Abstract
Dnmt1 is frequently overexpressed in cancers, which contributes significantly to cancer-associated epigenetic silencing of tumor suppressor genes. However, the mechanism of Dnmt1 overexpression remains elusive. Herein, we elucidate a pathway through which nuclear receptor SHP inhibits zinc-dependent induction of Dnmt1 by antagonizing metal-responsive transcription factor-1 (MTF-1). Zinc treatment induces Dnmt1 transcription by increasing the occupancy of MTF-1 on the Dnmt1 promoter while decreasing SHP expression. SHP in turn represses MTF-1 expression and abolishes zinc-mediated changes in the chromatin configuration of the Dnmt1 promoter. Dnmt1 expression is increased in SHP-knockout (sko) mice but decreased in SHP-transgenic (stg) mice. In human hepatocellular carcinoma (HCC), increased DNMT1 expression is negatively correlated with SHP levels. Our study provides a molecular explanation for increased Dnmt1 expression in HCC and highlights SHP as a potential therapeutic target.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world (1). HCCs are genetically heterogeneous tumors that commonly emerge in the presence of cirrhosis, that is often caused by viral hepatitis or other chronic liver diseases (2). Recent insights into the biology of HCC suggest that certain molecular alterations promote hepatocyte proliferation and survival (3). HCC is frequently advanced when detected and the 5-year survival rate is generally <5%. There is a major need for a better understanding of the molecular and cellular mechanisms leading to HCC. Addressing this need is likely to improve the early diagnosis and treatment of HCC.
Small heterodimer partner (SHP, NROB2) is a unique member of the nuclear receptor superfamily (4) that functions as a transcriptional repressor of genes critical to metabolic diseases (5–16). We recently observed hepatocyte hyperproliferation and spontaneous hepatoma formation in SHP-deficient mice (17). On the other hand, over-expression of SHP in SHP-transgenic mice-induced hepatocyte apoptosis which was attributed to its unexpected role in mitochondria function (18). In addition, diminished SHP expression by promoter hypermethylation occurred in human HCC specimens (19). An association of SHP expression with HCC patient survival was recently established (20), indicating that SHP may serve as a good prognostic factor for liver cancer. These studies suggest a role for SHP in hepatocarcinogenesis by regulating cellular growth and apoptosis signaling.
Tumor suppressor gene silencing by promoter hypermethylation is mainly controlled by three DNA methyltransferases (Dnmt1, Dnmt3α and Dnmt3b) (21). Dnmt1 is a critical gene to maintain CpG methylation and aberrant gene silencing in human cancer cells (22). Dnmt is frequently over-expressed in human cancers (23). Interestingly, a lack of correlation between Dnmt expression and DNA methylation status for several tumor suppressor genes has been reported in HCC (24). This suggests that other mechanisms and tumor suppressor genes may be involved in hepatocarcinogenesis. Thus far, several transcription factors have been identified to regulate the expression of Dnmts, including sp1/sp3, E2F and STAT3 (25–28). The expression of Dnmt1 is also regulated via estrogen-related receptor gamma (ERRγ) and SHP crosstalk (29). Despite these studies, the mechanism of Dnmt overexpression in cancers remains elusive.
In this report, we showed that SHP is a transcriptional repressor of the Dnmt1 expression via crosstalk with metal-responsive transcription factor-1 (MTF-1) through a zinc dependent mechanism. We revealed a feed forward cross-inhibition between SHP and MTF-1 in fine tuning the expression of Dnmt1 via zinc. The diminished SHP expression and subsequently the de-repression of Dnmt1 provide a molecular basis which, in part, explains the increased Dnmt1 expression in HCC.
MATERIALS AND METHODS
Cell lines, animals and human HCC specimens
Human cervix adenocarcinoma cells (Hela, ATCC CCL-2), human hepatoma cells (Huh7, Health Science Research Resources Bank JCRB0403; HepG2, ATCC HB-8065), human fetal kidney cell line (HEK293, ATCC CRL-1573), murine normal liver epithelial cell line (Nmuli, ATCC CRL1638), mouse hepatoma cell line Hepa-1 (ATCC CRL-1830), mouse embryonic fibroblast (MEF) cells prepared from wild-type (MTF-1+/+) and MTF-1 knockout (MTF-1−/−) mice were maintained in Dulbecco's-modified Eagle's medium (DMEM) with 100 U of penicillin G-streptomycin sulfate/ml and 10% heat-inactivated fetal bovine serum (FBS). The stably re-expressed Flag-MTF-1 cells (designated as M42) were described previously (30) and were maintained in DMEM supplemented with 10% FBS and 200 µg/ml hygromycin B. SHP+/+ [wild-type (wt)], SHP−/− (sko), SHP nontransgenic control (nc) and hepatocyte-specific SHP transgenic (stg) mice were described previously (18). Wild-type and sko were maintained on a pure C57BL/6 background and nc and stg were generated with a mixed C57BL6/129sv hybrid background. Protocols for animal use were approved by the Institutional Animal Care and use Committee at the University of Utah. HCC specimens were obtained through the Liver Tissue Cell Distribution Service (LTCDS) (Minneapolis, MN, USA).
Plasmids, siRNA, adenovirus and antibodies
The mouse Dnmt1 (Gene ID: 13433) promoter luciferase construct (Dnmt1Luc), mouse SHP (Gene ID: 23957) promoter luciferase construct (SHPLuc), Flag-MTF1 and Flag-SHP were described previously (29–31). Mouse SHP and non-specific small interfering RNAs (siRNAs) were purchased from Ambion. Both the green fluorescent protein (GFP) control adenoviruses and the GFP–SHP adenoviruses were described previously (18). The following antibodies were used for chromatin immunoprecipitation (ChIP) and western blots (WBs): M-280 sheep anti-rabbit or mouse IgG Dynabeads (Invitrogen Dynal As), rabbit normal IgG (Sigma, R-2004) and antibodies against Flag (Sigma, F-7425), β-actin (Sigma, A-1978), Dnmt1 (Cell signaling, #5032), histone H3 acetyl antibody (H3Ac) (Millipore, #06-599), histone H4 acetyl antibody (H4Ac) (Millipore, #06-866), histone H3 dimethyl Lys4 antibody (H3K4Me2) (Millipore, #07-030) and histone H3 dimethyl Lys9 antibody (H3K9Me2) (Millipore, #17-648).
DNA methyltransferase activity assay
The DNA methyltransferase activity was assessed using the EpiQuik™ DNA methyltransferase activity/inhibition assay kit (Epigentek, Brooklyn, NY, USA). Briefly, 12 µg of nuclear extracts or 0–2.5 units of purified Dnmt enzymes was incubated with 1.6 mM of adomet for 60 min at 37°C followed by the incubation with 1 µg/ml of capture antibody for 60 min at room temperature. Then each well was washed with wash buffer for four times and incubated with 0.2 µg/ml of detection antibody at room temperature for 30 min. Developing solution was added and incubated for 2–10 min away from light. Then reactions were stopped and read at 450 nm using a microplate spectrophotometer (Benchmark plus, Bio-Rad).
Dnmt1 amount assay
The Dnmt1 amount was assessed using the epiquik™ Dnmt1 assay kit. Briefly, 12 µg of nuclear extracts or 0–10 ng of purified Dnmt1 protein was incubated for 60 min at 37°C followed by the incubation with 1 µg/ml of affinity antibody for 60 min at room temperature. Then each well was washed four times and incubated with 0.2 µg/ml of detection antibody at room temperature for 30 min. Developing solution was added and incubated for 2–10 min away from light. Then reactions were stopped and read at 450 nm using a microplate spectrophotometer (Benchmark plus, Bio-Rad).
RT-PCR and real-time qPCR analysis
The method can be found in our recent publications (18,32). In brief, qPCR was performed with total RNA using the SYBR Green PCR master mix (Applied Biosystems). The melting-curve data were collected to check PCR specificity. Each cDNA sample was run as triplicates, and the corresponding no-reverse transcriptase (RT) mRNA sample was included as a negative control. The amount of PCR products was measured by threshold cycle (Ct) values and the relative ratio of specific genes to HPRT1 for each sample was then calculated. The sequences for the primers are available upon request.
Promoter activity assays
Detailed methods can be found in our recent publications (18,33–36). In brief, Hela, MEF or HEK293 cells were transfected with the plasmids as indicated in the figure legends. Empty vector DNA was added as needed so that the same amounts of expression vector DNA were present in each transfection. Transfection was carried out using Lipofectamine 2000 (Invitrogen). Luciferase activities were measured and normalized against renilla activities (Promega). Consistent results were observed in three independent triplicate transfection assays.
ChIP assay
Detailed methods can be found in our recent publications (18,29,33–36). M42 cells were transfected with the plasmids and chromatin crosslinked, and immunoprecipitations were performed with specific antibodies as indicated in the figure legends, with rabbit normal IgG as negative control (Upstate). Ethanol-extracted DNA was used as the template to amplify the Dnmt1 promoter or SHP promoter. The sequences for the primers are as follows: Dnmt1 promoter P1 forward 5′-TGTGAGTGCAATGCCCATAGA-3′ and reverse 5′-GGGAAGACCATGCTATGCCC-3′; SHP promoter P1 forward 5′-GACCTTGGTGCCCTGGTACA-3′ and reverse 5′-ATGCATACACGCTGACCCTG-3′. The control region from −1143 nt to −950 nt in the Dnmt1 promoter (forward 5′-TGGCAGAAGTGGTTCCTGG-3′ and reverse 5′-AAAGGCCTCCCGTCTAACCC-3′) and −1536 nt to −1343 nt in SHP promoter (forward 5′-TCTCTGCAGGGCTTTGCTCT-3′ and reverse 5′-TTCCAGCCAGGGTTTCTCTG-3′) are served as negative controls, respectively.
WBs in cells and liver tissues
Detailed methods can be found in our recent publication (37). Hepa-1 cells were transfected with Flag-SHP plasmids and cultured for 36 h. For Zn treatment, MEF cells were incubated for up to 6 h in medium containing ZnSO4 (100 µM). For WBs using mouse livers, 100 mg of liver fragments from 2-, 13- and 20-month-old SHP−/−mice or their respective controls were homogenized in lysis buffer and subjected to WBs.
Statistical analysis
Data are expressed as the mean ± SD. Statistical analyses were carried out using Student's unpaired t-test; P < 0.01 was considered statistically significant.
RESULTS
SHP-deficiency results in increased hepatic Dnmt1 expression
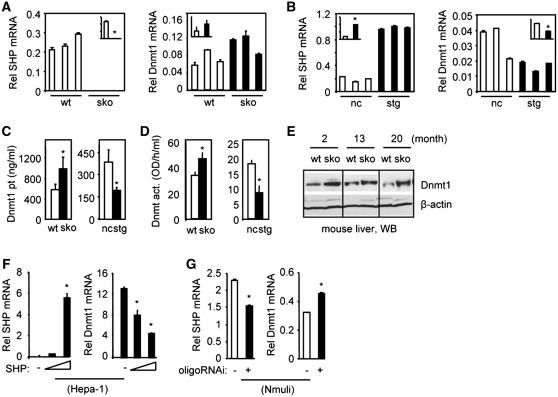
qPCR analysis of mRNA of three individual mice per genotype revealed a moderate, although statistically insignificant, up-regulation of hepatic Dnmt1 mRNA in 2-month-old SHP−/− (sko) mice relative to wt mice (Figure 1A) and a significant down-regulation of Dnmt1 mRNA in hepatocyte SHP-transgenic (stg) mice relative to non-transgenic (nc) mice (Figure 1B). The levels of Dnmt1 protein (Figure 1C) and Dnmt enzymatic activity (Figure 1D) corresponded to the changes of Dnmt1 mRNA in sko and stg mice, i.e. increased in sko mice and decreased in stg mice. In addition, the Dnmt1 protein was increased in the liver of 2-, 13- and 20-month-old sko mice as determined by WB (Figure 1E).
Figure 1.
SHP inhibition of Dnmt1 expression. (A and B) qPCR analysis of SHP and Dnmt1 mRNAs in the liver of wt and SHP−/− (sko), non-transgene (nc) and hepatocyte specific SHP transgenic (stg) mice (n = 3 per genotype). (C and D) The levels of Dnmt1 protein (pt) and enzymatic activity (act.) in the liver of wt and sko, nc and stg mice. (E) WB of Dnmt1 protein in wt and sko mice. (F) qPCR analysis of SHP and Dnmt1 mRNA in Hepa-1 cells over-expressing SHP from an expression plasmid. (G) qPCR analysis of SHP and Dnmt1 mRNA in Nmuli cells with SHP knockdown using an oligoSHP-siRNA (oligoRNAi). Statistical results represent mean ± SD of triplicate assays (*P < 0.01).
SHP inhibits Dnmt1 mRNA in mouse hepatocyte cell lines
Mouse hepatoma Hepa-1 cells lack SHP (17) but have high levels of Dnmt1 whereas the mouse normal hepatocyte Nmuli cells have higher levels of SHP but low levels of Dnmt1. Over-expression of SHP dose-dependently decreased Dnmt1 mRNA in Hepa-1 cells (Figure 1F). In contrast, knockdown of SHP in Nmuli cells with siRNA against SHP led to the induction of Dnmt1 mRNA (Figure 1G). The results indicate that Dnmt1 transcription is negatively regulated by SHP.
The expression of MTF-1 is repressed by SHP
MTF-1 is a transcription factor that activates the transcription of metallothionein genes in response to heavy metal load and other stresses such as hypoxia and oxidative stress (38,39).
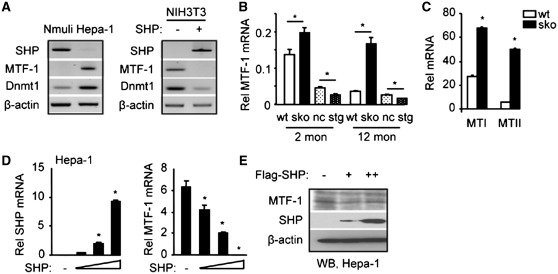
Examination of the correlation between SHP, MTF-1 and Dnmt1expression in vitro in Nmuli and Hepa-1 cell cultures revealed an inverse relationship between SHP and MTF-1/Dnmt1 (Figure 2A, left). Similarly, MTF-1 and Dnmt1 are expressed in NIH3T3 cells in which SHP is barely detectable and their expression was repressed when the cells were transfected with a SHP expression plasmid (Figure 2A, right). An inverse expression correlation between SHP and MTF-1 was also observed in vivo in mouse liver, in which MTF-1 mRNA was up-regulated in sko versus wt mice but down-regulated in stg versus nc mice at both 2 and 12 months of age (Figure 2B). The decreased MTF-1 mRNA in 12-month-old wt mice relative to 2-month-old mice was in agreement with the higher SHP level in the older mice (data not shown). The expression of MTF-1 target genes MTI and MTII was strongly induced by zinc in sko as compared to the wt mice (Figure 2C), confirming the activation of MTF-1 in sko mice.
Figure 2.
SHP inhibition of MTF-1 expression. (A) Semi-quantitative PCR analysis of SHP (45× cycles), MTF-1 (35× cycles) and Dnmt1 (28× cycles) expression in Nmuli and Hepa-1 cells (left), and in NIH3T3 cells with SHP over-expression (right). (B) qPCR analysis of hepatic MTF-1 mRNA expression in wt and sko, nc and stg mice at 2 and 12 months of age. *P < 0.01 versus corresponding control. (C) qPCR analysis of hepatic MTI and MTII mRNA in wt and sko mice at 2 months of age. *P < 0.01 versus corresponding control. (D) qPCR analysis of SHP and MTF-1 mRNA in Hepa-1 cells that over-expressed SHP from an expression vector. *P < 0.01 versus control (−) group without SHP expression. (B–D) Statistical results represent mean ± SD of triplicate assays. (E) WB of MTF-1 and SHP protein in Hepa-1 cells that were over-expressing Flag-SHP from an expression vector. The endogenous MTF-1 protein was detected using an anti-MTF-1 antibody and the over-expressed SHP protein was detected using an anti-Flag antibody.
The above observations suggest that SHP may inhibit the expression of MTF-1. Indeed, over-expression of SHP in Hepa-1 cells dose-dependently inhibited MTF-1 at both the mRNA (Figure 2D) and protein (Figure 2E) levels.
Zinc decreases SHP expression through MTF-1
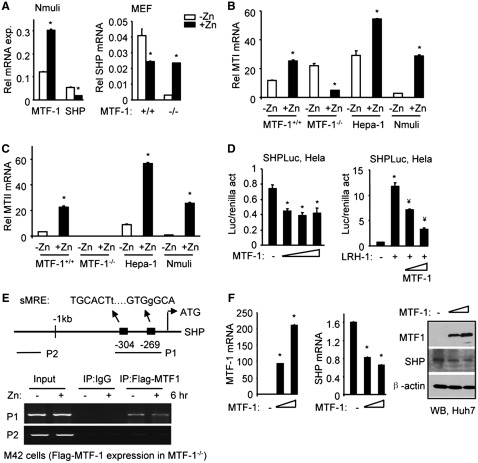
MTF-1 can be activated in response to heavy metal exposure (39,40). In Nmuli cells when MTF-1 was activated by treatment with exogenous zinc (Zn), a marked reduction in SHP mRNA was observed (Figure 3A, left). Zinc treatment also decreased SHP mRNA in MTF-1+/+ MEF cells (Figure 3A, right). Interestingly, the basal SHP mRNA was decreased in MTF-1−/− MEFs which was up-regulated by zinc. This suggests that, in the absence of MTF-1, zinc may regulate SHP through alternative mechanisms. MTI (Figure 3B) and MTII (Figure 3C) mRNAs were also markedly induced by zinc in MTF-1+/+, Hepa-1 and Nmuli cells, confirming the efficacy of zinc treatment.
Figure 3.
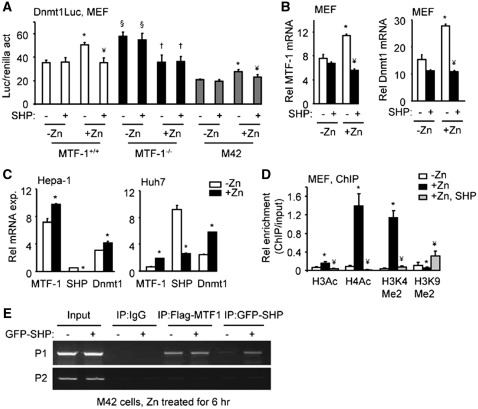
Zinc inhibition of SHP transcription requires MTF-1. (A) qPCR analysis of MTF-1 and SHP mRNA in Nmuli cells (left), or in MTF-1+/+ and MTF-1−/− MEF cells (right), without (−Zn) or with (+Zn, 100 µM ZnSO4) treatment for 6 hr. *P < 0.01, + Zn group versus −Zn group. (B and C) qPCR analysis of MTI and MTII mRNA in MTF-1+/+ MEFs, MTF-1−/− MEFs, Hepa-1 and Nmuli cells, without (−Zn) or with (+Zn, 100 µM ZnSO4) treatment for 6 hr. *P < 0.01, +Zn group versus −Zn group. (D) Transient transfection assays in Hela cells to determine the activity of the mSHP promoter luciferase (Luc) reporter in the absence or presence of the expression plasmid for MTF-1 (200 ng), without (left) or with LRH-1 (30 ng) co-transfection (right). *P < 0.01 versus control (−) group. ¥P < 0.01 versus LRH-1 alone. (A–D) Statistical results represent mean ± SD of triplicate assays. (E) Left: diagram showing putative MTF-1 binding sites (MREs) in the SHP promoter. Small letters indicate base pair mismatch from a consensus MRE. Right: ChIP assays to determine the physical association of MTF-1 with the SHP promoter in M42 cells in the without or with 100 µM ZnSO4 treatment for 6 hr. M42 cells were generated by stably re-expressing a Flag-MTF-1 in the MTF-1−/− MEF cells. (F) Left: qPCR analysis of MTF-1 and SHP mRNA in Huh7 cells that were overexpressed with a MTF-1 plasmid. Right: WB of MTF-1 and SHP protein in Huh7 cells that were overexpressed with a MTF-1 plasmid. *P < 0.01 versus control (−) group.
Liver receptor homolog-1 (LRH-1) is a known SHP activator (31). Further analysis of the SHP promoter showed that when co-transfected with a SHP promoter luciferase reporter, MTF-1 not only inhibited the basal SHP promoter activity (Figure 3D, left), but also repressed LRH-1 transactivation (Figure 3D, right).
Next, Flag-MTF-1 was stably expressed in MTF-1−/− MEF cells (designated as M42) for ChIP analysis using an anti-Flag antibody. A direct association of MTF-1 with putative metal-response elements in the SHP promoter (sMRE) was observed (Figure 3E), but the association was not affected by zinc treatment. In addition, the levels of endogenous SHP mRNA and protein were reduced dose-dependently by ectopic expression of MTF-1 (Figure 3F), demonstrating a direct inhibition of MTF-1 on SHP expression. GST pull down assays did not identify physical interactions between SHP and MTF-1 (data not shown). Overall, the data presented in Figures 2 and 3 demonstrate a reciprocal inhibition between SHP and MTF-1.
Zinc-induced Dnmt1 transcription requires MTF-1
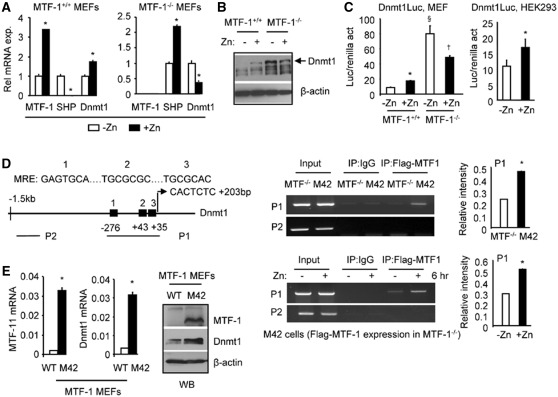
We tested the effect of zinc on the expression of MTF-1, SHP and Dnmt1 in MTF-1+/+ and MTF-1−/− MEF cells. Zinc (Zn) exposure resulted in a marked induction of MTF-1 mRNA in MTF-1+/+ MEFs, which was accompanied by a decreased SHP expression and increased Dnmt1 expression (Figure 4A, left). Zn treatment in MTF-1 deficient (MTF-1−/−) MEFs induced SHP expression, which corresponded with the attenuated expression of Dnmt1 in response to Zn (Figure 4A, right). SHP mRNA was induced by zinc in MTF-1−/− MEFs, suggesting an alternative mechanism that activates SHP in the absence of MTF-1. The increased expression of SHP may be responsible for the inhibition of Dnmt1 expression.
Figure 4.
Zinc activation of Dnmt1 expression requires MTF-1. (A) qPCR analysis of MTF-1, SHP and Dnmt1 mRNA in MTF-1+/+ and MTF-1−/− MEF cells without (−Zn) or with 100 µM ZnSO4 treatment for 6 hr (+Zn). *P < 0.01, +Zn group versus −Zn group. (B) WB of Dnmt1 protein in MTF-1+/+ and MTF-1−/− MEF cells in the without (−) or with 100 µM ZnSO4 treatment for 6 hr (+). (C) Left: Transient transfection assays in MTF-1+/+ and MTF-1−/− MEF cells to determine the activation of Dnmt1 promoter-luciferase reporter (Luc) in response to zinc treatment. Right: Transient transfection assays of the Dnmt1 promoter-luciferase (Luc) reporter in HEK293 cells treated with zinc. Cells were cultured in medium containing 100 µM ZnSO4 for 6 hr. *P < 0.01, +Zn group versus −Zn group; §P < 0.01, −Zn group in MTF-1−/− versus −Zn group in MTF-1+/+ cells; †P < 0.01, +Zn group versus −Zn group in MTF-1−/− cells. (A and C) Statistical results represent mean ± SD of triplicate assays. (D) Left: diagram showing putative MREs in the Dnmt1 promoter and the location of primers used for ChIP assays. Middle: ChIP assays to monitor the association of MTF-1 to the endogenous Dnmt1 promoter in MTF-1−/− and M42 cells in the absence of zinc (upper panel), or in M42 cells treated with 100 µM ZnSO4 for 6 hr (lower panel). Right: Quantification of the binding signals in ChIP assays. (E) Left: qPCR analysis of MTF-1 and Dnmt1 mRNA in MEF cells (WT and M42). Right: WB of MTF-1 and Dnmt1 protein in MEF cells (WT and M42).
Dnmt1 protein was elevated by zinc treatment in MTF-1+/+ cells compared to untreated cells (Figure 4B). Surprisingly, Dnmt1 protein showed a significant elevation in basal expression in MTF-1−/− cells without zinc, which was reduced by zinc exposure. This result correlated with the lower SHP levels in MTF-1−/− MEFs without zinc and the up-regulation of SHP by zinc (Figures 3A and 4A).
To confirm if the induction of MTF-1 by zinc is directly associated with the activation of the Dnmt1 promoter, MEFs were transfected with the Dnmt1 promoter in the absence or presence of zinc. The luciferase activity driven by the Dnmt1 promoter was induced by zinc treatment in MTF-1+/+ (Figure 4C, left), consistent with the induction of MTF-1 expression by zinc in MTF-1+/+ cells (Figure 4A, left). Elevated Dnmt1 promoter activity was observed in MTF-1−/− cells without zinc, which was decreased by zinc. The result was consistent with the alterations of Dnmt1 protein (Figure 4B) and SHP mRNA (Figure 3A, right) in MTF-1−/− cells. Similar activation of the Dnmt1 promoter by zinc treatment was observed in HEK293 cells that have endogenous MTF-1 (Figure 4C, right).
Several potential binding motifs for MTF-1 (MRE) were identified in the Dnmt1 promoter (26) (Figure 4D, left), suggesting that MTF-1 may be capable of inducing the transcription of Dnmt1. An association of MTF-1 with the endogenous Dnmt1 promoter was observed using P1 primers covering three putative MREs in M42 cells that expressed MTF-1, but not in MTF-1−/− cells (Figure 4D, top right), and this association was enhanced by zinc (Figure 4D, bottom right). In addition, both Dnmt1 mRNA and protein were markedly increased in M42 cells compared with the wt cells (Figure 4E). The data suggest that zinc induces Dnmt1 expression by increasing MTF-1 expression as well as its recruitment to the Dnmt1 promoter, which in turn activates Dnmt1 gene transcription.
SHP antagonizes zinc-mediated activation of Dnmt1
Activation of a transfected Dnmt1 promoter-luciferase reporter by zinc was decreased by SHP over-expression in MTF-1+/+ and M42 cells (Figure 5A). Basal Dnmt1 promoter activity was somewhat lower in M42. In MTF-1−/− cells, zinc treatment decreased Dnmt1-luc activity which was not further decreased by SHP over-expression. Furthermore, induction of MTF-1 and Dnmt1 mRNAs by zinc was repressed by over-expression of SHP (Figure 5B). In Hepa-1 (mouse HCC) and Huh7 (human HCC) cells, zinc also induced MTF-1 and Dnmt1 mRNAs whereas it reduced SHP mRNA (Figure 5C).
Figure 5.
SHP inhibition of MTF-1-dependent induction of Dnmt1. (A) Transient transfection assays of the Dnmt1 promoter-luciferase (Luc) reporter in MEF (MTF-1+/+, MTF-1−/− and M42) cells without (−Zn) or with 100 µM ZnSO4 treatment for 6 hr (+Zn) and in the absence (−) or presence (+) of SHP over-expression. *P < 0.01, +Zn group versus −Zn group without SHP; ¥P < 0.01, +Zn with SHP versus +Zn without SHP; §P < 0.01, −Zn group in MTF-1−/− versus −Zn group in MTF-1+/+ cells; †P < 0.01, +Zn group versus −Zn group in MTF-1−/− cells. (B) qPCR analysis of MTF-1 and Dnmt1 mRNA in MTF-1+/+ MEF cells without (−Zn) or with 100 µM ZnSO4 treatment for 6 hr (+Zn) and without (−) or with SHP over-expression (+). *P < 0.01, +Zn group versus −Zn group without SHP; ¥P < 0.01, +Zn with SHP versus +Zn without SHP. (C) qPCR analysis of MTF-1, SHP and Dnmt1 mRNA in Hepa-1 and Huh7 cells without (−Zn) or with 100 µM ZnSO4 treatment for 6 hr (+Zn). *P < 0.01, +Zn group versus −Zn group. (A–D) Statistical results represent mean ± SD of triplicate assays. (D) ChIP assays of histone modifications in the Dnmt1 promoter after zinc treatment and SHP over-expression. MEF cells were cultured in medium with (+Zn) or without (−Zn) 100 µM ZnSO4 for 6 hr. ChIP PCR products were amplified from input-positive controls, IgG-negative controls and antibody immunoprecipitates. Histograms show antibody/input ratios of PCR products quantified using qPCR, expressed as Relative (Rel) enrichment. The P1 primers covering the three MREs (see Figure 4D) were used for ChIP assays. Error bars represent SD from three independent measurements. *P < 0.01 versus −Zn group; ¥P < 0.01 versus +Zn group. (E) ChIP assays to determine the effect of SHP expression on the recruitment of MTF-1 to the Dnmt1 promoter. M42 cells were transfected with or without GFP-SHP plasmid for 36 hr and treated with 100 µM ZnSO4 for 6 hr. The chromatin was immunoprecipitated using anti-GFP or anti-Flag antibodies. PCR was used to determine the association of SHP and MTF-1 to the endogenous Dnmt1 promoter by using P1 and P2 primers.
We next used ChIP assays to examine histone modifications in the Dnmt1 promoter in cells treated with zinc and over-expressing SHP. Zinc treatment markedly increased active histone marks (H3Ac, H4Ac and H3K4Me2) whereas it decreased the inactive histone mark (H3K9Me2) in the Dnmt1 promoter (Figure 5D). These effects of zinc were largely abolished in the presence of SHP over-expression. These histone modifications of the Dnmt1 promoter by SHP correlate with Dnmt1 expression regulation by SHP. Thus, zinc and SHP counteract each others regulation of Dnmt1 promoter activity by rapidly modulating local chromatin configuration. SHP did not decrease recruitment of MTF-1 to the Dnmt1 promoter (Figure 5E), it is postulated that MTF-1 utilizes a zinc dependent mechanism to facilitate its binding to the Dnmt1 promoter and SHP employs a distinct mechanism not by direct interaction but by inhibiting the expression of MTF-1.
SHP expression inversely correlates with DNMT1 expression in human HCC
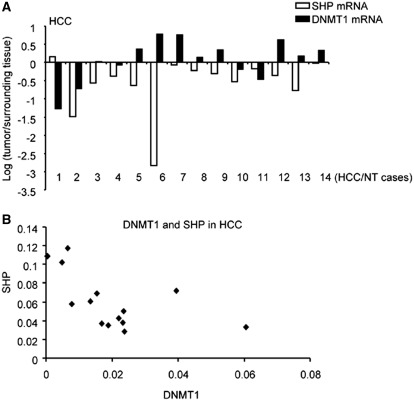
Our early studies reported decreased SHP mRNA due to promoter hypermethylation in human HCC specimens (19). Quantification of DNMT1 mRNA levels in 14 matched HCC specimens and paired non-neoplastic liver tissues (surrounding tissues) revealed elevated DNMT1 mRNA in a majority of these HCC (8/14) (Figure 6A). Increased DNMT1 mRNA negatively correlated with decreased SHP mRNA in HCC specimens (Figure 6B), consistent with a regulatory link between SHP and DNMT1 expression in HCC.
Figure 6.
Inverse correlation between DNMT1 and SHP mRNAs in human HCC. (A) qPCR analysis of DNMT1 and SHP mRNAs in 14 pairs of human HCC specimens (HCC) and their corresponding normal surrounding tissues (NT). The expression of DNMT1 and SHP in HCC was normalized to NT and the results were expressed as log 2. (B) Correlation between DNMT1 and SHP mRNA abundance in human HCC.
DISCUSSION
DNA methylation is an epigenetic mark critical for regulating the chromatin structure and gene transcription. DNA methylation is mainly catalyzed by three DNA methyltransferases (Dnmts) encoded by Dnmt1, Dnmt3α and Dnmt3b. Dnmt1, the maintenance DNA methyltransferase and a major Dnmt in adult cells, plays an important role in cancer progression associated with epigenetic silencing of tumor suppressor genes. Previous work has shown that the mouse Dnmt1 promoter is independently activated by Sp1/Sp3 (25) and E2F (26,41), and the human DNMT1 by signal transducer and activator of transcription-3 (STAT3) (28) transcription factors. Dnmt1 expression can also be modulated by Rb (41), AUF1 (42), BRCA1 (43) and small non-coding microRNAs miR-152 (44,45). In addition to its expression regulation, Dnmt1 protein stability is controlled by SET7 (46) and LSD1 (47), and its enzymatic activity is mediated by G9α (48), EZH2 (49), PML-RAR (50) and hNaa10p (51). A recent study demonstrated that p53 is a negative regulator of the Dnmt1 promoter activity (52). However, the mechanism of Dnmt1 overexpression in cancers remains largely unknown. The results in this study identified a zinc-mediated activation of Dnmt1 that is modulated by MTF-1 and SHP crosstalk.
An important finding of this study is our establishment of a regulatory link between zinc/MTF-1 and Dnmt1 in HCC. Although there is no report investigating the specific function of MTF-1 in the development of liver cancer, a recent study showed that MTF-1 protein levels were significantly elevated in breast, lung and cervical carcinomas (53), suggesting a role for MTF-1 in human tumor development and growth. MTF-1 was also proposed as a candidate lymphoma susceptibility gene (54), and loss of MTF-1 resulted in delayed tumor growth associated with increased matrix collagen deposition and reductions in vasculature density (55). Both tissue hypoxia and oxidative stress are well documented to be common features of most solid tumors (56). MTF-1 has been associated with hypoxic-induced placenta growth factor (PIGF) expression, an angiogenic factor expressed in many tumors (57). Up-regulation of MTF-1 and PIGF occurred in human intrahepatic cholangiocarcinoma due to loss of liver–intestine cadherin, which contributed to tumor differentiation and vascular invasion, and thus poor prognosis (58).
Interestingly, we found that the induction of Dnmt1 by MTF-1 requires the presence of zinc. Zinc not only increases MTF-1 expression, as seen by other studies (59,60), but more importantly, it also enhances the recruitment of MTF-1 to the Dnmt1 promoter and causes transcriptionally active configuration of the local chromatin. It should be noted that, in the 14 HCC specimens that we analyzed (Figure 6), the expression of MTF-1 was not significantly altered, and no strong positive correlation between MTF-1 and DNMT1 mRNA was observed (data not shown). This suggests that the disrupted intracellular zinc homeostasis, but not merely changes of MTF-1 mRNA, may be critical in promoting MTF-1-mediated activation of Dnmt1. Although it remains to be determined how zinc metabolism is altered during HCC growth, MTF-1 may play a role by activating Dnmt1 under aberrant metal conditions. Up-regulated Dnmt1 may further silence other tumor suppressors and stimulate HCC progression. It would be interesting to determine in future studies whether hypoxia or oxidative stress contribute to Dnmt1 expression regulation by MTF-1. In this regard, MTF-1 could be a potential therapeutic target that offers the opportunity to manipulate metal or redox homeostasis in tumor cells.
One intriguing observation is the cross-inhibition between SHP and MTF-1. SHP directly represses MTF-1 expression at the transcriptional level. Conversely, induction of MTF-1 by zinc inhibits SHP expression by binding to the SHP promoter and repressing the basal, as well as LRH-1-induced SHP promoter activity. Surprisingly, the basal level of SHP is decreased in MTF-1-deficient MEFs, which is induced by zinc. On the other hand, the induction of MTF-1 target gene MTI, as well as Dnmt1, is observed in MTF-1−/− cells, which is repressed by zinc. The changes in SHP may be responsible for the alterations of Dnmt1. It is postulated that MTF-1 may play a predominant role to control a zinc-dependent activation of Dnmt1 through inhibition of SHP. In the absence of MTF-1, zinc may turn on other zinc responsive genes that function as SHP activators, resulting in the elevation of SHP and reduction of Dnmt1.
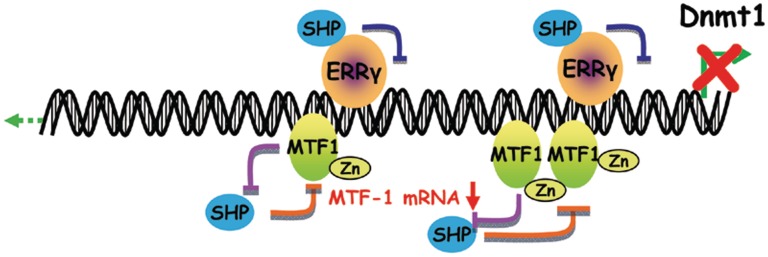
Recently we showed that SHP also inhibits Dnmt1 promoter transactivation by ERRγ in several cancer cells (29). The effect of SHP is through a direct protein–protein interaction with ERRγ to convert the local chromatin structure of the Dnmt1 promoter from a transcriptionally active mode to an inactive mode. Because SHP does not interact with the MTF-1 protein directly, SHP appears to repress Dnmt1 through decreasing MTF-1 expression. The induction of MTF-1 by zinc may activate Dnmt1 by repressing SHP, which represents a feed-forward inhibitory mechanism between SHP and MTF-1 that controls Dnmt1 expression. Thus, SHP modulates the expression of Dnmt1 by at least two distinct mechanisms (Figure 7).
Figure 7.
Schematic showing SHP inhibition of the Dnmt1 promoter through two distinct mechanisms. Our recent study showed that SHP inhibits ERRγ transactivation of the Dnmt1 promoter. The present study identified a zinc-mediated induction of Dnmt1 which is modulated by the cross-inhibition between MTF-1 and SHP.
In conclusion, we identified a second pathway through which SHP represses Dnmt1. SHP inhibition of Dnmt1 may affect global DNA methylation and alter methylation levels of tumor suppressors. Targeting SHP may prove a useful approach to demethylate and reactivate the silenced tumor suppressors to slow the progression of HCC.
FUNDING
T32CA092347 Multidisciplinary Cancer Research Training Program (MCRTP) (to Y.Z.) and National Institutes of Health (DK080440) (L.W., partial). Y.Z. is supported by T32CA092347 Multidisciplinary Cancer Research Training Program (MCRTP). This work is in part supported by National Institutes of Health (DK080440) to L.W. Funding for open access charge: Institutional start up fund.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
HCC specimens were obtained through the LTCDS under the NIH Contract # N01-DK-7-0004/HHSN267200700004C (Minneapolis, MN, USA).
REFERENCES
- 1.Seeff LB, Hoofnagle JH. Epidemiology of hepatocellular carcinoma in areas of low hepatitis B and hepatitis C endemicity. Oncogene. 2006;25:3771–3777. doi: 10.1038/sj.onc.1209560. [DOI] [PubMed] [Google Scholar]
- 2.Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25:3866–3884. doi: 10.1038/sj.onc.1209550. [DOI] [PubMed] [Google Scholar]
- 3.Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778–3786. doi: 10.1038/sj.onc.1209547. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim. Biophys. Acta. 2011;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Han Y, Kim CS, Lee YK, Moore DD. Resistance of SHP-null mice to bile acid-induced liver damage. J. Biol. Chem. 2003;278:44475–44481. doi: 10.1074/jbc.M305258200. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 9.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabbi-Anneni I, Cooksey R, Gunda V, Liu S, Mueller A, Song G, McClain DA, Wang L. Overexpression of nuclear receptor SHP in adipose tissues affects diet-induced obesity and adaptive thermogenesis. Am. J. Physiol. Endocrinol. Metab. 298:E961–E970. doi: 10.1152/ajpendo.00655.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman HB, Lai K, Evans MJ. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J. Lipid Res. 2009;50:193–203. doi: 10.1194/jlr.M800323-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, Lee YK. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47:1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 14.Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pellicciari R, Morelli A. The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology. 2004;127:1497–1512. doi: 10.1053/j.gastro.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, Wang L. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol. Cell. Biol. 30:1341–1356. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Park YY, Choi HS, Lee JS. Systems-level analysis of gene expression data revealed NR0B2/SHP as potential tumor suppressor in human liver cancer. Mol. Cells. 2010;30:485–491. doi: 10.1007/s10059-010-0136-6. [DOI] [PubMed] [Google Scholar]
- 21.Park HJ, Yu E, Shim YH. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006;233:271–278. doi: 10.1016/j.canlet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 23.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 24.Chan AO, Rashid A. CpG island methylation in precursors of gastrointestinal malignancies. Curr. Mol. Med. 2006;6:401–408. doi: 10.2174/156652406777435417. [DOI] [PubMed] [Google Scholar]
- 25.Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur. J. Biochem. 2002;269:2961–2970. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 26.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–3113. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinawath A, Miyake S, Yanagisawa Y, Akiyama Y, Yuasa Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem. J. 2005;385:557–564. doi: 10.1042/BJ20040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–1064. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang L. Nuclear receptor SHP inhibition of Dnmt1 expression via ERRγ. FEBS Lett. 585:1269–1275. doi: 10.1016/j.febslet.2011.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Kimura T, Huyck RW, Laity JH, Andrews GK. Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP, and Sp1. Mol. Cell. Biol. 2008;28:4275–4284. doi: 10.1128/MCB.00369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, Xu P, Pellicciari R, Wang L. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J. Biol. Chem. 285:24871–24881. doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, Andrews GK. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PloS One. 2010;5:e13158. doi: 10.1371/journal.pone.0013158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PloS One. 2009;4:e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song G, Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727–5735. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song G, Wang L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PloS One. 2009;4:e7829. doi: 10.1371/journal.pone.0007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J. Biol. Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Bonzo JA, Gonzalez FJ, Wang L. Diurnal regulation of the early growth response 1 (Egr-1) protein expression by hepatocyte nuclear factor 4alpha (HNF4alpha) and small heterodimer partner (SHP) cross-talk in liver fibrosis. J. Biol. Chem. 286:29635–29643. doi: 10.1074/jbc.M111.253039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews GK, Lee DK, Ravindra R, Lichtlen P, Sirito M, Sawadogo M, Schaffner W. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 2001;20:1114–1122. doi: 10.1093/emboj/20.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Wimmer U, Lichtlen P, Inderbitzin D, Stieger B, Meier PJ, Hunziker L, Stallmach T, Forrer R, Rülicke T, et al. Metal-responsive transcription factor-1 (MTF-1) is essential for embryonic liver development and heavy metal detoxification in the adult liver. FASEB J. 2004;18:1071–1079. doi: 10.1096/fj.03-1282com. [DOI] [PubMed] [Google Scholar]
- 40.Saydam N, Steiner F, Georgiev O, Schaffner W. Heat and heavy metal stress synergize to mediate transcriptional hyperactivation by metal-responsive transcription factor MTF-1. J. Biol. Chem. 2003;278:31879–31883. doi: 10.1074/jbc.M302138200. [DOI] [PubMed] [Google Scholar]
- 41.McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25:2727–2735. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- 42.Torrisani J, Unterberger A, Tendulkar SR, Shikimi K, Szyf M. AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability. Mol. Cell. Biol. 2007;27:395–410. doi: 10.1128/MCB.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shukla V, Coumoul X, Lahusen T, Wang RH, Xu X, Vassilopoulos A, Xiao C, Lee MH, Man YG, Ouchi M, et al. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 2010;20:1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 45.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 46.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl Acad. Sci. USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 48.Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 50.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 51.Lee CF, Ou DS, Lee SB, Chang LH, Lin RK, Li YS, Upadhyay AK, Cheng X, Wang YC, Hsu HS, et al. hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, Lu YY, Tang YA, Yang YC, Yang PC, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Amin K, Sato BG, Samuelsson SJ, Sambucetti L, Haroon ZA, Laderoute K, Murphy BJ. The metal-responsive transcription factor-1 protein is elevated in human tumors. Cancer Biol. Ther. 2010;9:469–476. doi: 10.4161/cbt.9.6.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura Y, Maruyama M, Mishima Y, Fujisawa H, Obata M, Kodama Y, Yoshikai Y, Aoyagi Y, Niwa O, Schaffner W, et al. Predisposition to mouse thymic lymphomas in response to ionizing radiation depends on variant alleles encoding metal-responsive transcription factor-1 (Mtf-1) Oncogene. 2005;24:399–406. doi: 10.1038/sj.onc.1208197. [DOI] [PubMed] [Google Scholar]
- 55.Haroon ZA, Amin K, Lichtlen P, Sato B, Huynh NT, Wang Z, Schaffner W, Murphy BJ. Loss of metal transcription factor-1 suppresses tumor growth through enhanced matrix deposition. Faseb J. 2004;18:1176–1184. doi: 10.1096/fj.03-1205com. [DOI] [PubMed] [Google Scholar]
- 56.Williams KJ, Cowen RL, Stratford IJ. Hypoxia and oxidative stress. Tumour hypoxia–therapeutic considerations. Breast Cancer Res. 2001;3:328–331. doi: 10.1186/bcr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green CJ, Lichtlen P, Huynh NT, Yanovsky M, Laderoute KR, Schaffner W, Murphy BJ. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 2001;61:2696–2703. [PubMed] [Google Scholar]
- 58.Takamura M, Yamagiwa S, Wakai T, Tamura Y, Kamimura H, Kato T, Tsuchiya A, Matsuda Y, Shirai Y, Ichida T, et al. Loss of liver-intestine cadherin in human intrahepatic cholangiocarcinoma promotes angiogenesis by up-regulating metal-responsive transcription factor-1 and placental growth factor. Int. J. Oncol. 2010;36:245–254. [PubMed] [Google Scholar]
- 59.Chung MJ, Hogstrand C, Lee SJ. Cytotoxicity of nitric oxide is alleviated by zinc-mediated expression of antioxidant genes. Exp. Biol. Med. (Maywood) 2006;231:1555–1563. doi: 10.1177/153537020623100916. [DOI] [PubMed] [Google Scholar]
- 60.Ostrakhovitch EA, Olsson PE, von Hofsten J, Cherian MG. P53 mediated regulation of metallothionein transcription in breast cancer cells. J. Cell. Biochem. 2007;102:1571–1583. doi: 10.1002/jcb.21381. [DOI] [PubMed] [Google Scholar]