Biol. 1975;39(pt 2):827–834. doi: 10.1101/sqb.1974.039.01.096. [DOI] [PubMed] [Google Scholar]
- 5.Stokrová J, Korb J, Riman J. Electron microscopic studies on the structure of 60-70S RNA of avian myeloblastosis virus. Acta Virol. 1982;26:417–426. [PubMed] [Google Scholar]
- 6.Höglund S, Ohagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 7.Laughrea M, Jetté L, Mak J, Kleiman L, Liang C, Wainberg MA. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303:152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 9.Russell RS, Hu J, Beriault V, Mouland AJ, Laughrea M, Kleiman L, Wainberg MA, Liang C. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type 1 RNA. J. Virol. 2003;77:84–96. doi: 10.1128/JVI.77.1.84-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 11.Paillart JC, Berthoux L, Ottmann M, Darlix JL, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore MD, Hu WS. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009;11:91–102. [PMC free article] [PubMed] [Google Scholar]
- 14.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 17.Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000;299:145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- 18.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 19.Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RR, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein structural studies and implications for genome recognition. J. Mol. Biol. 2001;314:961–970. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 20.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 21.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 22.Kerwood DJ, Cavaluzzi MJ, Borer PN. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry. 2001;40:14518–14529. doi: 10.1021/bi0111909. [DOI] [PubMed] [Google Scholar]
- 23.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 24.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 25.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007;81:7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 29.Aldovini A, Walker BD. Techniques in HIV Research. New York: Stockton Press; 1990. [Google Scholar]
- 30.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuragi JI, Panganiban AT. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J. Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamada M, Sato K, Kiryu H, Mituyama T, Asai K. Predictions of RNA secondary structure by combining homologous sequence information. Bioinformatics. 2009;25:i330–i338. doi: 10.1093/bioinformatics/btp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 38.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TEI, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 40.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 41.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 43.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc. Natl Acad. Sci. USA. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettit SC, Simsic J, Loeb DD, Everitt L, Hutchison CA, 3rd, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 46.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buxton P, Tachedjian G, Mak J. Analysis of the contribution of reverse transcriptase and integrase proteins to retroviral RNA dimer conformation. J. Virol. 2005;79:6338–6348. doi: 10.1128/JVI.79.10.6338-6348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernández-Luna MT, Miranda-Rios J. Riboswitch folding: one at a time and step by step. RNA Biol. 2008;5:20–23. doi: 10.4161/rna.5.1.5974. [DOI] [PubMed] [Google Scholar]
- 49.Turner KB, Hagan NA, Fabris D. Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein. J. Mol. Biol. 2007;369:812–828. doi: 10.1016/j.jmb.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 52.Freisz S, Lang K, Micura R, Dumas P, Ennifar E. Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site. Angew. Chem. Int. Ed. Engl. 2008;47:4110–4113. doi: 10.1002/anie.200800726. [DOI] [PubMed] [Google Scholar]
- 53.Ennifar E, Paillart JC, Bernacchi S, Walter P, Pale P, Decout JL, Marquet R, Dumas P. A structure-based approach for targeting the HIV-1 genomic RNA dimerization initiation site. Biochimie. 2007;89:1195–1203. doi: 10.1016/j.biochi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Bernacchi S, Freisz S, Maechling C, Spiess B, Marquet R, Dumas P, Ennifar E. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007;35:7128–7139. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ennifar E, Paillart JC, Bodlenner A, Walter P, Weibel JM, Aubertin AM, Pale P, Dumas P, Marquet R. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell. Nucleic Acids Res. 2006;34:2328–2339. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koh KB, Fujita M, Adachi A. Elimination of HIV-1 plasmid DNA from virus samples obtained from transfection by calcium-phosphate co-precipitation. J. Virol. Methods. 2000;90:99–102. doi: 10.1016/s0166-0934(00)00224-x. [DOI] [PubMed] [Google Scholar]
- 57.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.