Abstract
The 23S rRNA nucleotide m2G2445 is highly conserved in bacteria, and in Escherichia coli this modification is added by the enzyme YcbY. With lengths of around 700 amino acids, YcbY orthologs are the largest rRNA methyltransferases identified in Gram-negative bacteria, and they appear to be fusions from two separate proteins found in Gram-positives. The crystal structures described here show that both the N- and C-terminal halves of E. coli YcbY have a methyltransferase active site and their folding patterns respectively resemble the Streptococcus mutans proteins Smu472 and Smu776. Mass spectrometric analyses of 23S rRNAs showed that the N-terminal region of YcbY and Smu472 are functionally equivalent and add the m2G2445 modification, while the C-terminal region of YcbY is responsible for the m7G2069 methylation on the opposite side of the same helix (H74). Smu776 does not target G2069, and this nucleotide remains unmodified in Gram-positive rRNAs. The E.coli YcbY enzyme is the first example of a methyltransferase catalyzing two mechanistically different types of RNA modification, and has been renamed as the Ribosomal large subunit methyltransferase, RlmKL. Our structural and functional data provide insights into how this bifunctional enzyme evolved.
INTRODUCTION
Ribosomal RNAs are subjected to numerous post-transcriptional modifications in all three domains of life. Most of the modification sites are located close to functional centers, particularly the peptidyltransferase loop of the large ribosomal subunit and the decoding region of the small subunit (1,2). Collectively, these modifications are known to be important for several processes including ribosome maturation and fine-tuning of reactions during protein synthesis (3–5), although individually the functions of the modifications are still poorly understood.
The main types of modification in rRNAs are base methylation, 2′-O-ribose methylation and uridine isomeration (pseudouridylation). In Archaea and Eukaryota, most rRNA modifications are uridine isomerations and 2′-O-methylations, and the specificities of the modification enzymes depends on their being guided to their target nucleotides by small RNAs (6,7). In Bacteria, base methylations are the most frequent type of modification (8) and are added by methyltransferases that belong to a superfamily of enzymes characterized by their relatively well-conserved S-adenosyl-l-methionine (SAM)-binding domain (9). All the bacterial methyltransferases in this family find their rRNA target nucleotides without the help of guide RNAs (10). Consequently, the bacterial enzymes contain a variety of auxiliary domains such as PUA, THUMP, or TRAM (11–15) to recognize and differentiate between the different rRNA sequences and structures.
Ribosomal RNA modifications have been most extensively studied in the model Gram-negative bacterium Escherichia coli (8) where there are in total 36 modified nucleotides, 25 of which are located in 23S rRNA. With only a few exceptions, the enzymes responsible for adding these modifications have been identified (10,16,17) and representative structures have been solved for many of these enzymes or their orthologs. In E. coli, most of the enzymes have single nucleotide specificities, with the exceptions of the two pseudouridine synthases RluC and RluD that have multiple targets (18), and the methyltransferase RsmA that methylates at two adjacent adenosines in 16S rRNA (19,20).
It was previously shown that the E.coli gene ycbY encodes the methyltransferase specific for the m2G2445 modification (21). Nucleotide G2445 is located in 23S rRNA helix 74 adjacent to the highly conserved and heavily modified peptidyltransferase centre (22). The structural characteristics of YcbY have remained unsolved, primarily due to its unusual length of over 700 amino acids and the complexity of its structure with at least four different domains. YcbY is thus distinct from most other RNA modification enzymes, which are commonly between 200 and 400 amino acids with a single methyltransferase catalytic site and not more than two auxiliary domains.
Here, we present the crystal structures of E.coli YcbY and the two Streptococcus mutans proteins Smu472 and Smu776, which are respectively Gram-positive orthologs of the N- and C-halves of YcbY. The enzyme structures are viewed in terms of their modification functions which are defined by a combination of molecular genetics studies and rRNA analyses by MALDI mass spectrometry. We show that whereas E. coli YcbY is a bifunctional rRNA methyltransferase catalyzing the m2G2445 and m7G2069 modifications located on opposite sides of helix 74 (Figure 1), only the former modification is added in the S. mutans 23 S rRNA. The evolutionary implications of the apparent emergence of YcbY from the fusion of Smu472 and Smu776 orthologs are considered.
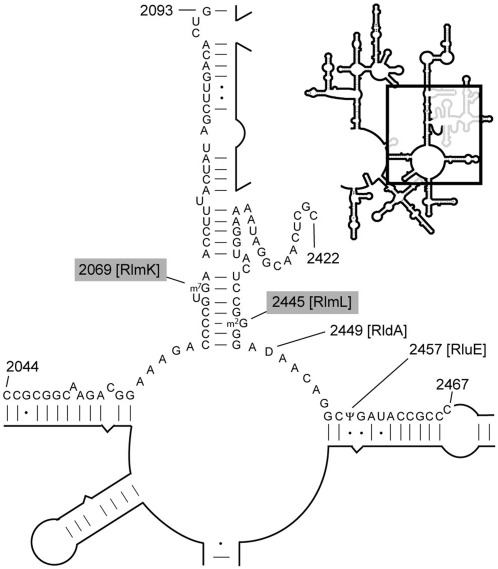
Figure 1.
Methylation sites in the peptidyl transferase region of E. coli 23S rRNA. The enlarged secondary structure (boxed in the outline) shows the rRNA region including the peptidyl transferase centre (55–56). The modifications at m7G2069 and m2G2445 are added by methyltransferases with RlmK and RlmL functions, respectively. The sequences from C2044 to G2093 and C2420 to C2467 containing these modified nucleotides were isolated by hybridization for MALDI–MS analyses.
MATERIALS AND METHODS
In silico analysis of YcbY and related proteins
Orthologs of YcbY were identified from a search of the KEGG database (23) (http://www.genome.jp/kegg) using the E. coli sequence (accession code b0948) as the query. Candidate sequences were fed into the protein–protein interaction database StringDB (24) (http://string.embl.de) to detect possible functional relationships. Evidence of gene fusion, as well as cooccurrence and coexpression of orthologs was identified using a high confidence cutoff level of 0.8. Bacterial species with YcbY orthologs possessed either one ORF with similar size and sequence to the E. coli protein or two separate ORFs that correspond to the N- and C-terminal of YcbY (YcbY-N and YcbY-C). These are exemplified here by the S. mutans proteins Smu472 (accession code SMU_472) and Smu776 (SMU_776). Multiple sequence alignments of all YcbY orthologs were made using the CLUSTALX software program (25), structural comparisons were carried out using DALI, and conserved domains were found in the COG database (26). The likelihood of gene fusion having occurred was tested in the fusion protein database FusionDB (27). Alignment graphics were generated using ESPript (28).
Cloning, purification, crystal preparation and structure determination
Detailed procedures for the crystallization of Smu472 and YcbY have been published previously (29,30). The cloning, purification and crystal screening procedures were similar for Smu776, although the crystallization conditions differed (Supplementary Table S1). The genes of all three proteins were amplified by PCR from the respective bacterial genomes and cloned into the plasmid vector pET28a. Recombinant proteins expressed from this vector in the E. coli strain BL21(DE3) were equipped with a 6 × His tag at their N-termini. Proteins were purified by passing through Ni2+-chelating columns followed by size exclusion chromatography. Protein crystals were obtained at 16°C using the hanging-drop or sitting-drop vapor-diffusion method. Addition of the SAM cofactor or the SAH product improved the reproduction and quality of the crystals and was essential in the case of YcbY. The detergent molecule, n-octanoylsucrose, was additionally required to produce YcbY crystals giving high-quality diffraction patterns. Glycerol was added as cryoprotectant for crystals of all three proteins; further details of protein crystallization and structure determination are given in Supplementary Table S1. Solved structures have been deposited at the Protein Data Bank under the files codes 2B78, 3LDG, 3LDF, 3V8V and 3V97.
Preparation of the ycbY knock-out strain and complementation
We made our own version of the E. coli ycbY knock-out strain in-house. This was done after PCR analysis of the Keio knock-out strain JW0931 (Keio Collection, obtained through the Coli Genetic Stock Center CGSC, Yale) showed that the ycbY gene had not been cleanly removed. Briefly, we excised ycbY from the chromosome of E. coli strain BW25113 using the procedure of Datsenko and Wanner (31). The ycbY gene was replaced with a kanamycin cassette flanked by FRT sites (Flp Recombination Target) in a one step site-specific recombination event, creating an in-frame deletion of the entire ycbY gene (32). The structure of the relevant chromosomal region of the BW25113 ΔycbY strain was tested by PCR and showed that the ycbY ORF had been removed without disturbing neighboring sequences. This ΔycbY strain made in-house was used in all experiments reported here. S. mutans wild-type strain UA140 was kindly provided by Prof. Li-Hong Guo at The Dental Hospital Beijing, China. The S. mutans genes were knocked-out using an in-frame deletion method as previously described (33).
For gene complementation experiments, the full-length ycbY gene was amplified from E. coli chromosomal DNA and placed at the SfiI restriction site under the control of the lac promoter in the expression vector pCA24N (34). The ycbY gene regions corresponding to the N-terminal methyltransferase domain (amino acids 1–383, YcbY-N) and the C-terminal methyltransferase domain (amino acids 390–702, YcbY-C) were cloned independently into the pCA24N vector, as were the smu472 and smu776 genes from S. mutans. The structures of all the inserts were confirmed by Sanger sequencing, and the plasmids were used to transform our E. coli BW25113 ΔycbY strain by standard methods (35).
Growth of strains for rRNA purification
Escherichia coli was grown with aeration at 37°C in 200 ml of LB medium (35) with kanamycin at 20 mg/l for the ΔycbY strain and/or chloramphenicol at 20 mg/l for strains transformed with the vector pCA24N and its derivatives. Plasmid-encoded genes were induced by adding IPTG to 1 mM when cultures reached an optical density A600 of 0.6; cells were kept at 37°C for an additional 3 h. The expression of recombinant methyltransferases was checked by SDS-PAGE (35). The wild-type E. coli strain BW25113 was grown without addition of any antibiotic, and was harvest by centrifugation upon reaching an optical density A600 of 0.6. Cells were lyzed, and rRNA was extracted and purified as previously described (36).
S. mutans strains were grown under anaerobic condition at 37°C in 100 ml of brain–heart infusion broth (Oxoid) with light shaking to an optical density A600 of 0.6. Cells were harvested by centrifugation and washed in 0.9% NaCl. Cell pellets were resuspended in 20 mM sodium acetate pH 4.5, 1 mM EDTA and 0.5% SDS, and the cells were lyzed using a FastPrep instrument (QBIOgen). After centrifugation, proteins were removed from the supernatant with phenol/chloroform and RNA was precipitated with ethanol/isopropanol in 300 mM sodium acetate pH 4.5.
Analyses of rRNA modifications
For analysis of rRNA by MALDI–MS, the 23S rRNA sequences from nucleotides C2044 to G2095 and C2422 to C2467 were isolated by hybridization (37) to complementary deoxynucleotides (Supplementary Table S2). Briefly, 100 pmol of total rRNA was heated in presence of 500 pmol of deoxynucleotide at 80°C for 5 min, followed by slow cooling to 45°C over 2 h. Unprotected rRNA regions were removed with nucleases and the hybridized rRNA sequences were isolated by gel electrophoresis. The isolated rRNA fragments were digested by RNase A or RNase T1 and analyzed by MALDI–MS (Voyager Elite, Perspective Biosystems) recording in reflector and positive ion mode (38). Further analyses by tandem mass spectrometry were carried out on all modified fragments, in some cases after the fragment had been equilibrated in heavy water (H2O containing > 95% 18O) to distinguish between y and c ions (39). Spectra were recorded in positive ion mode on a MicroMass MALDI Q-TOF Ultima mass spectrometer (40).
In the case of nucleotide G2069 analyses, sequences were additionally scanned by reverse transcriptase primer extension (41) after pretreating with sodium borohydride and aniline to cleave the rRNA chain at m7G modifications (42,43).
RESULTS AND DISCUSSION
Bioinformatics indicates that E. coli YcbY is a fusion of two separate methyltransferases
A search of database sequences showed that the YcbY protein and its orthologs are present in most Bacteria. In 943 sequenced bacterial species, almost all full-length orthologs of YcbY are found in members of the Gammaproteobacteria class of Gram-negative bacteria. Specifically, 213 out of 238 genomes in this group, which includes E. coli, have orthologs of the YcbY fusion (Supplementary Figure S1A). Outside this group, only three Gram-negative and three Gram-positive bacteria have YcbY as a fusion protein. Alphaproteobacteria do not possess orthologs of YcbY, while β- and δ-proteobacteria have a gene arrangement similar to the Gram-positive Firmicutes branch of Bacilli.
In the Bacilli, homologous regions of YcbY are evident as proteins encoded on two separate genes (Supplementary Figure S1B). In Bacillus subtilis, the model organism from this branch, ypsC and ywbD are orthologs of the S. mutans genes smu472 and smu776 and encode proteins that respectively show high degrees of similarity to YcbY-N and YcbY-C. Most bacteria in this branch (115 out of 135) possess both the smu472 and smu776 orthologs, and these genes tend to be located on widely separated regions of the genome.
Comparative searches within Clusters of Orthologous Groups (26) showed that the N-terminal half of YcbY (YcbY-N) groups together with Smu472 in COG0116, while the C-terminal half of YcbY (YcbY-C) segregates together with Smu776 in a separate cluster, COG1092. The N-terminal 70 residues of Smu776 constitute a PUA domain, which is absent from the corresponding location in the YcbY sequence. Both COG0116 and COG1092 have previously been annotated as methyltransferase families (26), and the sequence conservation of the corresponding regions in the full-length E. coli YcbY protein indicate two independent catalytic sites with methyltransferase activity.
Structures of Smu472 and Smu776 from S. mutans
Our structural study of YcbY-related proteins began with orthologs from S. mutans (44). The two S. mutans proteins Smu472 and Smu776 are about 400 amino acids in length and each possesses one distinctive methyltransferase domain plus auxiliary domains. The structure of Smu472 was solved using single-wavelength anomalous dispersion (SAD). The crystal belongs to space group P212121 with one molecule in the asymmetric unit, and diffracted to 1.97 Å. The N-terminus of Smu472 consists of a THUMP domain (12) followed by an FLD domain, which together appear to form the RNA-binding module. The C-terminus of Smu472 has a typical SAM-dependent methyltransferase fold (9) where residues 219–254 between αA and β2 of the methyltransferase domain make contact with the FLD domain. Clear density for SAH could be seen in the catalytic centre of the methyltransferase domain (Figure 2A).
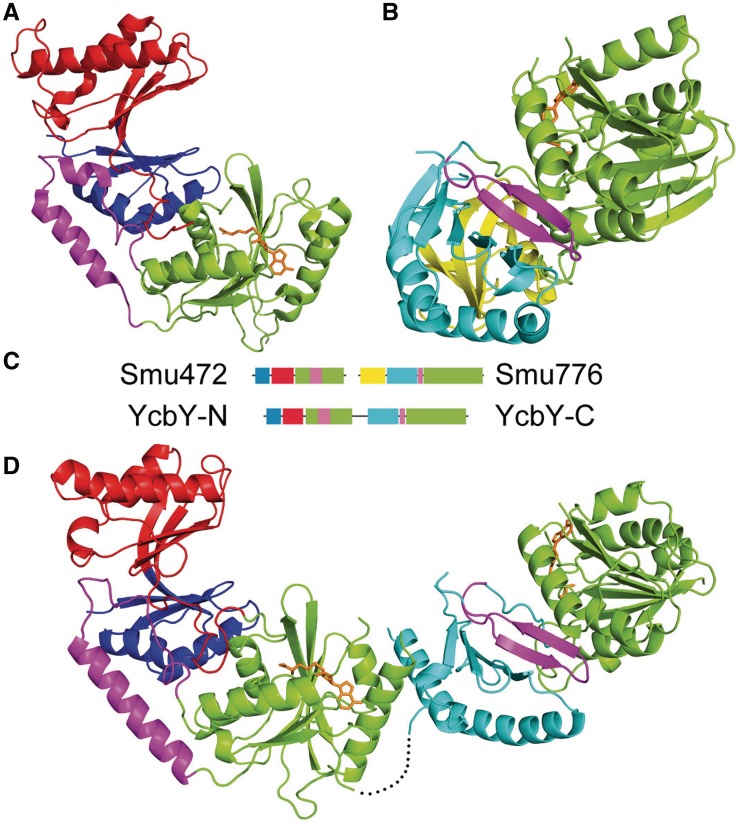
Figure 2.
Structures of Smu472, Smu776 and YcbY depicted as secondary structure cartoons. (A) Domain organization of the Smu472 monomer: THUMP (blue), FLD (red) and MTase I (green) interrupted by a short α-helical region (magenta). (B) Domain organization of the Smu776 monomer: PUA (yellow), EEHEE (cyan), β-hairpin (magenta), and MTase II (green). (C) Primary structure alignment illustrating the relative positions of the conserved domains with the insertion (magenta) that interrupts MTase I in Smu472 and YcbY-N and the short β-hairpin (magenta) that separates the EEHEE and MTase II domains in Smu776 and YcbY-C. (D) Domain organization of YcbY monomer with SAH in the two active sites; the domain colors are as above. The black dotted line represents the flexible loop (residues 383–392) linking the two halves of YcbY; the density of this sequence was missing in the final map. SAH molecules are shown in sticks (orange); all structural figures were generated using PyMOL (http://www.pymol.org) (57).
The Smu776 protein also formed crystals with one monomer per asymmetric unit, and its structure was solved using single isomorphous replacement (SIR). The Smu776 structure was resolved at 2.20 Å, and consists of an N-terminal PUA domain, a central EEHEE domain, and a C-terminal methyltransferase domain that contains a single SAH molecule (Figure 2B). The PUA domain is a small RNA-binding region found in many other RNA modification enzymes (45) as well as two functionally uncharacterized Smu776 orthologs in the PDB database (3K0B and 3LDU). The only other characterized structure with similar PUA and EEHEE domains is that of YccW/RlmI (15), which is also a member of the COG1092 methyltransferase family and is responsible for the E. coli 23S RNA nucleotide m5C1962 methylation (11). We predict that a β-hairpin between the EEHEE domain and the methyltransferase domain provides a scaffold for an aromatic residue that stacks upon and stabilizes the target base.
Structure of E. coli YcbY reveals two active sites
The E. coli YcbY crystals belonged to space group P21 with two molecules in one asymmetric unit, and diffracted to 2.20 and 2.60 Å in the SAH- and SAM-binding form, respectively. The YcbY structure was solved using the structures of Smu472 and Smu776 (minus the PUA domain in the latter) as starting search models. The 702 amino acid residues of YcbY are organized into an elongated structure with several domains that are essentially a combination of the corresponding regions in Smu472 and Smu776. Thus, the YcbY structure is arranged as an N-terminal THUMP domain followed by an FLD domain, a methyltransferase domain (MTase domain I), an EEHEE domain and then a second methyltransferase domain (MTase domain II) (Figure 2).
All the YcbY domains have high structural similarity with their Smu472 and Smu776 counterparts (Figure 3). The root mean square deviation (RMSD) between the N-terminal half (YcbY-N) and Smu472 is 2.07 Å, comparing 300 aligned Cα atoms; while the RMSD between the C-terminal half (YcbY-C) and Smu776 (minus its PUA domain) is 2.28 Å comparing 210 aligned Cα atoms. In the solved structure of YcbY, obvious densities for SAH molecules were seen within both MTase domains. The residues around the active site are conserved both in sequence and in structure (Figure 3C and D) supporting the contention from the bioinformatics analysis that YcbY has two catalytic sites. The N- and C-terminal halves of YcbY are linked by a highly flexible loop (residues 383–392) forming a hinge-like structure that would enable the distance between the two catalytic sites to be adjusted (Figure 2D).
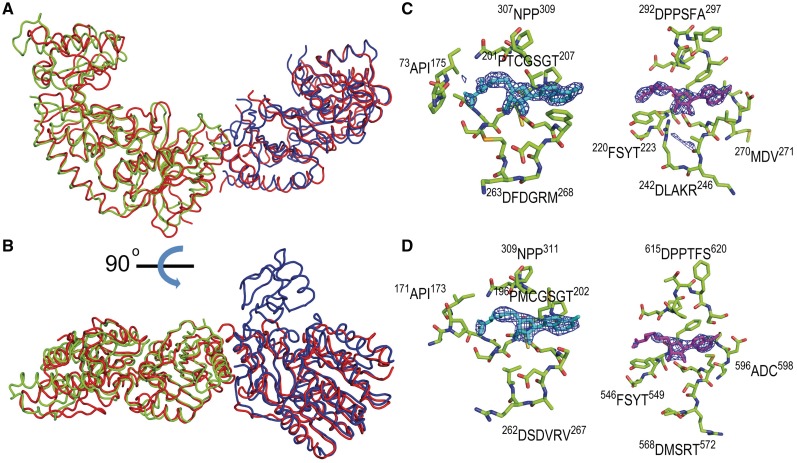
Figure 3.
Superimposition of the Smu472, Smu776 and YcbY structures. (A) The folds of Smu472 (green) and Smu776 (blue) have high structural similarity to the corresponding halves of YcbY (red). (B) Image rotated 90 degrees, showing the PUA domain of Smu776, which is missing in YcbY. (C) Stick representation of SAH molecules in the active sites of Smu472 (light blue, left) and Smu776 (magenta, right) and (D) in the corresponding sites of YcbY-N (left) and YcbY-C (right); conserved amino acid residues are shown with carbons in green, nitrogens in blue, oxygens in red and sulfurs in yellow. |Fo–Fc| electron density map was calculated with the protein model excluding ligand SAH molecules and contoured at 3.0 σ.
m2G2445 modification by YcbY is stoichiometric
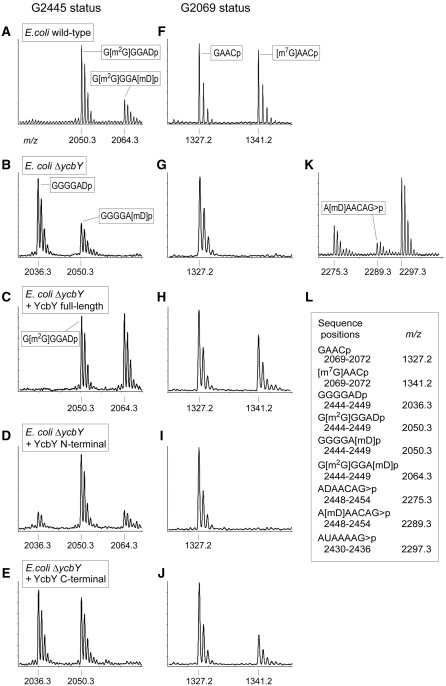
The 23S rRNA sequence C2422 to C2467 was isolated from the E. coli strains and was digested with RNase A to give a series of fragments that included GGGGADp (2444–2449) containing nucleotide G2445. This sequence also contains the dihydrouridine D2449, a modification that is catalyzed by the as-yet unidentified enzyme RldA (10) and adds 2 Da of mass. The D2449 modification together with the methyl group at m2G2445 gives the G[m2G]GGADp fragment a theoretical m/z value of 2050, and this fits with the major MALDI–MS peak from the wild-type rRNA (Figure 4A). Loss of YcbY gave rise to a new peak at m/z 2036, indicating that a methyl group was missing (Figure 4B), and this was then recovered by expressing an active copy of ycbY from a plasmid in the null mutant (Figure 4C). Tandem MS analyses showed that this methyl group was located on the base of nucleotide G2445 (Supplementary Figure S2) and that methylation was stoichiometric.
Figure 4.
MALDI–MS analyses of nucleotides m7G2069 and m2G2445 in E. coli 23S rRNA. The methylation status of nucleotide G2445 was assessed in the RNase A fragment GGGGADp (A–E). The m2G2445 modification in rRNA from the wild-type strain (A) is lost in the rRNA from the ycbY-null strain (B), and can be recovered by expression of an active copy of the entire ycbY gene (C) or the first part of the gene encoding the N-terminal half of YcbY (D). The small peak at m/z 2050 in (B) is due to a substoichiometric amount of a second methyl group attached to D2449 (also in the m/z 2064 peak), and was analyzed further in the RNase T1 fragment ADAACAG>p (K). The boxed sequences were confirmed by tandem MS. The methylation status of nucleotide G2069 was assessed in the RNase A fragment GAACp (F to J). The m7G2069 modification is substoichiometric in rRNA from the wild-type strain (F) and is lost in the rRNA from the ycbY-null strain (G). The modification was recovered by expression of an active copy of the entire ycbY gene (H) or the 3′-gene portion encoding the C-terminal half of YcbY (J). The N-terminal half of YcbY had no effect on methylation at G2069 (I), nor did the C-terminal half of YcbY affect methylation at G2445 (E). Sequences and theoretical mass/charges (m/z) of all the RNase fragments are shown in (L) and matched the empirically measured values to within 0.1 Da; >p designates a cyclic 2′–3′-phosphate.
These data clearly confirmed the m2G2445 modification function of YcbY previously reported by Lesnyak and coworkers (21). However, the MALDI–MS spectra were more complex than we had anticipated. The wild-type spectrum contains a minor additional peak at m/z 2064 (Figure 4A), and there was a small peak at m/z 2050 after removal of ycbY (Figure 4B). These minor peaks were due to a second methyl group (Figure 4K), the location of which was pinpointed to D2449 by tandem MS analysis (Supplementary Figure S3). In all the E. coli wild-type, knock-out and complemented strains, the methylation at nucleotide D2449 was consistently substoichiometric. No methylation has previously been reported at this position, and the enzyme catalyzing this reaction is unknown. We can, however, rule out any involvement of YcbY because the D2449 methylation remains evident in the null mutant (the m/z 2050 peak, Figure 4B).
YcbY adds the m7G2069 modification substoichiometrically
The second active site of YcbY indicated by the structural studies had a somewhat limited number of potential targets. At the onset of this study, the enzymes responsible for 25 of the 27 E. coli rRNA methylations had already been identified (10,16–17), and only m6A2030 and m7G2069 in 23S rRNA still remained to be linked to their respective enzymes. These had provisionally been dubbed RlmJ and RlmK, respectively (46). Both sites are relatively close to nucleotide G2445, although the secondary structural fold of 23S rRNA brings G2069 to within 11 Å on the other side of helix 74 (Figure 1).
The E. coli 23S rRNA sequence from C2044 to G2093 (Figure 1) was isolated and digested with RNase A giving rise to a unique tetramer, GAACp, containing nucleotide G2069. The tetramer from the wild-type rRNA produced peaks at m/z 1327 and m/z 1341 (Figure 4F) with the larger peak containing a methyl group attached to the base of nucleotide G2069 (Supplementary Figure S4). Reverse transcriptase primer extension on borohydride/aniline-treated rRNAs showed that the methyl group was at the N7-position of the guanine base (not shown). The methylation was completely missing in the rRNA from our ycbY-null strain (Figure 4G), but was recovered upon complementation of this strain with an active copy of ycbY (Figure 4H). Taken together, these observations conclusively showed that YcbY is responsible for addition of the m7G2069 modification.
The appreciable proportion of the GAACp fragment at m/z 1327 peak in rRNA from both the wild-type and the complemented strains indicates that methylation at G2069 was substoichiometric, and thus YcbY carries out this reaction less efficiently than the reaction at G2445. We note that the m7G2069 modification was completely missing in the Keio strain with truncated ycbY. This is consistent with the findings of the Suzuki group, who have also defined the m7G2069 modification function of YcbY in an independent study using both the Keio strain and a strain with a larger genetic deletion (47).
Dissecting the YcbY methylation functions
The two halves of the ycbY gene encoding the N-terminal methyltransferase domain (YcbY-N, amino acids 1–383) and C-terminal methyltransferase domain (YcbY-C, amino acids 390–702) were cloned into plasmid expression vectors and used independently to transform our ycbY-null strain. Analyses of the rRNAs from these strains using MALDI–MS showed that enzymatic activity was retained in both the recombinant halves of YcbY. Specifically, the YcbY-N construct restored the methylation activity at m2G2445 (Figure 4D), but not at m7G2069 (Figure 4I). Conversely, the YcbY-C construct partially restored m7G2069 (Figure 4J), but showed no methylation activity at m2G2445 (Figure 4E). Ambiguities in the spectra due to the additional methyl group at D2449 were resolved by tandem MS analyses and verified the methylation status of G2445 (e.g. Figures 4 and Supplementary Figure S2).
Functions of the Smu472 and Smu776 orthologs
Ribosomal RNAs were extracted from S. mutans cells, and were analyzed for modifications using MALDI–MS. Nucleotides G2069 and G2445 are highly conserved in bacterial 23S rRNAs (22), and the sequence of the RNase A fragment containing G2445 (GGGGAUp) is the same in E. coli and S. mutans rRNAs (Supplementary Figure S5). The S. mutans fragment was recorded at m/z 2048 (2 Da less than the E. coli fragment) indicating that nucleotide 2449 is a uridine and not dihydrouridine. There were no variants of higher mass, and thus the S. mutans sequence contains only one methyl group, which was localized to the base of G2445 and was shown to be stoichiometric (Supplementary Figure S5A). In a S. mutans smu472-null strain, the methylation was lost (Supplementary Figure S5B). Consistent with this, the m2G2445 methylation was recovered in the E. coli ycbY-null strain after transforming with a plasmid expressing an active copy of the S. mutans smu472 gene (Supplementary Figure S5C). Taken together, these results clearly demonstrate that the S. mutans Smu472 protein is an m2G2445 methyltransferase, and is thus functionally equivalent to the N-terminal half of the E. coli YcbY enzyme.
The sequence around nucleotide G2069 varies slightly in the bacterial species, and this nucleotide is recovered in the pentamer sequence GGAGCp after RNase A digestion of the S. mutans rRNA. This fragment flew at m/z 1688 without larger variants indicating that the sequence contained no modifications (Supplementary Figure S5E). Predictably, rRNAs from the smu472- and smu776-null mutant strains of S. mutans produced identical unmodified fragments (Supplementary Figure S5F). Moreover, expression of Smu472 or Smu776 in the E. coli ycbY-null strain failed to recover the G2069 methylation (Supplementary Figure S5G). Thus, despite the structural similarity, the function of the Smu776 protein is clearly distinct from that of the C-terminal half of the E. coli YcbY methyltransferase.
Recognition of dual rRNA targets by YcbY
In the assembled 50S structure, nucleotides G2069 and G2445 are folded within the subunit structure and appear inaccessible to the YcbY methyltransferase (48,49). Consistent with this, the methyltransferase has been shown to use naked 23S rRNA rather than assembled subunits as the substrate for both G2069 (47) and G2445 methylation (21,47). The distance spanning the two nucleotide targets through the minor groove of helix 74 is just over 10 Å, and thus well within the 44 Å distance between the active sites in the YcbY structure (Figure 5). The distance between the target nucleotides could be altered by opening of H74 through the helicase activity of the YcbY-C domain (47) and by flipping out the bases from a stacked conformation into the enzyme catalytic sites, as has been seen for other methyltransferases (50,51). Such changes in the rRNA substrate might be comfortably accommodated by domain movement around the flexible mid-section of YcbY (Supplementary Figure S7), making it feasible that recognition and modification of G2069 and G2445 occur through a single enzyme–rRNA-binding event.
Figure 5.
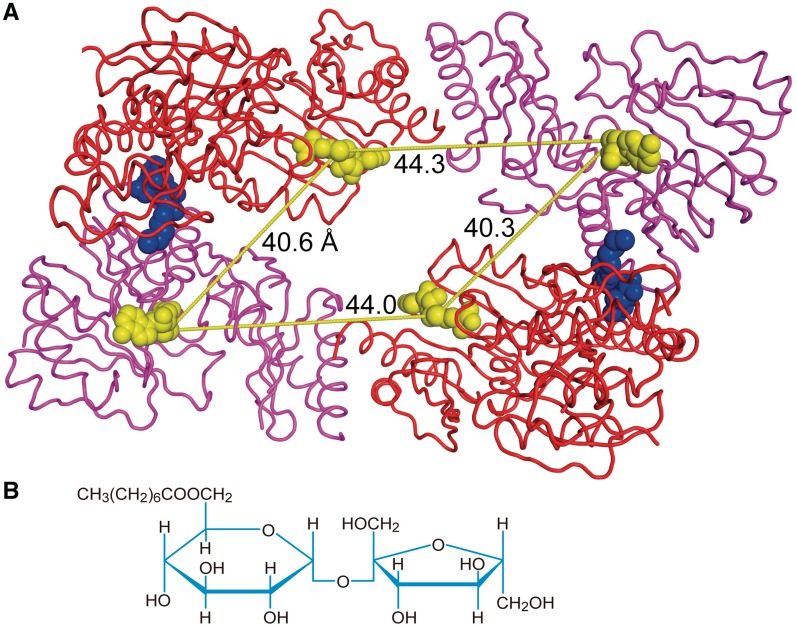
Crystal packing of YcbY as dimer. (A) The YcbY dimer in the asymmetric unit shown in ribbon mode. The upper N-terminal (red) and C-terminal halves (magenta) represent one monomer unit and interact in the crystal to the second (lower) monomer through binding of two n-octanoylsucrose detergent molecules (blue). SAH molecules (yellow) are shown in space filling mode. The intramolecular distances between the active sites (∼44 Å) are similar to the intermolecular distance between the sites in the dimer (∼40 Å). The consensus of data suggests that the monomeric form represents the true physiological state of YcbY. (B) Structure of the synthetic detergent n-octanoylsucrose.
Flexible altering the distance between the catalytic sites would necessitate that YcbY functions as a monomer. Several lines of evidence indicate that YcbY is a monomer in solution, despite the head-to-tail arrangement of two YcbY molecules in the asymmetric unit, with comparable inter- and intramolecular distances between the active sites (Figure 5). The dimer interface is formed by phylogenetically variable residues together with the detergent n-octanoylsucrose (Supplementary Figure S6), and only 10% of the total surface area is buried within the dimer, which would presumably be too little to maintain a stable interaction in solution. This was supported by gel filtration, where YcbY eluted at a position predicted for an elongated monomer, and also by analytical ultracentrifugation under physiological buffer conditions where molecular weight estimates of YcbY were close to those expected for a monomer (data not shown).
Evolutionary implications of rRNA recognition
The COG0116 family members Smu472 and YcbY-N possess N-terminal THUMP and FLD domains that are probably the RNA-binding modules. Similar domain arrangement are seen in a thiouridine synthase (PDB ID 2C5S) (52) and a cytidine deaminase (PDB ID 3G8Q) (53), both of which are RNA modification enzymes. The THUMP and FLD domains of YcbY form a continuous channel of β-sheet that might direct the RNA substrate into the catalytic site (Supplementary Figure S7), and these features are conserved on the surfaces of Smu472 and YcbY-N (Supplementary Figure S8). The m2G2445 methyltransferase function of these orthologs is also highly conserved, and loss of this modification has been linked with resistance to the peptidyl transferase inhibitor linezolid (54).
The relationship between Smu776, YcbY-C and the other members of the COG1092 family is more complicated. Despite similarities in their domain architecture and common SAM-binding motifs, COG1092 members are divided into five branches (15). The first branch is exemplified by the m5C methyltransferase YccW (RlmI) protein and the fourth branch contains Smu776 and YcbY-C, while the functions of members of the other branches of COG1092 remain unknown. The Smu776 surface displays several conserved regions that can be linked with functions in RNA recognition, cofactor binding and catalysis of methyl transfer (Supplementary Figure S8). In contrast to Smu472, the central cleft of Smu776 is too narrow to accommodate double strand RNA. The present evidence indicates that Smu776 is indeed an RNA methyltransferase that modifies another, yet-to-be-determined nucleotide target. The extra PUA region of Smu776, which has been lost in YcbY (Figure 3), is undoubtedly central to target recognition.
The evolutionary route that led to the present-day YcbY enzyme still remains to be mapped. Clearly, fusion occurred between an upstream COG0116 sequence that retained its m2G2445 methyltransferase function and a downstream COG1092 sequence, but the main unresolved question is whether this COG1092 sequence already functioned as an m7G2069 methyltransferase before fusion occurred. The report of separate genes in the Betaproteobacterium Neisseria meningitidis that function identically to YcbY-N and YcbY-C (47) might suggest that this was indeed the case. However, another scenario is suggested by the extensive array of separate orthologs in the Gram-positive Firmicutes (Supplementary Figure S1). The S. mutans COG1092 ortholog does not methylate nucleotide G2069 (Supplementary Figure S5) and there is no m7G2069 modification in any of the Gram-positive rRNAs that we have studied so far (unpublished data on bacteria including B. subtilis, Mycobacterium smegmatis and Streptomyces coelicolor). Feasibly, the COG1092 sequence of YcbY originally had another function, possibly as an m5C methyltransferase, and altered its specificity to m7G catalysis after the fusion event and being guided to nucleotide 2069 by the interaction of the COG0116 sequence at nucleotide G2445 on the other side of helix 74.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8, Supplementary Tables 1–2, Supplementary Reference [58].
FUNDING
National Basic Research Program of China 973 (No. 2011CB911103 to X.D.S.); the Danish Research Agency (FNU-rammebevillinger 09-064292/10-084554 to S.D.); the Nucleic Acid Center of the Danish Grundforskningsfond (to S.D.). Funding for open access charge: Danish Research Agency (FNU-rammebevillinger 10-084554).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Finn Kirpekar, Simon Rose and Raymond P. Cox are thanked for invaluable help with MS studies, strain analysis and culturing of Streptococcus mutans. Nelson Enrique Arenas Suarèz is thanked for comments on the manuscript.
REFERENCES
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grosjean, H. (2009) DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Biosciences, Austin, TX.
- 4.Green R, Noller HF. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry. 1999;38:1772–1779. doi: 10.1021/bi982246a. [DOI] [PubMed] [Google Scholar]
- 5.Lapeyre B. Fine-Tuning of RNA Functions by Modification and Editing. NY: Springer Verlag; 2005. Conserved ribosomal RNA modification and their putative roles in ribosome biogenesis and translation; pp. 85–91. [Google Scholar]
- 6.Dennis PP, Omer A, Lowe T. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 7.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 10.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YgdE is the 2'-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 2009;72:1147–1158. doi: 10.1111/j.1365-2958.2009.06709.x. [DOI] [PubMed] [Google Scholar]
- 11.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Koonin EV. THUMP–a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 13.Lee TT, Agarwalla S, Stroud RM. Crystal structure of RumA, an iron-sulfur cluster containing E. coli ribosomal RNA 5-methyluridine methyltransferase. Structure. 2004;12:397–407. doi: 10.1016/j.str.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Hallberg BM, Ericsson UB, Johnson KA, Andersen NM, Douthwaite S, Nordlund P, Beuscher AE, Erlandsen H. The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J. Mol. Biol. 2006;360:774–787. doi: 10.1016/j.jmb.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 15.Sunita S, Tkaczuk KL, Purta E, Kasprzak JM, Douthwaite S, Bujnicki JM, Sivaraman J. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J. Mol. Biol. 2008;383:652–666. doi: 10.1016/j.jmb.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 16.Basturea GN, Dague DR, Deutscher MP, Rudd KE. YhiQ is RsmJ, the methyltransferase responsible for methylation of G1516 in 16S rRNA of E. coli. J. Mol. Biol. 2012;415:16–21. doi: 10.1016/j.jmb.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofengand J, Del Campo M. Modified nucleotides of Escherichia coli ribosomal RNA. In: Böck A, Curtis R, Kaper JB, Neidhardt T, Nyström T, Squires C, editors. EcoSal - Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2004. [Google Scholar]
- 19.Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli. Nature. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 20.Poldermans B, Roza L, Van Knippenberg PH. Studies on the function of two adjacent N6, N6-dimethyladenosines near the 3'-end of 16S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30S interactions. J. Biol. Chem. 1979;254:9094. [PubMed] [Google Scholar]
- 21.Lesnyak DV, Sergiev PV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m2G methyltransferase: I. The ycbY gene encodes a methyltransferase specific for G2445 of the 23S rRNA. J. Mol. Biol. 2006;364:20–25. doi: 10.1016/j.jmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 27.Suhre K, Claverie JM. FusionDB: a database for in-depth analysis of prokaryotic gene fusion events. Nucleic Acids Res. 2004;32:D273. doi: 10.1093/nar/gkh053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouet P, Courcelle E, Stuart DI. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 29.Wang KT, Li LF, Zhang XY, Liang YH, Wei SC, Su XD. Preparation, crystallization and preliminary X-ray crystallographic analysis of Smu.776 from caries pathogen Streptococcus mutans. Progr. Biochem. Biophys. 2007;34:176–179. [Google Scholar]
- 30.Wang KT, Ma L, Nan J, Su XD, Li L. Purification, crystallization and preliminary X-ray crystallographic analysis of 23S RNA m(2)G2445 methyltransferase RlmL from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:1484–1486. doi: 10.1107/S1744309110035074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006. 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt J, Tsang P, Zheng L, Shi W, Qi F. Construction of a counterselection-based in-frame deletion system for genetic studies of Streptococcus mutans. Oral Microbiol. Immunol. 2007;22:95–102. doi: 10.1111/j.1399-302X.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- 34.Saka K, Tadenuma M, Nakade S, Tanaka N, Sugawara H, Nishikawa K, Ichiyoshi N, Kitagawa M, Mori H, Ogasawara N, et al. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 2005;12:63–68. doi: 10.1093/dnares/12.1.63. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2001. [Google Scholar]
- 36.Desmolaize B, Rose S, Warrass R, Douthwaite S. A novel Erm monomethyltransferase in antibiotic-resistance isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 2011;80:184–194. doi: 10.1111/j.1365-2958.2011.07567.x. [DOI] [PubMed] [Google Scholar]
- 37.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirpekar F, Douthwaite S, Roepstorff P. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA. 2000;6:296–306. doi: 10.1017/s1355838200992148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 40.Kirpekar F, Krogh TN. RNA fragmentation studied in a matrix-assisted laser desorption/ionisation tandem quadrupole/orthogonal time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2001;15:8–14. doi: 10.1002/1097-0231(20010115)15:1<8::AID-RCM185>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 41.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 42.Douthwaite S, Garrett RA, Wagner R. Comparison of Escherichia coli tRNAPhe in the free state, in the ternary complex and in the ribosomal A and P sites by chemical probing. Eur. J. Biochem. 1983;131:261–269. doi: 10.1111/j.1432-1033.1983.tb07258.x. [DOI] [PubMed] [Google Scholar]
- 43.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su XD, Liang Y, Li L, Nan J, Bröstromer E, Liu P, Dong Y, Xian D. A large-scale, high-efficiency and low-cost platform for structural genomics studies. Acta Crystallogr. D: Biol. Crystallogr. 2006;62:843–851. doi: 10.1107/S0907444906024395. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Arellano I, Gallego J, Cervera J. The PUA domain - a structural and functional overview. FEBS J. 2007;274:4972–4984. doi: 10.1111/j.1742-4658.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 46.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Kimura S, Ikeuchi Y, Kitahara K, Sakaguchi Y, Suzuki T, Suzuki T. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 2012;40:4071–4085. doi: 10.1093/nar/gkr1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl Acad. Sci. USA. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 51.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl Acad. Sci. USA. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterman DG, Ortiz-Lombardía M, Fogg MJ, Koonin EV, Antson AA. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 53.Randau L, Stanley BJ, Kohlway A, Mechta S, Xiong Y, Soll D. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009;324:657–659. doi: 10.1126/science.1170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng J, Lupien A, Gingras H, Wasserscheid J, Dewar K, Légaré D, Ouellette M. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 2009;19:1214–1223. doi: 10.1101/gr.089342.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noller HF. Structure of ribosomal RNA. Ann. Rev. Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL Molecular Graphics System. 2002 Version 1.2r3pre, Schrödinger, LLC. [Google Scholar]
- 58.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.