Abstract
The symmetry of i-motif tetramers gives to cytidine-rich oligonucleotides the capacity to associate into supramolecular structures (sms). In order to determine how the tetramers are linked together in such structures, we have measured by gel filtration chromatography and NMR the formation and dissociation kinetics of sms built by oligonucleotides containing two short C stretches separated by a non-cytidine-base. We show that a stretch of only two cytidines either at the 3′- or 5′-end is long enough to link the tetramers into sms. The analysis of the properties of sms formed by oligonucleotides differing by the length of the oligo-C stretches, the sequence orientation and the nature of the non-C base provides a model of the junction connecting the tetramers in sms.
INTRODUCTION
The formation of hemiprotonated C•C+ pairs has been reported 50 years ago (1). At slightly acidic pH, the pairing of the neutral and protonated cytidines of C-rich oligonucleotides results in the formation of a four-stranded structure, the i-motif, including two parallel duplexes mutually intercalated in a head to tail orientation (2). I-motif structures reported by NMR (3) or X-ray (4) show that C•C+ pair intercalation and stacking produces a rigid core of long-lived base pairs. The symmetry of i-motif tetramers gives to C-rich oligonucleotides the self-associative properties allowing the formation of supramolecular structures (sms) with potential technological applications (5–7). This property has been first mentioned in a study reporting on the association of C7 oligonucleotides into i-motif wires (8). Using AFM microscopy, a recent publication has described the self-assembly of poly C into nanospheres (9). Oligonucleotides containing two cytidine stretches separated by a single non-cytidine base associate into sms (10). The model derived from this study shows that C7GC4 for example forms a building block including the cytidines of the longest C stretch associated into an i-motif tetramer and two overhanging shorter C stretches at each end available for self-association. The end symmetry gives to this structure the capacity to build up sms by successive block addition and by association of short assemblies. In order to characterize the number of C•C+ pairs required to link i-motif tetramers into sms and to determine how much the sms stability depends on the number of C•C+ pairs involved in the junction between tetramers, we have investigated the formation and dissociation of sms formed by oligonucleotides with cytidine tracts as sort as possible. We observe that oligonucleotides with a C2 tract at the 3′- or 5′-end can associate into sms. However in all the conditions tested, the sms are too large to be investigated by NMR. Nevertheless, the comparison of the properties of sms built by oligonucleotides differing by the length of the oligo-C stretches, by the sequence orientation and by the nature of the non-C residue gives indirect information about the conditions required for tetramer association. A model of the junction linking i-motif tetramers into sms is presented.
MATERIALS AND METHODS
Oligonucleotide synthesis and sample preparation
The oligonucleotides were synthesized and purified as previously described (11). The oligonucleotide concentrations were determined using the A260 values computed according to a nearest neighbor model (12).
Formation and dissociation rate measurements of sms
The formation of sms was always measured with samples initially melted at 100°C and rapidly cooled at the incubation temperature. To measure the sms dissociation kinetics, we used concentrated samples with a maximal sms proportion. The kinetics was initiated by diluting the samples in such a way as to allow complete sms dissociation at equilibrium. In order to avoid evaporation during incubation, the samples (50–200 ml) were always prepared in vials closed by airtight twisted caps with o-ring. They were incubated in a mastercycler Eppendhorf® PCR incubator
Gel filtration chromatography
The sample composition during sms association and dissociation was followed by gel filtration chromatography using GPC 100 and GPC 1000 columns (250 × 4.6 mm I.D.) provided by Eprogen®. Except otherwise stated, the chromatography were always performed at room temperature in 0.4 M NaCl, 10 mM Na–acetate and 10 mM Na–phosphate, pH 4.6. The volume of the injection loop was 25 µl. Systematic addition of thymidine at a final concentration of ∼5 µM to the injected samples provided a reference marker. The GPC-1000 column, whose permeation and exclusion volumes correspond to oligonucleotides containing 50 and 104 residues respectively, was calibrated as previously reported (10). The GPC-100 was calibrated with i-motif tetramers and dimers, with monomeric C-rich oligonucleotides, un-fractioned tRNA, CC and TC dinucleotides, and thymidine. The comparison of the calibration curves obtained using i-motif structures and C-rich non-structured monomers shows that the elution volume of i-motif structures is somewhat larger than that of the monomers of identical molecular weights (Supplementary Figure S1). This indicates that the hydrodynamic radii of i-motif structures are smaller than those of monomers containing the same nucleotide number. The molecular weights of i-motif multimers were therefore estimated accordingly to the calibration curves obtained with i-motif structures. When this was required, the multimer stoichiometry was unambiguously determined by measuring the equilibrium multimer and monomer concentrations versus the oligonucleotide concentration.
Electrophoresis
The sms were also analyzed by a native polyacrylamide (PA) electrophoresis run on 6% and 10% gels containing 20 mM Robinson–Britton buffer, pH 4.6. During the electrophoresis the gel temperature was kept constant to 14°C with a waterbath. The gels were stained by Stains-All (Sigma), prepared as a 0.01% solution in a 50% formamide.
NMR method
The NMR experiments were performed using a 500 MHz Varian Inova spectrometer with the jump and return sequence for water suppression (13). The magnetization transfer experiments performed to investigate the conformational exchange of the duplexes intercalated in the tetramers of C3TC2, C4TC2 and C3TC3 have been described (14).
Multimer dissociation constant
The dissociation constant of a multimer may be expressed as a function of αeq, the monomer equilibrium fraction of an oligonucleotide solution at concentration [M0] by: Kdis = s αeqs [M0]s−1/(1 − αeq) were s in the multimer stoichiometry. We will characterize the multimers stability by Fi, a parameter whose dimension is independent of the stoichiometry which is equal to the free monomer concentration for which αeq = 0.5. Fi is related to the multimer dissociation constant by: Fi = (Kdis/s)[1/(s−1)] and it will be designated as the reduced dissociation constant.
RESULTS
We will consider as an sms all the species formed by association of at least two i-motif tetramers. The oligonucleotides investigated are sorted into four groups according to their capacity to associate into sms.
i-motif tetramer that cannot associate into sms
The oligonucleotides of this group associate into fully intercalated tetramers independently of the salt concentration. The group includes oligonucleotides with a thymidine at the 5′- or 3′-end (TCn and CnT, n = 2 and 5 (2,11); C2TC2 (14) and C2TC2T and short homopolymers: Cn (n = 2 to 5). The chromatograms of 2–5 mM solutions of these oligonucleotides show the equilibrium between the monomer and tetrameric structures. As previously shown (15), the salt concentration influences the intercalation topology of Cn tetramers but the association of these sequences into sms was negligible in the chromatograms of C3 or C5 recorded in 0.05, 0.4 or 1 M NaCl solutions.
This is also the case of oligonucleotides with a CT- sequence at the 5′-end (CTC2 for example) or with a TC 3′-end as in C5TC. It is noteworthy that at the extreme concentration of 25 mM, the chromatogram of C2TC2 shows that ∼4% of the sample is associated in structures larger than tetramer.
C7 and C12 assembly into supramolecular structures
These two oligonucleotides have the same properties. A few minutes after melting at 100°C and fast cooling at 20°C, we observe that 1.5 mM C7 or C12 solutions become opalescent and precipitate. The chromatogram of a C7 solution displayed in Figure 1 shows two very weak components. According to the column calibration (Supplementary Figure S1), one is eluted as a tetramer; the other may be either the unstructured monomer, or/and an i-motif dimer formed by the association of two C7 hairpins. The same solution injected immediately after melting at 100°C shows an intense (monomer + dimer) peak. The comparison of both chromatograms establishes that a fraction of ∼95% of the sample is missing on the chromatogram of the non-melted sample and therefore that C7 associates into extremely large sms that are trapped on the column. It is remarkable that the sms of C7 are also trapped in a GPC-1000 column whose exclusion size corresponds to structures including about 104 nucleotides. The behavior of C7 depends on the NaCl concentration. We do not observe precipitation in a 1.5 mM C7 solution incubated in a buffer containing only 10 mM Na–acetate and 10 mM Na–phosphate, pH 4.6. In this buffer, the sms and tetramer fractions are 10% and 90% respectively at equilibrium, the sms half formation time is ∼30 min and the elution time of sms is centered on a position corresponding to the assembly of 20 tetramers.
Figure 1.
C7 association into multimers and sms. Left panel: GPC-100 chromatograms of 1.5 mM C7 solutions. The peak eluted at 8.3 min is that of thymidine, a marker used for normalization. (A) The solution incubated at 20°C shows two weak peaks. One corresponds to a tetramer (T) and the other to non-resolved (dimer+monomer) species. (B) The same sample injected in the column immediately after melting at 100°C shows an intense (dimer+monomer) peak. The comparison of both chromatograms reveals that in the non-melted sample, 95% of the oligonucleotide is incorporated in large sms that are retained on the chromatography column. Right panel: sms (red), tetramer (cyan) and (monomer + dimer) (green) equilibrium fractions in C7 solutions versus the concentration of the incubated samples. The sms proportion was determined by the comparison of chromatograms recorded before and after melting as in the left panel. The exclusion and permeation times are indicated by dashed lines.
C2TCn (n = 3, 4, 5) and C5PurC2 (Pur = A or G)
These oligonucleotides associate into sms. The chromatograms indicate common properties: (i) the formation and dissociation times of sms are comparable to the elution times, 4–6 min, in the GPC-100 column; (ii) sms dissociation induced by sample dilution results in the formation of tetramers. Representative chromatograms of C2TC5 solutions are displayed in Figure 2. The solutions were incubated and injected at the concentrations indicated in the figure. The chromatogram of samples whose concentration is <1 µM shows only two species eluted as a tetramer (Te) and a monomer (M). The stoichiometry of Te was confirmed by the observation that the Te concentration varies as the power of four of the M concentration (Figure 2, right panel). The tetramer reduced dissociation constant is 1.3 × 10−7 M. It is noteworthy that incubation during days is required to reach the equilibrium between M and Te. The chromatograms recorded with concentrated C2TC5 solutions show a peak corresponding to sms and a broad shoulder indicated by arrows in Figure 2. The elution times of both peaks decrease continuously when the sample concentration increases.
Figure 2.
C2TC5 association into tetramer and sms. The solutions were incubated and injected in the GPC-100 column at the concentrations indicated. The chromatograms recorded at low C5TC2 concentration show two components identified to a tetramer, Te, and a monomer, M, by the slope of four of the log–log plot of concentration [Te] versus [M] (right panel). When the oligonucleotide concentration increases, the continuous shift of the sms and Te peaks (arrows) indicates an exchange situation and therefore that the sms formation and dissociation times are comparable to the elution time. The dashed line shows the exclusion time.
Supplementary Figure S2 shows that all the oligonucleotides of this family exhibit the same characteristic elution profile. For these oligonucleotides, we observe that the chromatogram of a solution incubated at concentration C and diluted by a factor of 10 immediately before injection is similar (except for the Te/M ratio) to that of a sample incubated at concentration C/10. This observation indicates that the sms formation and dissociation times are comparable to or shorter than the elution time. According to this interpretation, the elution time of Ten, the sms formed by association of n tetramers, is averaged by exchange between Ten and [Te(n−m) + Tem] during the migration. The broad component designated by arrows in Figure 2 and Supplementary Figure S2 is attributed to Te, the tetramers involved in sms formation. The elution time of Te depends on the fraction of the time during which it is associated to sms and on the sms elution time. For this reason, the elution peak of Te is shifted in a direction indicating an apparent larger molecular weight when the oligonucleotide concentration increases. Chromatography performed as a function of temperature support this interpretation (Supplementary Figure S3). The chromatograms of a C5AC2 solution incubated at 20°C and injected in the column at controlled temperatures show that at 0°C the sms are eluted in the exclusion volume. At 20°C the sms elution time is that expected for the assembly of three tetramers. Sms are dissociated into tetramer during the chromatography at 42°C. At 56°C the solution is eluted as a monomer. In relation with exchange between different species and with the structural heterogeneity of these samples, the NMR spectra of 0.5–1 mM C2TCn and C5PurC2 solutions have extremely broad lines unfavorable to structural investigations.
CnTC2 (n = 3, 4, 5), C3TC3, C4TC4 and C5AC3
These oligonucleotides associate into sms with the following common properties: (i) the sms formation and dissociation times are much longer than the elution time in the chromatography column, (ii) sms dissociation does not releases tetramers but monomers.
C5TC2
The evolution during incubation at 20°C of the composition of a C5TC2 solution is displayed in Figure 3. The chromatogram recorded immediately after melting shows two poorly resolved components labeled M and D. The measure of the equilibrium concentration of M and D as a function of the oligonucleotide concentration establishes that peak D increases as the square of peak M and thus identifies M to a monomer and D to a dimer (Supplementary Figure S4). The calibration curves of Supplementary Figure S1 show that the anomalously long elution time of the dimer, by comparison to that of the unstructured monomer, seems to be a common feature in relation with the compact character of i-motif. The dimer reduced dissociation constant estimated from Supplementary Figure S4 is 8 × 10−8 M at 20°C and 9 × 10−6 M at 42°C. The chromatograms recorded after several hours show the slow apparition of sms peaks whose elution times correspond to the assemblies of 2, 3 and 4 tetramers. Lastly the chromatograms show larger sms and the decrease of the species including a small number of tetramers. C5TC2 association into sms was also followed by NMR (Supplementary Figure S5). It must be reminded that the imino proton peak of non-paired cytidines is broadened out by exchange with water at pH 4.6 (16). Hence, the narrow imino proton peaks (∼15.3 ppm) of the first spectrum recorded after melting demonstrate the early formation of a structured hemiprotonated species corresponding to that detected as a dimer by chromatography. The spectrum recorded at equilibrium shows extremely broad NMR lines indicative of large sms. The proportion of short sms is always too small to be detected by NMR. Supplementary Figure S6 show that the sms half formation time decreases approximately as the power of −1 of the oligonucleotide concentration in contrast to the tetramer half formation time that decreases as the power of −2. The half formation times of C5TC2 and C3TC3 sms decrease when the temperature is raised with comparable activation energies of −190 kJ/M (Supplementary Figure S7).
Figure 3.
Evolution during incubation at 20°C of the composition of a 0.3 mM C5TC2 solution initially melted. Left panel: The GPC-100 chromatograms are normalized to the same integrated area. The vertical scale of the chromatograms drawn in heavy lines are multiplied by a factor of five. On the chromatogram recorded at t = 0, the oligonucleotide is eluted as a monomer (M) and as a dimer (D). The anomalous elution order of D and M reflects probably the hydrodynamic radius difference between the unstructured monomer and the compact i-motif (Supplementary Figure S1). The chromatograms recorded versus time show the early formation of a tetramer (Te), the transient apparition of species including 2, 3 and 4 tetramers and the accumulation of non-resolved sms. Right panel: Evolution of the sms (red), tetramer (blue) and (monomer + dimer) (green) fractions as a function of the incubation time. The dotted line shows the exclusion time.
C4TC2, C3TC2 and C3TC3
The sms, tetramer and (dimer + monomer) fractions measured as a function of the oligonucleotide concentrations are comparable to those reported above for C5TC2 (Supplementary Figure S8). Nevertheless, these oligonucleotides show specific points that are described below.
The evolution of the species observed in a 1 mM C3TC3 solution during incubation at 42°C gives a valuable indication about the sms formation pathway (Figure 4). The sms fraction first increases with an initial formation rate about one-half that of the tetramer, reaches a maximum value of 20% and slowly decreases to zero while the tetramer fraction increases towards a limit value of ∼75%.
Figure 4.
Evolution versus time at 42°C of the sms (red), tetramer (cyan) and monomer + dimer (green) fractions in a 1 mM C3TC3 solution initially monomeric. Sms accumulation in the early stage of the kinetics and final disappearance is typical of kinetics trapping.
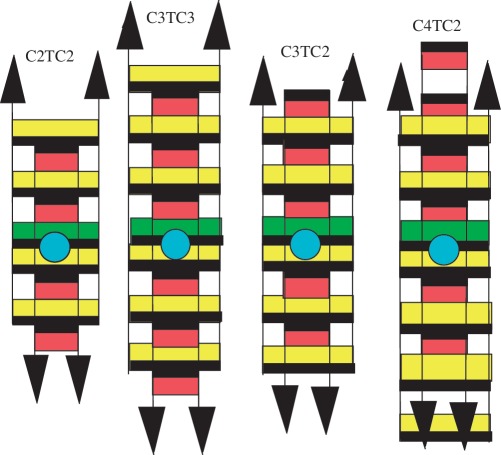
The evolution in opposite directions of the tetramer and sms fractions demonstrates that the sms are not built by association of this tetramer species. The NMR spectra of C4TC2, C3TC2 and C3TC3 recorded during sms built-up present a common characteristic features providing key information about the intercalation topology of the corresponding tetramers. The spectra of C3TC3 are exemplary (Figure 5). They show two thymidine imino proton peaks of comparable intensities (11.2–10.95 ppm) that increase as a function of the time as the tetramer fraction measured by chromatography. These thymidine imino protons were therefore assigned to the tetramer. Magnetization transfer experiments show that selective saturation of either the 11.2 or the 10.95 ppm peak results in saturation of both protons (Figure 5). This demonstrates that the thymidine exchanges within the tetramer between two non-equivalent environments. The inter-conversion rate between the two environments of [C3TC3]4 was determined by measuring the magnetization transferred between the T imino protons as a function of the length of the saturation pulse (14). We find that it is equal to 1 s at 0° and to 50 ms at 20°C. It is noteworthy that the NMR properties of the thymidine imino proton of [C3TC3]4, [C3TC2]4 and [C4TC2]4 are identical to those previously described for the thymidines of [C2TC2]4 (14). This gives a strong argument to assume that these tetramers have an intercalation topology similar to that of [C2TC2]4 (Figure 6) and that thymidine exchange reflects, as in [C2TC2]4, duplexe inter-conversion within the tetramers.
Figure 5.
Evolution versus time after initial melting of the composition of a 0.85 mM C3TC3 solution as detected by NMR and chromatography at 20°C. Right side: sms (red), tetramer (cyan) and (monomer + dimer) (green) fractions measured on chromatograms recorded as a function of the time. Left side: NMR spectra of the imino proton region. In the T imino proton region, the intensity of two narrow exchangeable peaks (11.2 and 10.95 ppm) increases as that of the tetramer peak on the chromatograms. The bottom spectrum is a magnetization transfer experiment performed at equilibrium showing the difference between a reference spectrum and a spectrum selectively irradiated during 150 ms at the position indicated by the arrow. The magnetization transfer between the two T imino proton peaks establishes that the oligonucleotide adopts two non-equivalent conformations that exchange together.
Figure 6.
The intercalation topologies of the i-motif tetramers of C3TC3, C3TC2 and C4TC2. Their NMR spectra have characteristics indicating that as in [C2TC2]41 (2), the tetramers include two intercalated duplexes, one (yellow) with a T•T pair (green) stacked on the 5′-adjacent C•C+ pair and the other (red) whose thymidines are looped out in the i-motif wide grove (blue circle). A black heavy line marks the face of the bases oriented in the 5′-direction. The magnetization transfer detected between the T imino protons of each duplex establishes that concerted opening/closing of the T•T pairs switches the duplex conformations. Full intercalation topology of the tetramers of C3TC3 and C3TC2 prevents association into sms. This implies that the sms building block is a minor tetrameric species formed by a reaction parallel to that leading to the fully intercalated tetramer.
Sms dissociation and lifetimes
We have shown above that the sms of C2TCn and C5PurC2 oligonucleotides dissociate in <6 min at 20°C into tetramers. The lifetime of the sms of C5AC3, 12 ± 3 h at 20°C, shows the dramatic effect of an additional cytidine.
The dissociation pathways of the sms of CnTC2 and C3TC3 have common characteristics. The chromatograms recorded as a function of the time after dilution of sms solutions show that the decrease of the sms fraction is approximately exponential and that the (dimer + monomer) peak increases consecutively to sms dissociation. The tetramer fraction decreases much more slowly (Supplementary Figure S9). The lifetimes of the tetramer of C3TC3 (Supplementary Figure S10) and CnTC2 are at least 10 times longer than those of the corresponding sms. It is therefore very intriguing to note that sms disruption does not result in accumulation of tetramer. This may indicates either that the sms dissociate into monomers or that sms dissociation releases a short-lived tetramer that dissociates and does not accumulate. The sms lifetimes measured at 20°C and 42°C increase with the number of cytidines in the sequences (Supplementary Figure S11).
Evaluation of the sms length
The size of sms measured at equilibrium by gel filtration chromatography on GPC-1000 column (Figure 7) and by electrophoresis on PA gels (Figure 8) are in fairly good agreement. At the concentration of 1 mM, the equilibrium distributions of the sms of C5TC2 and C3TC3 are centered on molecular weights corresponding to the association of 20 and 15 tetramers respectively. Considering that the distance between sequentially adjacent i-motif C•C+ pairs is 6.3 Å, the lengths of these structures should be ∼70 nm. The sms eluted in the rising edge of the chromatograms are about 10 times larger. The size of sms increases with the sample concentration, as shown with the example of a 9 mM C3TC3 solution whose sms distribution at equilibrium corresponds to the assembly of 170 tetramers (Figure 7). Due to exchange during chromatography, only averaged molecular weight are accessible at 20°C for the sms of C2TCn and C5PurC2 oligonucleotides. In 1–3 mM solutions, the elution times of the sms formed by these oligonucleotides correspond to structures including 5–8 tetramers (Supplementary Figure S2). In a 3 mM solution at equilibrium, the sms of C5AC3 (Figure 7) are about three times larger than those of C5AC2.
Figure 7.
GPC-1000 chromatograms at 20°C of a 9 mM C3TC3 solutions and of 3 mM C5TC2 and C5GC3 solutions at equilibrium. The top horizontal scale, drawn according to the column calibration, indicates the elution positions of nucleic acids containing the indicated base numbers. The non-resolved tetramer, dimer and monomer in equilibrium with the sms are eluted in the peaks at 8.2 min. The exclusion and permeation times are indicated by dotted lines.
Figure 8.
Electrophoresis on 10% (upper panel) and 6% (lower panel) PA gels of 1 and 0.3 mM C3TC3 (lanes 3 and 4 respectively) and C5TC2 (lanes 6 and 7) oligonucleotide solutions at equilibrium after incubation at room temperature. The other lanes are used for calibration with markers. Lane 1: Glu-tRNA (76 nt), Lane 2: Un-fractioned-brewer yeast tRNA (∼76 nt) and 5 s RNA contaminant (140 nt). Lanes 5 and 10: ladder ranging from 100 bp (200 nt) to 1000 bp by 100 bp increments, Lanes 8 and 9: 0.3 and 0.1 mM solutions of C5TC associated into i-motif tetramers (28 nt). Lane 11: dT9, dT24 and dT57. Lane 12: 9-mer duplex and 26-mer hairpin. In agreement with the gel filtration analysis, the C3TC3 lanes shows a band corresponding to the tetramer (28 nt) and a smear indicating structures including 100–500 nt i.e. 4–25 tetrameric repeats. The sms of C5TC2 include up to 30 tetramers. Very weak bands corresponding to [C5TC2]4 and to assemblies including two and three tetramers are detectable in lane 6.
DISCUSSION
It emerges from this study that stretches of only two cytidines, either at the 3′-or 5′-oligonucleotide end, give to i-motif tetramers self-associative capacity and that the sms formation/dissociation kinetics is influenced by the sequence polarity. However, in the concentration range required for NMR studies, the sms are always too large to allow structural investigations. Our model of the junction linking the tetramers in sms is therefore based on the comparison the properties of sms built by different oligonucleotides.
The sms formation pathway
A characteristic feature of C-rich oligonucleotides is their ability to associate, via parallel reactions, into a variety of i-motif structures including fully or partially intercalated tetramers and hairpin dimers. The formation rate of i-motif structures depends on the stoichiometry and on the oligonucleotide concentration. For solutions in the millimolar concentration range, the formation of dimer is faster than that of tetramer (17,18). Tetramers with full or partial intercalation topologies are built at comparable rates and their lifetimes increase by one or two magnitude orders for each additional stacked C•C+ pair (19). For these reasons, the thermodynamically stable tetramers are those with optimal intercalation topology.
The formation rate of C5TC2 sms increases as the oligonucleotide concentration by contrast to that of [C5TC2]4, which increases as the square of the oligonucleotide concentration (Supplementary Figure S6). This suggests that sms formation does not proceed via consecutive incorporation of monomer, a process that should have stronger concentration dependence, but, by stepwise association of tetrameric building blocks and possibly by association of preformed short sms. The equilibrium of the sms length is reached when spontaneous sms dissociation counterbalances elongation. The sms formation rate may be extremely slow (Supplementary Figures S6 and S7). However, this does not seems to be an intrinsic property of sms as shown by the examples of C7, C2TCn and C5purC2 oligonucleotides (Figure 2, Supplementary Figures S2 and S3). It appears instead that the slow formation rate of CnTC2 and C3TC3 sms is due to competitive production of i-motif dimers or of fully intercalated tetramers that limit the availability of the sms building blocks and by that way controls the sms formation rates. The identification of the building block tetramer has therefore a crucial importance to understand how i-motif tetramers associate into sms.
The sms building blocks
The magnetization transfer experiments performed on the tetramers observed in C3TC3, C4TC2 and C3TC2 solutions indicate structures similar to that of [C2TC2]4. However, as shown in Figure 6, the structures built on this model for C3TC3 and C3TC2 are fully intercalated and cannot associate into sms. This demonstrates that the sms are built by minor species, intrinsically less stable but which are stabilized into sms. We note that this conclusion is in line with the interpretation of the dissociation process of the sms of CnTC2 and C3TC3 suggesting that sms dissociation releases an unstable tetramer (see above). This also gives the following interpretation for the evolution after initial melting of the composition of C3TC3 solutions at 42°C (Figure 4). Due to its long lifetime, the fully intercalated tetramer of C3TC3 displayed in Figure 6 is the thermodynamically stable specie at 42°C. Nevertheless, the building block and the fully intercalated tetramer are created at comparable rates. This induces the transient accumulation of kinetically trapped sms. At last, sms dissociation contributes to increase the fully intercalated tetramer fraction.
The comparison of the association kinetics of CnTC2 and C2TCn oligonucleotides into sms is particularly instructive. What is the feature related to the sequence polarity that could account for the differences observed in sms formation and dissociation? Structural investigations by NMR have established that T•T pairs intercalated in i-motif adopt always the same structural arrangement (14,17,19,20). Let us consider the example of [C2TC2]4 (Figure 6). The tetramer is formed by two intercalated parallel duplexes. One whose tymidines (T3) are unstacked and the other whose tymidines (T3) are paired and stacked to the sequentially adjacent pair in the 5′-direction. The intercalation order, C5/C1/C4/C2/T3C2/C4/C1/C5, shows that pairs T3•T3 and C2•C2+ are intercalated into the other duplex in the empty space between C2•C2+ and C4•C4+.
The building block models described below are based on the following assumptions: (i) the thymidines in the building blocks are paired and intercalated with the same topology than the T•T pair intercalated in [C2TC2]4; (ii) the effects of the sequence polarity on the sms formation and dissociation kinetics originate from the stacking directionality of the T•T pair on the 5′-adjacent C•C+ pair. Using the example of C3TC2 and C2TC3, Figure 9 shows that for CnTC2 oligonucleotides, stacking of Tn + 1 to Cn allows the formation of a i-motif core including the 2x n C•C+ pairs of the longest C stretches. At each end two empty intervals are available for intercalation of the two terminal C•C+ pairs of another identical building block. In the building block model of C2TCn, stacking of the thymidine to the 5′-adjacent C•C+ pair leaves a single empty interval available for intercalation of the terminal C•C+ pairs of another building block. According to this model, the number of intercalation sites in the junctions between the corresponding building blocks account for the lifetime differences of CnTC2 and C2TCn sms. The slow formation rate of CnTC2 and C3TC3 sms is due to the weak proportion of the building block species (Figure 9) in competition with the more stable fully intercalated tetramers (Figure 6). It is also remarkable that the building blocks of C2TCn oligonucleotides have a larger number of stacked bases and therefore probably longer lifetimes than the building blocks of CnTC2. We suggest that this is exactly the reason why the sms of C2TCn oligonucleotides dissociate into long-lived tetramers in contrast to CnTC2 sms that release unstable tetramers rapidly dissociated into monomers.
Figure 9.
Building blocks models selected for their capacities to self-associate into sms with optimal intercalation topologies. The cytidines are yellow and the thymidines are green. The purines, that are presumed unpaired, are blue. Black lines indicate the face of the bases oriented in the 5′-direction. The cytidines of the longer C stretches are associated in i-motif cores. In the building blocks of C3TC2 and C2TC3, the thymidines are paired and stacked on the 5′-adjacent C•C+ pairs, as this is observed for the thymidines intercalated in i-motif structures (14,17,18,20). Note that this model applies to C2TCn and C2TCn oligonucleotides. For each species, a section of a second identical building block is displayed with its bases in front of the position favorable for mutual intercalation into sms. The junction between the tetramer of C3TC2 involves intercalation of two bases. A single intercalation position is available to connect together the tetramers of C2TC3 and C5purC2. The difference of the number of intercalated bases in the junctions is consistent with the longer lifetimes of CnTC2 sms.
The C1′–C1′ distance in pur•pur pairs (21) is certainly incompatible with pur•pur insertion in i-motif. We can therefore consider that the purines are unstacked in the building block of C5PurC2 sms. Figure 9 shows that the tetramer built according to this assumption, by the cytidines of the longest C stretch, has also a single empty interval available for association into sms. The structural similarity of the terminal segments of the building blocks of C2TCn and C5PurC2 sms is consistent with the similarity of their formation/dissociation kinetics. According to this model the building block of C5purC3 sms should provide two intercalation sites. This prediction is in agreement with the observation that the half dissociation time of C5AC3 sms is at least 120 times longer than that of the sms of C5AC2.
Junction between cytidine homopolymers i-motif in sms
In contrast to short Cn oligonucleotides (n ≤ 5) that associate into fully intercalated tetramers, C7 and C12 aggregate into extremely large insoluble assemblies. We suggest that the presence of a non-C base in the oligonucleotide sequences prevent the formation of tetramers with staggered strands and that on the contrary, long Cn oligonucleotides associates into i-motif with staggered strands. Supplementary Figure S12 shows how staggered strands connecting together several i-motif cores can generate branched structures leading to the formation 2D networks or of 3D assemblies.
CONCLUSION
In summary, we have shown that C stretches as short as CC can link i-motif tetramers into sms. The sms grow in competition with i-motif tetramers or dimers and their formation rate is controlled by the availability of the building block involved in sms formation. The sms stability increases with the number of cytidines in the shorter C stretch. The comparison of the sms lifetimes with those of i-motif tetramers (19) shows that incorporation of tetramers into a larger structure has a stabilizing effect. The presence of a single non-C residue in the oligo-C sequences prevents the formation of structures with staggered strands, an effect certainly favorable to sms elongation into linear structures. Due to sugar backbone stretching induced by systematic intercalation, the i-motif is a stiff structure. Association of i-motif tetramers into sms can potentially form unbendable rods with a particularly large persistence length. This prediction is testable since the size of the sms examined in present work, typically 600 × 16 × 10 Å, is accessible to atomic force microscopy measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–12.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Aude Laisné for assistance in chromatography measurement and Denis Pompon for his enthusiasm and interest about this project. Jean-Louis Mergny is acknowledged for help in electrophoresis experiments.
REFERENCES
- 1.Langridge R, Rich A. Molecular structure of helical polycytidylic acid. Nature. 1963;198:725–728. doi: 10.1038/198725a0. [DOI] [PubMed] [Google Scholar]
- 2.Gehring K, Leroy JL, Guéron M. A tetrameric DNA structure with protonated cytidine·cytidine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 3.Guéron M, Leroy JL. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 4.Kang CH, Berger I, Lockshin C, Ratliff R, Moyzis R, Rich A. Crystal structure of intercalated four-stranded d(C3T) at 1.4 A resolution. Proc. Natl Acad. Sci. USA. 1994;91:11636–11640. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TN. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 6.Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, LaBean TN. Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures. Angew. Chem. Int. Ed. 2007;45:735–739. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Mao CJ. Putting a brake on an autonomous DNA nanomotor. J. Am. Chem. Soc. 2009;126:8626–8627. doi: 10.1021/ja047991r. [DOI] [PubMed] [Google Scholar]
- 8.Ghodke HB, Krishnan R, Vignesh K, Kumar GV, Narayana C, Krishnan Y. The I-tetraplex building block: rational design and controlled fabrication of robust 1D DNA scaffolds through non-Watson Crick. Angew. Chem. Int. Ed. 2007;46:2646–2649. doi: 10.1002/anie.200604461. [DOI] [PubMed] [Google Scholar]
- 9.Zikich D, Liu K, Sagiv L, Porath D, Kotlyar A. I-motif nanospheres: unusual self-assembly of long cytosine strands. Small. 2011;7:1028–1034. doi: 10.1002/smll.201002213. [DOI] [PubMed] [Google Scholar]
- 10.Laisné A, Pompon D, Leroy J-L. [C7GC4]4 Association into supra molecular i-motif structures. Nucleic Acids Res. 2010;38:3817–3826. doi: 10.1093/nar/gkq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leroy JL, Guéron M. Solution structures of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. doi: 10.1016/s0969-2126(01)00138-1. [DOI] [PubMed] [Google Scholar]
- 12.Cantor CR, Warshaw MM. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 13.Plateau P, Guéron M. Exchangeable proton NMR without base-line distortion, using strong-pulse sequences. J. Amer. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 14.Nonin S, Leroy J-L. Structure and conversion kinetics of a bi-stable DNA i-motif: broken symmetry in the [d (5mCCTCC)]4 tetramer. J. Mol. Biol. 1996;261:399–414. doi: 10.1006/jmbi.1996.0472. [DOI] [PubMed] [Google Scholar]
- 15.Leroy J-L, Snoussi K, Guéron M. Investigation of the CH-O hydrogen bonds in the DNA i-motif via the equilibrium between alternative topologies. Magnetic Resonance in Chemistry. 2001;39:171–176. [Google Scholar]
- 16.Leroy J-L, Gehring K, Kettani A, Guéron M. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization and proton exchange properties. Biochemistry. 1993;32:6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- 17.Leroy J-L. T.T pair intercalation and duplex interconversion within i-motif tetramers. J. Mol. Biol. 2004;333:125–139. doi: 10.1016/s0022-2836(03)00945-8. [DOI] [PubMed] [Google Scholar]
- 18.Canalia M, Leroy J-L. [5mCCTCTCTCC]4: an i-motif tetramer with intercalated T*T pairs. J. Am. Chem. Soc. 2009;131:12870–12871. doi: 10.1021/ja903210t. [DOI] [PubMed] [Google Scholar]
- 19.Leroy J-L. The formation pathway of i-motif tetramers. Nucleic Acids Res. 2009;37:4127–4134. doi: 10.1093/nar/gkp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canalia M, Leroy J-L. Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC) Nucleic Acids Res. 2005;33:5471–5481. doi: 10.1093/nar/gki843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.