Abstract
Sperm chromatin remodeling after oocyte entry is the essential step that initiates embryogenesis. This reaction involves the removal of sperm-specific basic proteins and chromatin assembly with histones. In mammals, three nucleoplasmin/nucleophosmin (NPM) family proteins–NPM1, NPM2 and NPM3–expressed in oocytes are presumed to cooperatively regulate sperm chromatin remodeling. We characterized the sperm chromatin decondensation and nucleosome assembly activities of three human NPM proteins. NPM1 and NPM2 mediated nucleosome assembly independently of other NPM proteins, whereas the function of NPM3 was largely dependent on formation of a complex with NPM1. Maximal sperm chromatin remodeling activity of NPM2 required the inhibition of its non-specific nucleic acid-binding activity by phosphorylation. Furthermore, the oligomer formation with NPM1 elicited NPM3 nucleosome assembly and sperm chromatin decondensation activity. NPM3 also suppressed the RNA-binding activity of NPM1, which enhanced the nucleoplasm–nucleolus shuttling of NPM1 in somatic cell nuclei. Our results proposed a novel mechanism whereby three NPM proteins cooperatively regulate chromatin disassembly and assembly in the early embryo and in somatic cells.
INTRODUCTION
Chromatin structure is significantly modified during spermatogenesis, resulting in the generation of highly condensed sperm chromatin. The core histones, H2A, H2B, H3 and H4, are largely removed and replaced with sperm-specific basic polypeptide protamines to form tight toroidal complexes. Sperm chromatin is rapidly reorganized in the oocytes after fertilization. During this process, protamines are efficiently removed and the canonical chromatin components, histones are deposited to form chromatin that can participate in zygote formation. This process is mediated by factor(s) present in the oocyte cytoplasm. In Xenopus, protamine removal and histone deposition are both mediated by an oocyte-specific acidic protein, nucleoplasmin, originally identified as a factor involved in chromatin assembly in Xenopus egg extracts (1,2). Huge amounts of histones are stored in Xenopus oocytes to ensure that DNA replication-coupled chromatin assembly occurs after fertilization and that subsequent cell division occurs without de novo transcription. Other important roles of nucleoplasmin during early embryogenesis include neutralization of the basic charge of histones and inhibition of their non-specific binding to DNA. Treatment of mouse embryonic carcinoma cell line F9 with nucleoplasmin induces the expression of oocyte-specific genes, suggesting that nucleoplasmin has a role in nuclear reprogramming by regulating global chromatin structure (3). Furthermore, ectopic expression of human NPM2, an ortholog of nucleoplasmin, in 293T cells induces the expression of pluripotency-associated genes (4). In mouse, the expression of NPM2 is restricted in growing oocytes (5). Targeted disruption of the mouse NPM2 gene results in abnormal nuclear structure formation in oocytes and early embryonic cells, despite its dispensable function in sperm chromatin decondensation (5). Therefore, NPM2 plays an important role in the regulation of chromatin structure during early embryogenesis. Other family proteins, including NPM1 and NPM3, may play compensatory roles in NPM2-knockout mouse eggs. Human NPM1/nucleophosmin/B23 also has a potential role in sperm chromatin decondensation (6), both NPM1 and NPM3 are expressed in mouse oocytes (5), and inhibition of NPM3 expression using an antisense oligonucleotide prevents sperm chromatin remodeling in mouse fertilized eggs (7). These observations suggest that NPM family proteins play important roles in sperm chromatin remodeling.
Proteins in the NPM family share a conserved N-terminal core domain containing eight-stranded β-sheets (8). Crystal structural analyses of the N-terminal domains of nucleoplasmin, Drosophila nucleoplasmin-like protein dNLP, Xenopus NO38/NPM1 and human NPM1 and NPM2, demonstrated that the core domain forms a pentameric ring, while two pentamers associate in a head-to-head fashion to form a decamer (9–13). Other features of this family are acidic stretches extending from the core domains, which are implicated in efficient histone chaperone activity (14). Unlike other NPM proteins, NPM1 has an RNA-binding domain that is required for efficient nucleolar localization at the C-terminal end (15). Proteins in the NPM family are likely to form hetero-oligomers because of the significant conservation found in the core domains.
We have been studying the molecular mechanism of adenovirus (Ad) chromatin assembly and disassembly. Ad DNA is complexed with viral basic core proteins, V, VII and Mu, in the mature virions to produce the highly condensed Ad chromatin structure. Upon entry into host cell nuclei, Ad chromatin is decondensed after a change in the mode of interaction between the core proteins and DNA, and DNA is used as a template for early gene expression (16). Ad chromatin does not function as a template for replication and transcription in vitro because of its condensed structure, indicating that Ad chromatin remodeling is crucial for the initiation of transcription and replication. We previously identified host factors, Template Activating Factor-I (TAF-I) and NPM1/Nucleophosmin/B23 as host factors involved in Ad chromatin remodeling (17,18). TAF-I and NPM1 directly associate with viral basic core proteins to remodel the viral chromatin structure (16,19). Since the major core protein VII is an arginine-rich basic polypeptide and shows limited sequence similarity to sperm-specific basic proteins (20), adenovirus chromatin remodeling is an advantageous model system to understand sperm chromatin remodeling. Consistent with this, we previously demonstrated that both TAF-I and NPM1 have potential sperm chromatin decondensation activity as they mediate adenovirus chromatin remodeling (6,21).
The three NPM proteins are expressed in oocytes and eggs, so that NPM proteins may share overlapping roles due to the formation of hetero-oligomers via the conserved core domain. However, the oligomerization status and biochemical activities of the three human NPM proteins have yet to be established. This report characterized the sperm chromatin decondensation and nucleosome assembly activities of homo- and hetero-oligomers of the three human NPM proteins.
MATERIALS AND METHODS
Plasmids
Plasmids for NPM1 expression were previously described (6,18,22). Human and mouse NPM3 genes were cloned by PCR using cDNA prepared from HeLa cells and NIH3T3 cells by using oligonucleotide sets, 5′-aaaacgcgtcatatggccgccggtactgcagc-3′ and 5′-aaaggatccctagggcctgcccccctgct-3′, and 5′-agctagcatatgttcagcatggcggccggcgc-3′ and 5′-agctggatcctaaggcctgcccctgtgct-3′, respectively. The cDNA was digested with Nde I and Bam HI, and subcloned into the same sites of pET14b (Novagene). To add N-terminal Flag-tag, the cDNA cut out from pET14b-NPM3 by Nde I and Hind III was subcloned into the same sites of pBS-Flag vector. To construct pGEX6P1-NPM3 and pEGFPC1-Flag-NPM3, the cDNA was cut out from pBS-Flag-NPM3 by digestion with Bam HI and subcloned into the same sites of pGEX6P1 (GE Healthcare) or pEGFPC1 (Clonetech). The expression of His-tagged NPM3 was not successful in Escherichia coli using pET14b-NPM3, so the full length NPM3 cDNA was synthesized to optimize the sequence for the E. coli expression system. The optimized NPM3 sequence was subcloned into the Nde I and Bam HI sites of pET14b. Similarly, the human NPM2 cDNA optimized for expression in E. coli was synthesized and subcloned into the Nde I and Bam HI sites of pET14b. The NPM2 cDNA was cut out from pET14b vector by Nde I and Hind III, and subcloned into the same sites of pBS-Flag vector. Then, the Flag-tagged NPM2 cDNA was cut out by Bam HI digestion and subcloned into the Bam HI sites of pGEX6P1 and pEGFPC1 vectors. The sequences of synthesized cDNAs (Operon Biotechnologies) for human NPM2 and NPM3 will be provided upon request. For construction of the chimeric NPM1–3Ch protein, cDNAs for NPM1(1–120) and NPM3(146–178) were independently amplified using primer sets 5′-cacttagtagctgtttccgaagaagaatct-3′ and T7 terminator primer, and 5′-ttcttcggaaacagctactaagtgctgtcc-3′ and T7 promoter primer, respectively, and appropriate pET14b vectors as templates. These two fragments were used as templates for second PCR using T7 promoter and terminator primers to amplify the full length cDNA. The cDNA was subcloned into the Nde I- and Bam HI-digested pET14b. To purify the NPM1–NPM3 complex, the cDNA for NPM3 containing N-terminal His-tag was cut out from pET14b-NPM3 vector by Nco I and Bam HI, and subcloned into the same sites of pETDuet-1 vector (Novagene). The NPM1 cDNA amplified by PCR with 5′- AGCGGATCCCATGGAAGATTCGATGGACAT-3′ and T7 terminator primer, and pET14b-NPM1 as template was digested with Bam HI and subcloned into the Bgl II site of pETDuet-1 vector containing the NPM3 cDNA. To construct phosphomimetic NPM2 proteins, oligonucleotide sets 5′-GCAGACGTTCGCGATAAAGACCCAGTCAAGAAA-3′ and 5′-TGGGTCTTTATCGCGAACGTCTGCACGGATTTC-3′ for NPM2 S196D, and 5′-GAGCAGGCTCCGGTGAAACAGGTG-3′ and 5′-CACCGGAGCCTGCTCTTCCAGACT-3′ for S159D, and 5′-ATGCAGCATATGAACCTGGACGACGCTGACGACGACGAAGAAAAAGCGGTTACT-3′ for 5D were used for PCR. NPM2 2D and 7D mutants were constructed using appropriate primer sets described above. The mutant cDNA was amplified by PCR and digested with Nde I and Bam HI, and subcloned into the same sites of pET14b.
Cell culture
HeLa cells and HEK293T cells were maintained in DMEM containing 10% FBS. HeLa cell lines expressing either EGFP–Flag (EF)–NPM1 or EF–NPM3 were established by transfection of pEGFPC1–Flag–NPM1 or pEGFP–Flag–NPM3 vectors. Cells stably expressing EGFP-tagged proteins were selected by 50 mg/l of G418. The colonies were isolated and the expression of EGFP-tagged proteins was examined by fluorescence microscope and western blotting with anti-Flag antibody.
Mouse oocytes and sperms preparation
MII-stage oocytes were prepared from the oviducts of 8-weeks-old ICR mice (SLC Japan, Inc.) supraovulated with equine chronic gonadotropin and human chronic gonadotropin. MII oocytes were separated from cumulus cells by hyaluronidase, and MII oocytes and cumulus cells were separately collected and used for western blotting. Sperms were isolated from testis of 12–14-weeks-old B6 mice. Sperms were washed with nuclei wash buffer (40 mM HEPES–KOH pH 7.2, 250 mM sucrose, 75 mM KCl, 0.5 mM spermidine and 0.15 mM spermine), permeabilized with the same buffer containing 0.33 mg/ml l-α-palmitoyl-lysophosphatidylcholine (SIGMA), 0.1% Nonidet P-40 and 10 mM dithiothreitol on ice for 10 min. The cells were washed with nuclei wash buffer, suspended in the same buffer containing 50% glycerol at a concentration of 1 × 105 nuclei/µl, frozen in liquid N2, and kept at −80°C until use.
Protein purification, chemical cross linking experiments, limited trypsin digestion
Recombinant proteins were expressed in E. coli and purified as described previously (18). His-tagged NPM proteins were separated by SDS–PAGE and recovered from the gel. The proteins were eluted, precipitated with acetone and denatured in 6 M Guanidine–HCl containing 50 mM Tris–HCl pH 7.9, and 10 mM DTT. Denatured proteins were dialyzed in 50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 50 mM KCl, 0.5% NP-40 and 20% Glycerol for 3 h, and then the dialysis buffer was changed to buffer H (20 mM HEPES–NaOH pH 7.9, 50 mM NaCl, 0.5 mM EDTA, 10% Glycerol and 0.5 mM PMSF) and dialyzed for 6 h.
To purify the His–NPM3–NPM1 complex, extracts were prepared from E. coli BL21 strain transformed with pETDuet-NPM3/NPM1 vector. The complex was purified with His select Nickel Affinity Gel (Sigma Aldrich) according to the manufacturers’ instruction. The eluted His–NPM3/NPM1 complex was then loaded on to a Mono Q column (0.1 ml, GE Healthcare) and purified with salt gradient from 0.1 to 0.6 M NaCl. The peak fraction containing both NPM1 and NPM3 was dialyzed as above. The samples (10 µg of NPM1 in 20 µl) were then separated by Superose 6 PC 3.2/30 column chromatography at a flow rate of 30 µl/min. Fractions (50 µl each) were collected and proteins were analyzed by SDS–PAGE and western blotting. Gel Filtration Calibration Kits (GE Healthcare) were used as molecular weight standards.
Phosphorylation of His-tagged NPM2 was performed with total cell extracts prepared from asynchronous and mitotic HeLa cells. Synchronization of HeLa cells and extracts preparation were performed as described previously (23). His–NPM2 (40 µg) was incubated with cell extracts (50 µg of proteins) at 37°C for 1 h in 20 mM Tris–HCl pH 7.9, 50 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 mM ATP and phosphatase inhibitors (10 mM β-d-glycerophosphate, 1 mM NaF and 1 mM Na3VO4). Then, His–NPM2 protein was purified with His select Nickel Affinity Gel as described above. The purified proteins were dialyzed in buffer H containing phosphatase inhibitors.
Chemical crosslinking experiments were performed with glutaraldehyde (Sigma). The proteins in buffer H were mixed with 0.05% glutaraldehyde and incubated at room temperature for 10 min. The reaction was terminated by adding SDS-sample loading buffer and boiling at 95°C for 10 min. The samples were separated by SDS–PAGE followed by silver staining or western blotting.
Blue Native (BN)–PAGE was performed with NativePAGETM Novex Bis–Tris Gel System (Invitrogen) according to the manufacturers’ instruction. Native MarkTM Unstained Protein Standard (Invitrogen) was used as molecular weight markers for BN–PAGE.
For trypsin digestion assay, recombinant proteins (500 ng) in buffer H were mixed with Trypsin (Sigma) and incubated at 37°C for 5 min. The reaction was stopped by adding SDS-sample loading buffer and boiled at 95°C for 5 min. Proteins were separated by 15% SDS–PAGE and visualized by Coomassie Brilliant Blue (CBB) staining or western blotting.
NCP assembly and DNA-binding assays
Nucleosome core particle (NCP) assembly assays were performed using 147-bp DNA fragment containing 5S rRNA gene sequence and core histones prepared as described previously (24). For DNA-binding assays shown in Figure 4, 196-bp DNA fragment was mixed with recombinant NPM2 proteins in 20 mM Tris–HCl pH 7.9, 100 mM NaCl, 10% Glycerol, 0.1 mg/ml of BSA. The complexes were loaded on 6% PAGE in 0.5 × TBE and the DNA was visualized with GelRed (Biotium) staining. Supercoiling assays were performed as described previously (6) using pCAGGS vector.
Figure 4.
Effect of phosphorylation on NPM2 sperm chromatin remodeling activity. (A) Phosphorylation of recombinant NPM2. Purified His–NPM2 was mixed and incubated without or with asynchronous (+Asyn) or mitotic (+M) HeLa cell extracts followed by purification with the Ni-NTA resin. Dialyzed proteins (200 ng) were separated on 10% SDS–PAGE and visualized with CBB and Pro-Q Diamond staining (lanes 1–3 and 4–6, respectively). The 45-kDa marker protein (ovalbumin) was detected by Pro-Q Diamond staining. Positions of molecular weight markers are indicated at the left side of the panel. (B) DNA-binding activity of recombinant NPM2. The 196-bp DNA fragment (100 ng) were mixed with increasing amounts (100, 200, 400 and 800 ng for each protein) of control NPM2 (lanes 2–5) or NPM2 phosphorylated by asynchronous (lanes 6–9) or mitotic (lanes 10–13) extracts. The mixtures were analyzed by native PAGE in 0.5× TBE and DNA was visualized with GelRed staining. (C) Phosphorylation of NPM2 mutants by extracts. NPM2 wild type (wt), S159D, S196D, S159/196D (2D), 5D and 7D mutant proteins (500 ng) were incubated with asynchronous (lanes 1–7) or mitotic (lanes 8–14) cell extracts (5 µg of proteins) and SDS-sample buffer was added. Proteins were analyzed by SDS–PAGE and Pro-Q diamond or CBB staining (top and bottom panels, respectively). Positions of NPM2 proteins are indicated by arrow heads at the right side of the panels. (D) Sperm chromatin decondensation by NPM2 proteins. Mouse sperm nuclei were incubated in the absence or presence of NPM2 wt, 2D, 5D and 7D (3 μg) and the size of sperm nuclei (n > 30) were estimated by Image J software and averaged. Results are means ± SD and statistical P-values were calculated and indicated by asterisk for P < 0.05. (E) Histone transfer activity of phosphorylated NPM2. Histone transfer assay was performed as in Figure 3F with increasing amounts (100, 200, 300 and 400 ng for each protein) of NPM2 wt, S159D, S196D, 2D, 5D and 7D. The mixtures were separated on 6% PAGE in 0.5× TBE and DNA was visualized with GelRed staining. Positions of free DNA and NCP are indicated at the right side of the panel.
Sperm chromatin decondensation
Mouse sperm nuclei were isolated as above and sperm nuclei decondensation assays were performed as described previously (21). A total of 2 × 104 sperm nuclei were incubated with NPM proteins and incubated at 37°C. Sperm DNA stained with 4′,6-diamidino-2-phenylindole (DAPI) was observed under fluorescence microscope equipped with a color, cooled CCD camera (DP71, Olympus). Sperm size was measured by Image J software (http://rsb.info.nih.gov/ij/).
Antibodies
Anti-NPM3 serum was generated in rabbits using the GST–Flag–NPM3 protein as an antigen. Anti-NPM3 antibody was purified with His–NPM3-immobilized HiTrap NHS-activated HP column (GE Healthcare). Purified anti-NPM3 antibody does not cross react with other NPM proteins (data not shown). Anti-Flag (M2, Sigma), anti-His-tag (HIS1, Sigma) and anti-NPM1 (Invitrogene) were commercially available. Anti-PCNA and anti-mouse NPM2 antibodies are generous gifts from Dr A. Verreault and Dr M. Matzuk, respectively.
Cell extracts preparation and immunoprecipitation
Exponentially growing HeLa cells were disrupted in gentle sonication in buffer A (50 mM Tris–HCl pH 7.9, 0.1% Triton X-100 and 0.5 mM PMSF) containing 150 mM NaCl. The extracts were recovered by centrifugation with 15 000 rpm for 15 min and mixed with anti-NPM1 or anti-NPM3 antibody, and incubated at 4°C for 2 h. As a control, normal rabbit IgG (Santacruz Biotechnology) was used. The NPM proteins were recovered by the addition of protein A sepharose (GE Healthcare). Proteins were separated on 12.5% SDS–PAGE and analyzed by western blotting.
For immunoprecipitation of EGFP–Flag (EF) tagged NPM proteins, EF, EF–NPM1, EF–NPM2 and EF–NPM3 were transiently transfected to HeLa cells with Gene Juice (Novagene). Twenty four hours after transfection, the cell extracts were prepared as above and immunoprecipitation was performed with anti-Flag M2 agarose beads (Sigma). The proteins bound to the beads were washed extensively by buffer A containing 150 mM NaCl, and eluted with the same buffer containing Flag peptide (Sigma). The proteins were separated by SDS–PAGE and analyzed by western blotting.
Glycerol density gradient assay
Nuclear extracts (50 µl) prepared from HeLa cells were loaded on 15–35% glycerol density gradient in 20 mM HEPES–NaOH pH 7.9, 50 mM NaCl and 0.5 mM EDTA (2.1 ml). The samples were centrifuged at 35 000 rpm for 14 h at 4°C in S55S rotor (Hitachi Koki). Fractions (100 µl) were collected from the top and analyzed by SDS–PAGE and western blotting.
Immunofluorescence and FRAP analysis
To examine the localization of endogenous NPM1 and NPM3, HeLa cells grown on cover slips were fixed with 3% paraformardehyde and permeabilized in PBS containing 0.5% Triton X-100. Cells were then incubated with anti-NPM1 and NPM3 antibodies diluted with PBS containing 0.5% non-fat dry milk. Localization of proteins was visualized with secondary antibodies conjugating with AlexaFluor dyes (Molecular Probes). During final wash with PBS containing 0.1% Triton X-100, To-Pro-3 DNA staining dye (Molecular Probes) was added and incubated for 15 min at room temperature. Localization of proteins and DNA was observed under confocal microscope (LSM EXCITER; Carl Zeiss Microimaging, Inc.).
HeLa cells stably expressing EF–NPM1 or EF–NPM3 were subjected to FRAP analyses. Cells were grown on 35-mm glass base dishes (IWAKI). The dish was set on an inverted microscope (LSM EXCITER; Carl Zeiss Microimaging, Inc.) in an air chamber containing 5% CO2 at 37°C, and the mobility of EGFP–tagged NPM proteins was analyzed by photobleaching with a Plan-Apochromat 63×/1.4 oil objective. EGFP signal in small nucleoli was bleached using 85% transmission of a 488-nm Ar laser (50 iterations), and images were collected (512 × 512 pixels, zoom 4.1, scan speed 13, pinhole 1 airy unit, LP505 emission filter and 1.0% transmission of a 488-nm Ar laser with 0.7% output power) every 0.5 s. The fluorescence intensity of the bleached area was measured using ZEN2008 software (Carl Zeiss Microimaging, Inc.). Background intensity was first subtracted from all the intensities measured and relative fluorescence intensity (RFI) was calculated as following: RFI = (Ia/Ia0) × (Ib0/Ib), where Ia is the fluorescence intensity of bleached area at each time points, Ia0 is the initial fluorescence intensity of bleached area, Ib is the fluorescence intensity of non-bleached nucleoli at each time point, and Ib0 is the initial fluorescence intensity of non-bleached nucleoli. Recovery curves were fit to a single exponential using the FRAP analysis module of the ZEN software (Carl Zeiss Microimaging, Inc.).
RESULTS
Association between NPM proteins in HeLa cells
We first examined the localization of human NPM proteins in HeLa cells. Enhanced green fluorescent protein (EGFP)-tagged NPM proteins were expressed in HeLa cells, and their localization was examined using fluorescent microscopy (Figure 1A). As previously reported (23,25,26), EF-tagged NPM1 and NPM3 preferentially localized in the nucleoli, whereas EF–NPM2 was distributed throughout the nucleus. The level of the EF–NPM3 signal was higher than that of NPM1 in both the nucleoplasm and cytoplasm. Furthermore, the expression pattern of endogenous NPM1 and NPM3 in HeLa cells was confirmed by immunofluorescence using specific antibodies (Figure 1B). Endogenous NPM1 and NPM3 were localized in the nucleolus: the staining patterns of NPM1 and NPM3 suggested that both are mainly found in the granular component of the nucleolus.
Figure 1.
Association between human NPM proteins. (A) Localization of EGFP-tagged NPM proteins. EF, EF–NPM1, EF–NPM2 and EF–NPM3 were transiently expressed in HeLa cells and the localization of EGFP proteins was examined by fluorescent microscopy. Bottom panels are phase contrast images. Bar at the bottom indicates 10 µm. (B) Localization of endogenous NPM1 and NPM3 in HeLa cells. HeLa cells were subjected to immunofluorescence analysis using anti-NPM1 and anti-NPM3 antibodies. DNA was stained with TO-PRO-3. Images were observed under confocal microscopy. Bar at the bottom indicates 10 µm. (C) Glycerol density gradient of HeLa cell nuclear extracts. Nuclear extracts from HeLa cells were fractionated by 15–35% glycerol density gradient. Fractions collected from the top were analyzed by western blotting with anti-nucleolin (NCL), anti-B23 and anti-NPM3 antibodies. (D) Immunoprecipitation with anti-NPM proteins. HeLa cell extracts were subjected to immunoprecipitation with control Ig (lanes 6 and 9), anti-NPM1 (lanes 7 and 10), and anti-NPM3 (lanes 8 and 11) antibodies. Bound proteins were eluted with SDS-sample buffer (30 µl) and 3 (lanes 6–8) or 9 (lanes 9–11) µl of samples were separated on SDS–PAGE and analyzed by western blotting with anti-NPM1 and anti-NPM3 antibodies (top and bottom panels, respectively). As standard, recombinant His-NPM1 (50, 25, 12 ng for lanes 1–3) and His–NPM3 (5, 2.5, 1.2 ng for lanes 1–3), and increasing amounts of input extracts (lanes 4–5) were also loaded. (E) Immunoprecipitation of transiently expressed NPM proteins. EF, EF–NPM1, -NPM2 and -NPM3 were transiently expressed in HEK293T cells and cell extracts were prepared. Immunoprecipitation with anti-Flag antibody beads was performed. Input (lanes 1–4) and immunoprecipitated (lanes 5–8) proteins were analyzed by western blotting with anti-NPM1, -NPM3 and -Flag antibodies. Positions of NPM proteins are indicated at the right side of the panel. Blank arrow head shows the protein likely to be corresponded to NPM3 degradation product. (F) Expression level of NPM proteins in mouse MII oocytes. MII oocytes (30 and 60 cells for lanes 7 and 8, respectively) and cumulus cells (3 × 103 and 10 × 103 cells for lanes 9 and 10) were prepared from mice and analyzed by western blotting with anti-NPM1, -NPM3 and -NPM2 antibodies. Recombinant human NPM1 and mouse NPM3 were loaded on the same gel as standards (lanes 1–6). Positions of NPM proteins are indicated by arrow heads.
NPM1 and NPM3 are expressed ubiquitously. We then examined the interaction between endogenous NPM1 and NPM3 in cell extracts. Nuclear extracts prepared from HeLa cells were fractionated using a 15–35% glycerol density gradient; fractions were collected from the top, the proteins in each fraction were analyzed by western blotting (Figure 1C). The NPM3 antibody generated in this study detected two NPM3 bands in the extracts (see the input lane of bottom panel). By comparing with the His-tagged recombinant protein (Figure 1D, lanes 1–5), the bottom band was likely to be a degradation product of full-length NPM3. Both NPM1 and NPM3 were broadly distributed and cofractionated in fractions 8–11. Since NPM1 and NPM3 (32.5 and 19 kDa, respectively) were recovered in higher molecular weight fractions than nucleolin (NCL, 76.6 kDa), both these proteins were mainly included in large protein complexes. To further test whether NPM1 and NPM3 formed a complex, immunoprecipitation assays were performed using antibodies specific for NPM1 and NPM3 (Figure 1D). Immunoprecipitation assays clearly demonstrated that NPM1 and NPM3 were associated with each other in extracts (lanes 6–11). Using recombinant proteins (migrating slower than endogenous proteins because of a His-tag) as standards (lanes 1–3), the amount of NPM3 (only full-length NPM3 (top band) was considered) in the input extracts derived from HeLa cells was rapidly estimated to be 1/10–1/20 of that of NPM1. Immunoprecipitation conducted using anti-NPM3 antibody indicated that NPM3 coprecipitated with higher amounts of NPM1 (lanes 8 and 11).
NPM2 expression is restricted in oocytes and early embryos (5), where both NPM1 and NPM3 are also expressed. We then examined the cellular interaction between NPM2 and other NPM proteins (Figure 1E). EF–tagged NPM proteins were expressed in HEK293T cells, and immunoprecipitation assays were conducted. EF–NPM1 coprecipitated endogenous NPM1 and NPM3 (lane 6). In contrast, the coprecipitation of endogenous NPM proteins with EF–NPM2 was barely detected (lane 7). EF–NPM3 coprecipitated with endogenous NPM1, but not with endogenous NPM3 (lane 8). These results raised the possibilities that NPM2 does not form a stable oligomer with other NPM proteins in oocytes and fertilized eggs and that NPM3 does not efficiently form a homo-oligomer in solution.
Three NPM proteins are expressed in mammalian oocytes (7), but their stoichiometry is unclear. To address this point, we examined the expression level of NPM proteins in mouse MII oocytes with specific antibodies using recombinant proteins as standards (Figure 1F). Because the anti-NPM1 antibody used in this study recognizes the C-terminal region (amino acids 259–294) of NPM1 and this region is completely identical in human and mouse NPM1, the amount of NPM1 in mouse MII oocytes was estimated using the human recombinant NPM1 protein as the standard. We used recombinant His-tagged mouse NPM3 as the standard for NPM3. The amounts of NPM1 and NPM3 were estimated to be ∼1 ng/60 oocytes (0.51 fmol/oocyte, top panel lane 8) and 0.1 ng/60 oocytes (0.09 fmol/oocyte, middle panel lane 8), respectively. Consistent with the previous report (5), NPM2 was abundantly expressed in oocytes, and its expression level was much higher than that in cumulus cells (lanes 7–10). We could not precisely determine the expression level of NPM2 in mouse oocytes by using the mouse NPM2 antibody and recombinant human NPM2 proteins generated in this study because the similarity between mouse and human NPM2 is not sufficiently high (the two proteins are 65% identical). However, it was recently reported that the amount of NPM2 is ∼75 pg (3.2 fmol)/oocyte (27). Taken together, these results support the idea that three NPM proteins are simultaneously expressed in oocytes and may play overlapping roles in sperm chromatin remodeling after fertilization.
Oligomerization of human NPM proteins
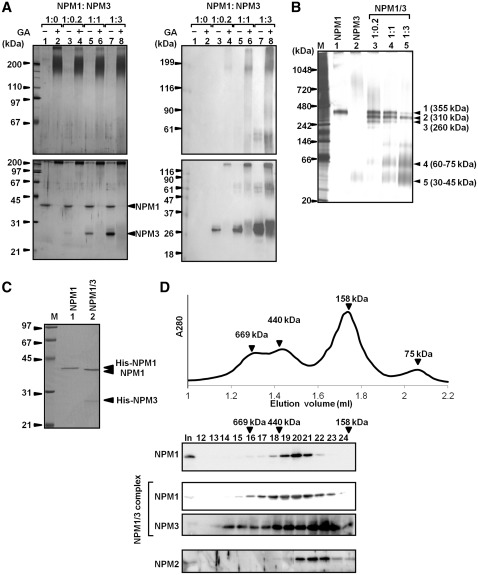
We next examined the oligomer formation ability of human NPM proteins. Bacterially expressed human NPM proteins were purified, and their oligomerization ability was tested using a chemical crosslinking assay (Figure 2A). NPM1 was subjected to SDS–PAGE and bands with 40 kDa and >200 kDa were detected even in the absence of chemical crosslinking (lane 1), indicating that NPM1 formed a stable oligomer. After glutaraldehyde (GA) treatment, ∼200- and 160-kDa protein bands were observed for NPM1 and NPM2, respectively (lanes 2 and 4). In addition, bands >200 kDa were also detected. This result suggested that human NPM1 and NPM2 both formed stable pentamers and that some of the pentamers formed decamers in solution. Conversely, the band corresponding to a pentamer was not detected when NPM3 was subjected to GA treatment (lanes 5 and 6). Instead, a band similar to that of the NPM3 monomer and a minor population of a 55–60 kDa bands were detected. To further confirm the oligomerization status of NPM proteins, the proteins were analyzed by BN–PAGE (Figure 2B). BN–PAGE was originally developed for the separation of mitochondrial membrane proteins and has been used to determine the native mass and oligomeric status of proteins (28). By comparison with the molecular standards separated on the same gel (Figure 2B, right panel), the native masses of NPM1, NPM2 and NPM3 were estimated to be 358, 231 and 42 kDa, respectively. In parallel, gel filtration analysis revealed that His–NPM1 and His–NPM2 were fractionated at the positions corresponding to approximately 370 and 270 kDa, respectively (Figure 6D). In addition, we examined the mass of NPM3 by gel filtration, but it distributed broadly and we could not estimate native mass (data not shown). From these results and previous structural analyses (9–11,13), we concluded that both human NPM1 (34.7 kDa including the His-tag) and NPM2 (26.3 kDa including the His-tag) formed stable pentamers in solution and that two pentamers are assembled into decamers. On the other hand, NPM3 (21.5 kDa including the His-tag) is suggested to from dimers in solution. Consistent with this data, preliminary results from electrospray ionization mass spectrometry analyses suggested that NPM3 formed a dimer at low salt concentrations (data not shown). NPM1 and NPM2 treated with GA and separated by SDS–PAGE were mainly detected at around pentamer positions, while these proteins were suggested to form decamers by BN–PAGE and gel filtration assays. This could be due to the assumption that the pentamer–pentamer interface is not efficiently crosslinked by GA, and two pentamers are dissociated under SDS–PAGE conditions.
Figure 2.
Oligomer formation of human NPM proteins. (A) Chemical crosslinking experiments. His-tagged NPM1, NPM2 and NPM3 (200 ng each) were treated without (lanes 1, 3 and 5) or with (lanes 2, 4 and 6) 0.05% glutaraldehyde (GA) and the fixed proteins were separated on 7.5% and 12.5% SDS–PAGE (top and bottom panels, respectively). Proteins were analyzed by western blotting with anti-His-tag antibody. Positions of molecular weight markers are indicated at the left side of the panels. (B) BN–PAGE analysis of NPM proteins. Recombinant His-tagged NPM1 (34.5 kDa), NPM2 (26.3 kDa) and NPM3 (21.5 kDa) (500 ng) were separated by 4–16% BN–PAGE and visualized with CBB staining. Lane M indicates molecular weight markers. The mobility of marker proteins was plotted as a function of their molecular weights (Mw) (right panel). The marker proteins were linearly separated and the masses of NPM proteins were estimated as shown at the right side of the gel. (C) Limited proteolysis of human NPM proteins. His-tagged NPM1, NPM2 and NPM3 were treated without or with increasing amounts of trypsin and incubated at 37°C for 5 min. Then the proteins were separated on 15% SDS–PAGE followed by CBB staining (Red) or western blotting with anti-His-tag antibody (Green). Positions of bands corresponding to the NPM core are indicated by arrow heads. CBB-stained gel and western blotting images are merged and shown in the top panel. (D) Alignment of the amino acid sequences of human NPM N-terminal domains. Amino acid sequences of human NPM1, NPM2 and NPM3 were aligned by ClastalW2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and conserved amino acids are highlighted by asterisks at the bottom of sequences. The conserved core domains of NPM proteins are indicated on blue background. The position of acidic ‘A1 tract’ is shown by red line at the top of the sequences.
Figure 6.
Oligomer formation between NPM1 and NPM3. (A) Crosslinking experiments. Recombinant His–NPM1 was mixed with increasing amounts of His–NPM3 (1:0 for lanes 1 and 2, 1:0.2 for lanes 3 and 4, 1:1 for lanes 5 and 6, and 1:3 for lanes 7 and 8), denatured in the buffer containing guanidine hydrochloride, and renatured by extensive dialysis. The mixtures were subjected to chemical crosslinking experiment with 0.05% GA. The mixtures treated without (lanes 1, 3, 5 and 7) or with (lanes 2, 4, 6 and 8) GA were separated on 7.5% and 12.5% SDS–PAGE (top and bottom panels, respectively) and visualized with silver staining (left panel) or western blotting with an anti-NPM3 antibody (right panel). Positions of molecular weight markers are indicated at the left, and those of free NPM1 and NPM3 are indicated at the right side of the panel. (B) BN–PAGE analysis of the NPM1–NPM3 complex. NPM1, NPM3 and NPM1–NPM3 complexes as in A were separated on 4–16% BN–PAGE and visualized with silver staining. Lane M is molecular weight markers. Masses of bands shown at the right side of the panel were estimated as in Figure 2B. (C) NPM1/His–NPM3 preparation. His-tagged NPM3 and NPM1 were coexpressed in E. coli and purified as described in ‘Materials and Methods’ section. His–NPM1 and NPM1–His–NPM3 complex were separated on 12.5% SDS–PAGE and visualized with CBB staining. Positions of His–NPM1, non-tagged NPM1 and His–NPM3 are indicated at the right side of the panel. (D) Gel filtration analysis of NPM proteins. His–NPM1 (5 µg), NPM1/His–NPM3 (5 µg NPM1) and His–NPM2 (5 µg) in 20 µl were loaded on Superose 6 PC 3.2/30 column and fractionated. Fractions 12–24 were analyzed by SDS–PAGE and western blotting. Molecular masses were estimated from the elution profile of marker proteins as shown at the top of the panel.
The N-terminal regions of NPM family proteins are folded into a compact domain containing an eight-stranded β-barrel (sequences colored by light blue in Figure 2D). Unlike other NPM proteins, NPM3 does not form a stable pentamer. Therefore, we tested whether NPM3 is folded into a monomeric structure similar to that found in other NPM proteins by conducting a limited proteolysis assay using trypsin (Figure 2C). Full-length His-tagged NPM1, NPM2 and NPM3 were digested with increasing amounts of trypsin, and digested proteins were analyzed by SDS–PAGE followed by Coomassie Brilliant Blue (CBB) staining (colored with red) or western blotting using the anti-His-tag antibody (green). CBB-stained gels clearly showed the appearance of distinct protein bands of approximately 20 kDa corresponding to the core domain after the trypsin treatment of His-tagged NPM1 and NPM2 (lanes 1–8). Moreover, distinct bands of approximately 20 kDa were also detected when His–NPM3 was treated with trypsin (lanes 9–12). Western blotting with anti-His-tag antibody indicated that these peptides did not contain N-terminal ends. Based on these observations, it is strongly suggested that the N-terminal domain of NPM3 is folded into a typical NPM core structure. Trypsin treatment of NPM3 produced distinct bands <20 kDa, as detected by western blotting with anti-His-tag antibody, those of which were not apparent with NPM1 and NPM2 (lanes 10–12). Given the molecular weight and the positions of lysine and arginine residues of the NPM3 sequence from the N-terminal His-tag, these fragments were likely to correspond to fragments digested within the loop structures between the β-sheets (i.e. K76, R87 and K102, Figure 2D). These loop structures are known to be involved in the monomer–monomer interactions in the pentamer or in pentamer–pentamer interactions in the decamer (11,13). These results support the hypothesis that NPM3 is folded into a NPM core structure although the loop regions were susceptible to trypsin because of their lack of pentamer/decamer formation ability.
Sperm chromatin remodeling by human NPM proteins
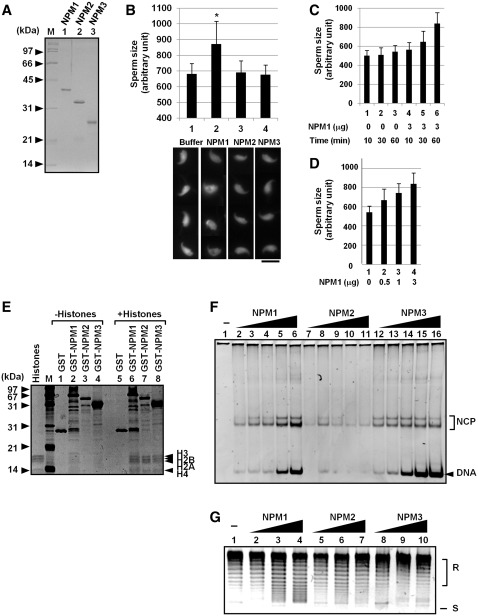
Three NPM proteins are simultaneously expressed in oocytes (Figure 1F). Next, we examined whether NPM proteins may have sperm chromatin remodeling activity. Sperm chromatin remodeling consists of sperm chromatin decondensation by protamine removal and nucleosome assembly. Therefore, we examined both of these activities using NPM proteins in vitro. To examine the sperm chromatin remodeling activity of NPM proteins, mouse sperm nuclei were prepared and incubated in the absence or presence of NPM proteins. The size of DAPI-stained nuclei was measured by Image J software. As shown in Figure 3B, sperm nuclear size significantly increased in the presence of NPM1, but not in the presence of NPM2 or NPM3. This NPM1 activity depended on its concentration and incubation time (Figure 3C and D). Although mouse NPM2 was previously reported to induce mouse sperm chromatin decondensation by incubation for 24 h (27), we did not observe a significant increase in sperm size within 1 h. These results suggest that an additional factor is required for NPM2 and NPM3 to induce efficient protamine removal, including complex formation with other NPM family proteins or post-translational modifications.
Figure 3.
Sperm chromatin remodeling by human NPM proteins. (A) Purified proteins. His-tagged NPM proteins were expressed in E. coli and purified. Proteins (200 ng each) were separated by 12.5% SDS–PAGE and visualized with CBB staining. Positions of molecular weight markers are indicated at the left side of the panel. (B) Sperm chromatin decondensation by NPMs. Mouse sperm nuclei (1 × 105) were incubated for 60 min without or with 3 µg of purified NPM1, NPM2, or NPM3. Sperm nuclei were fixed with formardehyde and stained with DAPI. Bar at the bottom of the panels indicates 10 µm. Four typical sperm nuclei are shown for each sample. Size of the sperm nuclei (n > 30) were estimated by Image J software and averaged. Results are means ± SD and statistical P-values were calculated and indicated * for P < 0.05. (C and D) Sperm chromatin decondensation by NPM1. Sperm nuclei were incubated in the presence or absence of NPM1 and incubated for 10–60 min as indicated at the bottom of the graph (C) or for 60 min (D). Size of sperm nuclei was analyzed as in (B). (E) Histone-binding activity of human NPM proteins. GST, GST-tagged NPM1, NPM2, and NPM3 (1 µg each) were mixed without (lanes 1–4) or with core histones (1 µg, lanes 5–8) purified from HeLa cells in the buffer containing 150 mM NaCl. The mixtures were recovered by glutathione sepharose beads. The proteins were separated by 15% SDS–PAGE and visualized with silver staining. Positions of molecular weight markers and core histones are shown at the left and right side of the panel, respectively. (F) Histone transfer assay. Increasing amounts of His-tagged NPM1, NPM2 and NPM3 (lanes 2–6, 7–11 and 12–16, respectively) (50, 100, 200, 300 and 400 ng for each protein) were incubated with core histones (100 ng) and incubated. Then the 147-bp DNA fragment (30 ng) were added and further incubated at 37°C for 30 min. The mixtures were separated by 6% PAGE in 0.5× TBE. DNA was visualized by staining with GelRed. Positions of free DNA and nucleosome core particle (NCP) are indicated at the right side of the panel. (G) Supercoiling assay. His-tagged NPM1, NPM2 and NPM3 (100, 300, and 1000 ng for lanes 2–4, 5–7 and 8–10) were incubated with core histones (100 ng). Then plasmid DNA (100 ng) treated with topoisomerase I was added to NPM-histone mixture and incubated. Plasmid DNA was purified and analyzed by 1% agarose gel electrophoresis in 1× TBE and visualized with GelRed staining. Positions of relaxed (R) and supercoiled (S) DNA are indicated at the right side of the panel.
We then examined the nucleosome assembly activity of human NPM proteins. We first examined the histone-binding activity of NPM proteins using GST-tagged human NPM proteins and core histones purified from HeLa cells (Figure 3E). GST alone did not precipitate histone proteins (lane 5), whereas four core histones efficiently precipitated with GST–NPM proteins (lanes 6–8). This indicated that all NPM proteins were similarly associated with histones under the assay conditions used. Xenopus nucleoplasmin (NPM2) is mainly associated with H2A–H2B dimers in egg extracts, but we observed that all NPMs were similarly associated with both histones H2A–H2B and H3–H4 in vitro (data not shown). We then sought to determine the nucleosome assembly activity of NPM proteins. Core histones were preincubated with increasing amounts of His-tagged NPM proteins (Figure 3A), mixed with a 147-bp DNA fragment, and incubated. The mixtures were separated by native polyacrylamide gel electrophoresis (Figure 3F). When histone alone was incubated with DNA, the DNA was present in large aggregates that could not enter the gel (lane 1). In contrast, when histones were preincubated with increasing amounts of NPM1, free DNA and NCP bands increased in a dose-dependent manner (lanes 2–6). The activity of NPM2 was also detected when its concentration was low, but DNA and NPC bands were decreased when NPM2 concentration was increased (lanes 7–11). This suggests that NPM2 randomly associates with DNA and/or DNA–histone complexes. When histones were preincubated with NPM3, DNA–histone aggregation decreased, and free DNA and NCP bands were clearly detected (lanes 12–16). To further verify the nucleosome assembly activity of NPM proteins, a supercoiling assay using relaxed plasmid DNA was conducted (Figure 3G). NPM1 mediated nucleosome assembly, and supercoil was introduced in plasmid DNA in a dose-dependent manner (lanes 2–4). In the presence of low concentration of NPM2 (lanes 5 and 6), supercoil was slightly introduced in plasmid DNA, and this weak activity was decreased in the presence of higher amounts of NPM2 (lane 7). In addition, we could not detect efficient nucleosome assembly activity of NPM3 (lanes 8–10) using supercoiling assay. Thus, we concluded that NPM3 binds to histones and inhibits DNA–histone aggregation, although this NPM3 activity is insufficient to form chromatin on long DNA as a histone chaperone. This is consistent with an earlier observation that the histone chaperone activity of NPM3 is significantly lower than that of NPM1 (29).
Effect of NPM2 phosphorylation on its sperm chromatin remodeling activity
We focused on the effect of post-translational modifications on NPM2 activity as a stimulatory mechanism for NPM2. The activity of Xenopus NPM2 was strictly regulated by phosphorylation during oogenesis and after fertilization (30). Mouse NPM2 is likely to be highly phosphorylated in metaphase II stage eggs and pronuclear stage zygotes (31). Thus, mammalian NPM2 function is also likely to be regulated by phosphorylation. Mitotic cell extracts prepared from HeLa cells and asynchronous HeLa cell extracts were used as kinase sources to mimic metaphase II stage eggs. A phosphorylated protein-specific dye, Pro-Q Diamond, was used to show that NPM2 proteins incubated with asynchronous and mitotic extracts were phosphorylated (Figure 4A, lanes 5 and 6). Based on the intensity of Pro-Q Diamond stained gel, NPM2 was most efficiently phosphorylated by mitotic extracts. This suggests that either kinases present in asynchronous and mitotic extracts differentially phosphorylated NPM2 or that the activity of a specific kinase in mitotic extracts was higher than that in asynchronous extracts. Because the histone transfer activity of recombinant NPM2 seemed to be inhibited by its non-specific DNA binding, we first examined the effect of NPM2 phosphorylation on its DNA-binding activity (Figure 4B). The 196-bp DNA fragment was incubated with increasing amounts of control or phosphorylated NPM2 proteins, and the complexes were separated by native PAGE. As expected, NPM2 bound DNA directly (lanes 2–5), and this DNA-binding activity was efficiently suppressed by phosphorylation with mitotic extracts (lanes 10–13). Next, we tried to identify the phosphorylation site(s) affecting NPM2 activity. Human NPM2 has typical cdc2 consensus serine residues at amino acids 159 and 196. In addition, N-terminal phosphorylation sites present in Xenopus nucleoplasmin/NPM2 are conserved in the human NPM2 protein. These sites were substituted by aspartic acid (D) to mimic phosphorylation status. NPM2 S159D, S196D, S159/196D (2D), 5D (S4, S5, S7, S8 and T9 were substituted by D) and 7D (combination of 2D and 5D) were incubated in the presence of interphase and mitotic extracts, and phosphorylation status was examined by Pro-Q Diamond staining (Figure 4C). NPM2 was efficiently phosphorylated by mitotic extracts (lane 2 and 9) and the phosphorylation was significantly reduced when two cdc2 consensus sites were substituted with D (lanes 10–12). N-terminal phosphorylation sites could be slightly phosphorylated by mitotic extracts (lanes 13 and 14), although these sites were not major phosphorylation sites in mitotic HeLa cell extracts. To examine the effect of phosphorylation of NPM2 on its activity, 2D, 5D and 7D mutants were tested for sperm chromatin decondensation activity (Figure 4D). Wild-type and 2D NPM2 proteins did not show significant sperm chromatin decondensation activity. In contrast, both 5D and 7D mutants slightly increased sperm nuclear size although the activity was lower than that of NPM1. The effect of NPM2 phosphorylation on its histone transfer activity was also examined (Figure 4E). NPM2 wild type did not efficiently transfer histones onto DNA fragments (lanes 1–5). The substitution of S159 and S196 with aspartic acids enhanced NCP formation (lanes 6–17). In addition, phosphomimetic mutation at the N-terminal region enhanced the NCP formation activity of NPM2 independently on S159 and S196 mutations (lanes 18–25). These results indicate that the phosphorylation of NPM2 at the N-terminal region plays a crucial role in sperm chromatin decondensation and chromatin assembly activities. Moreover, a kinase present in oocytes, but not in HeLa cells, and the cdc2 kinase are suggested to be required to fully activate the sperm chromatin remodeling activity of NPM2.
Oligomeric NPM3 may have histone chaperone activity
The low histone chaperone activity of NPM3 was presumably due to its lack of pentamer/decamer formation. To test this possibility, we constructed a chimeric protein in which the N-terminal core domain of NPM1 was fused with the C-terminal acidic region of NPM3 (NPM1–3Ch) (Figure 5A). SDS–PAGE clearly showed that both C-terminal truncated NPM1 (NPM1ΔC3) and NPM1–3Ch formed stable oligomers that could not be dissociated by SDS–PAGE sample buffer, and bands of approximately 100 and 70 kDa being detected in addition to monomer bands (lanes 3 and 4). Sperm nuclear size increased in the presence of NPM1, but not in the presence of NPM3 (Figure 5B). Sperm nuclear size increased and its structure was almost disrupted in the presence of the NPM1–3Ch protein. The supercoiling assay demonstrated that NPM3 and NPM1ΔC3 protein had a weak nucleosome assembly activity (Figure 5C, lanes 5–10). In contrast, the NPM1–3Ch protein showed a distinct nucleosome assembly activity (lanes 11–13). These results strongly suggest that the C-terminal region of NPM3 functions as a histone chaperone domain when it is included in the oligomer.
Figure 5.
NPM3 functions as a histone chaperone when it forms an oligomer. (A) Purified recombinant proteins. His-tagged NPM1, NPM3, NPM1ΔC3 and NPM1–3Ch (lanes 1–4, respectively, 200 ng each) were separated by 12.5% SDS–PAGE and visualized with CBB staining. Because the oligomers of NPM1–3Ch and NPM1ΔC3 were not completely disrupted under SDS–PAGE condition, two bands, 18- and 70-kDa bands for NPM1ΔC3 and 25 and 100-kDa bands for NPM1–3Ch, were detected. The protein amounts of NPM1ΔC3 and NPM1–3Ch were estimated by the sum of two bands. The proteins used are schematically represented at the right side of the panel. The NPM1–3Ch protein is a fusion of NPM1(1–120) and NPM3(146–178). (B) Sperm chromatin decondensation activity. Mouse sperm nuclei were incubated in the absence or presence of NPM proteins (3 μg) at 37°C for 60 min, fixed, and stained with DAPI. Three typical sperm nuclei are shown. (C) Supercoiling assay. Core histones were incubated without or with increasing amounts of NPM1 (lanes 2–4), NPM3 (lanes 5–7), NPM1ΔC3 (lanes 8–10) and NPM1–3Ch (lanes 11–13) (100, 300 and 1000 ng for each protein). Then the topoisomerase I-treated plasmid DNA (100 ng) was added and incubated. DNA was purified and separated on 1% agarose gel electrophoresis in 1× TBE. Positions of supercoiled (S) and relaxed (R) plasmid DNA are shown at the right side of the panel.
Effect of NPM3 on NPM1 function
NPM3 alone could not form homopentamers/decamers, but it was associated with NPM1 in cell extracts (Figure 1), suggesting that NPM3 formed a pentamer only with NPM1. To test this, purified NPM1 was mixed with increasing amounts of NPM3, denatured in guanidine hydrochloride and renatured by extensive dialysis. The oligomerization status of the renatured proteins was analyzed using a chemical crosslinking assay (Figure 6A). Proteins were separated by SDS–PAGE and visualized with silver staining and western blotting with an anti-NPM3 antibody (left and right panels, respectively). After GA treatment in the presence of NPM1, NPM3 was included in large complexes with molecular masses of approximately 180–200 kDa and >200 kDa (lanes 4, 6 and 8 in the right panel). The same proteins were also analyzed by BN–PAGE and visualized with silver staining (Figure 6B). NPM1 or NPM3 alone was detected at the position of approximately 350 kDa or 40 kDa, respectively (lanes 1 and 2). When the NPM1–NPM3 complexes were resolved by BN–PAGE, five main bands (bands 1–5 shown at the right side of the panel) were detected. The intensity of band 2 in addition to bands 4 and 5 increased when NPM3 ratio in the complex increased (lanes 3–5). The bands 4 and 5 corresponded to the NPM3 dimers and trimers that failed to associate with NPM1. The size of band 2 was estimated to be 310 kDa. i.e. similar to the size of the assembly of two His–NPM1 (34.5 kDa):His–NPM3 (21.5 kDa) (4:1) pentamers (319 kDa). These results suggest that NPM1 and NPM3 preferentially form pentamers at a ratio of 4:1, and two NPM1:NPM3 pentamers form a decamer in solution. However, it should be noted that the an oligomer with different stoichiometry was also detected on BN–PAGE (band 3). To further confirm oligomer formation between NPM1 and NPM3, His-tagged NPM3 and non-tagged NPM1 were coexpressed and purified with the Ni–NTA resin. Under this condition, all NPM1 molecules should be complexed with His-tagged NPM3. Purified proteins were further fractionated by Mono Q chromatography to remove excess His–NPM3 in the complex. The CBB-stained gel revealed that the ratio between NPM1:His–NPM3 in the complex was very similar to 4:1 (Figure 6C, lane 2). The complex was then subjected to gel filtration, and fractions were analyzed by western blotting (Figure 6D). As described previously, the masses of NPM1 and NPM2 were estimated to be 370 and 270 kDa, respectively. However, NPM1 was broadly distributed when complexed with His–NPM3 despite the masses of NPM1 peak fraction being similar to that of NPM1 alone. Taken together, we conclude that NPM1 and NPM3 preferentially form a 4:1 pentamer and two pentamers are assembled into decamers, and that NPM3 may have a role in destabilizing the NPM1 pentamer/decamer formation.
Next, we examined the function of the NPM1–NPM3 complex in sperm chromatin remodeling. As shown in Figure 7A, both NPM1 alone and the NPM1–NPM3 complex, but not NPM3, efficiently decondensed mouse sperm nuclei in incubation time-dependent manner. Similarly, the NPM1–NPM3 complex mediated nucleosome assembly, as did NPM1 (Figure 7B, lanes 2–4 and 8–10), while NPM3 did so less efficiently (lanes 5–7), indicating that NPM3 cooperates with NPM1 in the first and second steps of sperm chromatin remodeling during fertilization.
Figure 7.
Sperm chromatin remodeling activity of the NPM1–NPM3 complex. (A) Sperm chromatin decondensation activity of the NPM1–NPM3 complex. Sperm nuclei in the absence or presence of His–NPM1, NPM1–His–NPM3 complex (NPM1/3), His–NPM3 prepared as in Figure 6C were incubated for 30 or 60 min. Sperm DNA was fixed and stained with DAPI, and observed by fluorescent microscopy. Three typical sperms (from 60 min incubation samples) are shown for each sample. Sperm nuclear size (n > 30) was estimated by Image J and averaged. Results are means ± SD. (B) Nucleosome assembly activity of the NPM1–NPM3 complex. Increasing amounts of His–NPM1, NPM3, and the NPM1–His–NPM3 complex (NPM1/3) (200, 600 and 2000 ng, for lanes 2–4, 5–7 and 8–10) were mixed with core histones (100 ng). Topo I-treated plasmid DNA (100 ng) was added and further incubated. DNA was purified, separated on 1% agarose gel and visualized with GelRed staining. Positions of relaxed (R) and supercoilied (S) DNA are indicated at the right side of the panel.
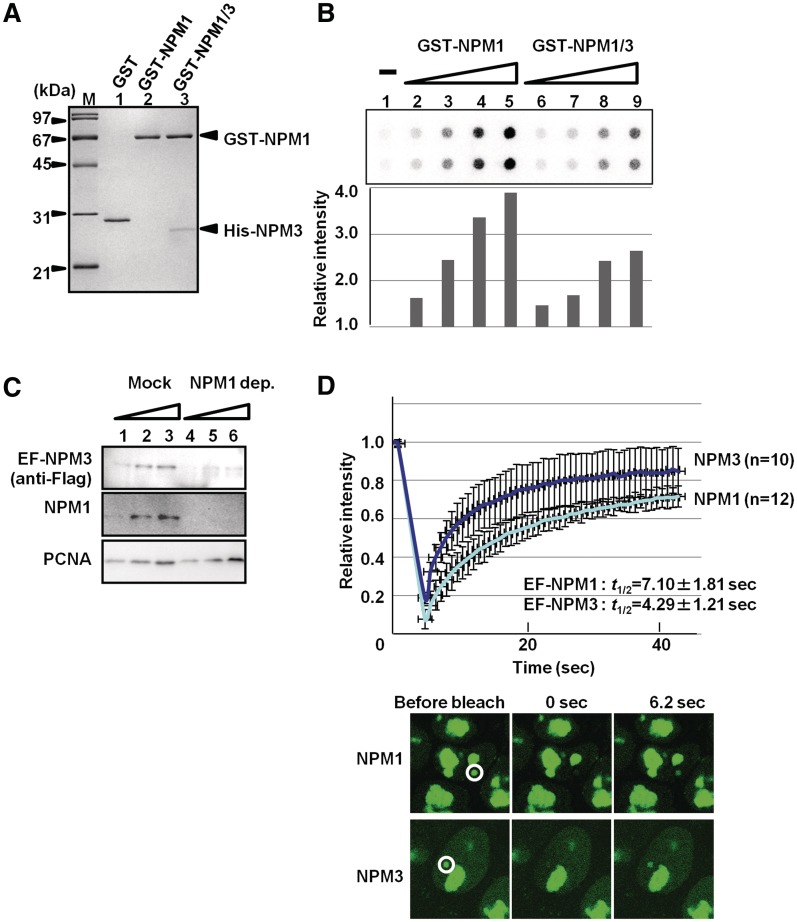
NPM1 and NPM3 are ubiquitously expressed, and their expression level is higher in actively growing cells than in quiescent cells. We therefore investigated the biological significance of NPM1 and NPM3 complex formation in growing cells. We previously demonstrated that RNA binding of NPM1 is required for efficient histone chaperone activity in the nucleolus (22). Thus, we tested the RNA-binding activity of the NPM1–NPM3 complex using a filter-binding assay (Figure 8B). The NPM1–NPM3 complex was prepared using GST–NPM1 and His–NPM3 (Figure 8A). GST–NPM1 alone or GST–NPM1 mixed with a 3-fold molar excess of His–NPM3 was denatured, and the proteins were refolded by extensive dialysis. Refolded proteins were further purified with glutathione sepharose to remove free His–NPM3 (lane 3). GST–NPM1 bound to radiolabeled RNA was retained on the membrane in a GST–NPM1 dose-dependent manner (lanes 2–5). The activity of the GST–NPM1/His–NPM3 complex was lower than that of GST–NPM1 alone (lanes 6–9). These results indicated that NPM3 incorporation into the NPM1 pentamer greatly impacted the RNA-binding activity of NPM1, although this incorporation did not completely abolish the RNA-binding activity of NPM1.
Figure 8.
Effects of NPM3 incorporation into the NPM1 pentamer on the NPM1 functions in somatic cells. (A) Purified GST–NPM1/His–NPM3 oligomer. GST–NPM1 was first purified and denatured in the absence or presence of three times molar excess of His–NPM3, the mixtures were dialyzed to refold the proteins and the excess His–NPM3 was removed by purification of the proteins with glutathione sepharose. GST, GST–NPM1 and GST–NPM1/His–NPM3 (lanes 1–3, respectively, 200 ng of GST proteins) were separated on 12.5% SDS–PAGE and visualized with CBB staining. (B) RNA-binding activity of the NPM1–NPM3 complex. Increasing amounts of GST–NPM1 and GST–NPM1/His–NPM3 were mixed with 32P-labeled total RNA purified from HeLa cells. The mixture was filtrated through nitrocellulose membrane. The membrane was extensively washed and the radio-active RNA retained on the membrane was detected by BAS2500 image analyzing system (top panel). Experiments were performed with doublet and the average radioactivity of the retained RNA is graphically shown at the bottom. The radio-active RNA incubated in the absence of NPM proteins (lane 1) retained on the membrane was set as 1.0 and relative amounts of retained RNA were measured. Two independent experiments demonstrated similar results. (C) Depletion of NPM1 from the extracts. NPM1 was immune-depleted from the extracts prepared from HeLa cells stably expressing EF–NPM3 using an anti-NPM1 antibody. Increasing amounts (1, 3 and 10 µg of proteins) of mock- and NPM1-depleted extracts (lanes 1–3 and 4–6, respectively) were separated by SDS–PAGE and analyzed by western blotting with anti-Flag, anti-NPM1 and anti-PCNA antibodies. (D) FRAP analysis of NPM1 and NPM3. HeLa cells stably expressing either EF–NPM1 or EF–NPM3 were grown on the glass-base dishes. FRAP analyses were performed as described in ‘Materials and Methods’ section. Small nucleoli as shown by white circle in the left panels were breached by a 488-nm laser and the fluorescent recovery at the bleached nucleoli was measured every 0.5 s. The fluorescence at the bleached area relative to the initial fluorescence (1.0) was calculated and plotted as a function of time. The data are represented as mean values ± SD from 12 and 10 experiments for NPM1 and NPM3, respectively. The t1/2 of fluorescence recovery (7.10 and 4.29 s for NPM1 and NPM3, respectively) was estimated by curve fitting as described in ‘Materials and Methods’ section. Typical FRAP images of EF–NPM1 and EF–NPM3 before bleaching, and 0 and 6.2 s after bleaching are shown at the bottom.
Given that the RNA-binding activity of NPM1 was important for its nucleolar localization and cellular mobility (32) and that NPM3 incorporation into the NPM1 pentamer decreased RNA-binding activity (Figure 8B), we hypothesized that NPM3 increases NPM1 mobility in the cell. To examine this, fluorescence recovery after photobleaching analysis was performed using HeLa cells that stably expressed EF–NPM1 or EF–NPM3. We confirmed that EF–NPM3 was efficiently depleted along with NPM1 from the nuclear extracts derived from HeLa cells that stably expressed EF–NPM3 (compare lanes 1–3 and 4–6 in Figure 8C). This result allowed us to test the mobility of a NPM1 oligomer containing NPM3 because NPM1 and NPM3 formed an oligomer (Figure 6) and most EF–NPM3 was complexed with NPM1 in cell extracts (Figure 8C). Thus, the observed mobility of EF–NPM3 localized in the nucleoli could represent the NPM1 complex containing EF–NPM3 molecule in the pentamer. For this experiment, the EGFP signal within a small nucleolus was targeted for bleaching with a 488-nm laser line. The intensity of the EGFP signal was measured every 0.5 s (Figure 8D). The recovery rate of NPM1 was rapid and the half time for fluorescence recovery (t1/2) was calculated to be 7.10 ± 1.81 s. Under the same experimental condition, the t1/2 of NPM3 was calculated to be 4.29 ± 1.21 s. These results strongly suggested that NPM3 incorporation into the pentamer increased the mobility of the NPM1 pentamer in the cell by restricting the RNA-binding activity of NPM1.
DISCUSSION
In this study, we characterized the sperm chromatin remodeling activity of three human NPM proteins. Sperm chromatin remodeling is divided into at least two reactions; the first step is chromatin decondensation by protamine removal, and the second step is assembly of nucleosomes with core histones. We systematically analyzed the function of human NPM proteins during both steps of sperm chromatin remodeling. We first demonstrated that NPM1 and NPM2 formed homo-pentamers in solution, whereas NPM3 mainly formed dimers (Figure 2). NPM3 showed significantly lower sperm chromatin decondensation and nucleosome assembly activities in vitro because of its inability to form a proper oligomer. Importantly, NPM3 efficiently mediated sperm chromatin remodeling when included in a pentamer containing NPM1 (Figure 7). The sperm chromatin remodeling activity of recombinant NPM2 was significantly lower than that of NPM1 (Figure 3). However, NPM2 activity was enhanced by phosphorylation at N-terminal region (Figure 4). These results suggest that all three NPM proteins can potentially remodel sperm chromatin. Furthermore, we also found that NPM3 enhanced the dynamics of NPM1 in somatic growing cell nuclei (Figure 8).
Oligomer formation by human NPM proteins
Unlike NPM1 and NPM2, NPM3 did not form stable homopentamers/decamers in solution (Figure 2). The NPM3 crystal structure has not been reported until date; therefore, it is difficult to understand why NPM3 cannot form a homopentamer. The trypsin digestion assay suggested that NPM3 forms a compact typical NPM core structure to form dimers or trimers in solution (Figures 2 and 6). NPM3 has conserved sequences such as the AKDE sequence and K-loop required for pentamer–pentamer interaction (Figure 2D) (10,11). Thus, dimer or trimer formation could be mediated by these sequences. However, given that the expression level of NPM3 was significantly lower than that of NPM1 in HeLa cells and mouse oocytes (Figure 1D and F) and NPM3 was efficiently depleted with NPM1 from cell extracts (Figure 8C), NPM3 mainly functions with NPM1 in cells. Crosslinking experiments and BN–PAGE data (Figure 2) suggested that NPM1 and NPM3 preferentially form a decamer consisting of two NPM1:NPM3 (4:1) pentamers. If a NPM3 molecule can form contacts with two NPM1 molecules, a NPM1–NPM3 (3:2) complex should be formed in addition to the (4:1) complex. However, we could not detect a clear band corresponding to the NPM1–NPM3 (3:2) complex by BN–PAGE (Figure 6B), suggesting that only one monomer–monomer interaction surface is likely to be active in NPM3. In addition, gel filtration analysis revealed that the NPM1–NPM3 complex was less stable than NPM1 alone (Figure 6D).
Nucleic acid-binding activity of human NPM2
We found that human NPM2 has potential DNA and RNA-binding activities (Figure 4B and data not shown) that are regulated by phosphorylation with mitotic extracts (Figure 4). In germinal vesicles (GV), the phosphorylation status of mouse NPM2 was found to be significantly lower than that of NPM2 in MII stage eggs (31). NPM2 is localized in the nucleoplasm and also in the nucleolus-like bodies (NLB) in GVs, but it is also distributed throughout the cell upon entry into the MII stage (5,27,31). This localization change is extremely similar to that found with NPM1 in growing somatic cells. NPM1 is mainly localized in the interphase nucleolus but becomes distributed throughout the cell during mitosis when the RNA-binding activity of NPM1 is suppressed by phosphorylation with cyclin B/cdc2 kinase (23). A NPM1 mutant mimicking mitotic phosphorylation status cannot stably localize in the nucleolus, even in interphase cells (32). Based on the relationship between the RNA-binding activity and nucleolar localization of NPM1, it is strongly suggested that NPM2 localization in NLBs is regulated by its nucleic acid-binding activity. The C-terminal basic clusters of mouse NPM2 were shown to be necessary for NLB localization in and formation of GVs (27). In addition, NPM2 possesses nuclear remodeling activity (3,4). Thus, we propose that the NLB localization of NPM2 in GVs is important for restricting any unfavorable remodeling of maternal chromatin.
Sperm chromatin remodeling by human NPM proteins
NPM1 and the NPM1–NPM3 complex efficiently mediated sperm chromatin decondensation and nucleosome assembly, whereas NPM2 activity was lower than those of NPM1 and the NPM1–NPM3 complex. Mouse and human NPM2 proteins do not possess the typical ‘A1 tract’, an acidic amino acid cluster, in the core domain, which is conserved in most NPM family proteins (Figure 2D). The A1 tract of Xenopus nucleoplasmin/NPM2 was shown to be crucial for sperm chromatin decondensation activity (33). In addition, the human NPM2 core domain alone did not efficiently bind to histones (9). This could be the reason why human NPM2 did not show efficient sperm chromatin remodeling activity. However, it is reasonable to speculate that NPM2 has sperm chromatin decondensation activity because sperm chromatin decondensation was reduced in NPM2 knock out mouse eggs (5,27), and the expression level of NPM2 was higher than those of NPM1 and NPM3 (Figure 1F). The N-terminal flexible region and the C-terminal basic region of nucleoplasmin (Xenopus NPM2) are phosphorylated in Xenopus eggs, and the phosphorylation of these two tail domains was found to be necessary for efficient sperm chromatin remodeling (30,34). Potential phosphorylation sites in the N-terminal tail domain and the C-terminal basic region are conserved in human NPM2 and phosphomimetic mutation at these sites significantly increased the sperm chromatin remodeling activity of NPM2 (Figure 4D and E). Thus, it is likely that the NPM2 activity is regulated by phosphorylation during oogenesis and after fertilization. Since the activity of phosphomimetic NPM2 mutants was lower than that of NPM1, it is possible that additional phosphorylation sites are involved in the stimulation of NPM2 activity.
Sperm chromatin decondensation occurs in NPM2 knockout mice eggs (5,27). It is therefore clear that additional factors are necessary for sperm chromatin remodeling. As shown in this study, NPM1 and NPM3 are good candidate sperm chromatin decondensation factors in fertilized eggs. Furthermore, NAP/SET family proteins (Nap1-like proteins and SET/TAF-I) are ubiquitously expressed and have potential sperm chromatin decondensation activity (21,35). Moreover, the histone chaperone HIRA and the chromatin remodeling protein CHD1 cooperatively mediate nucleosome assembly with histone H3.3 in the paternal genome (36,37). Given that sperm chromatin decondensation depends on the concentration of decondensation factor(s), the determination of the stoichiometry between these candidate factors should help elucidate the mechanism of this process.
NPM3 function in growing somatic cells
Our results clearly indicate that NPM3 can mediate sperm chromatin remodeling when included in a pentamer with NPM1 (Figure 7). Moreover, we found that NPM3 regulates NPM1 mobility in growing somatic cells (Figure 8). However, the exact cellular function of NPM3 has not been clarified. Transient expression of NPM3 inhibits the ribosome biogenesis function of NPM1 in a transformed monkey cell line (26). These results suggest that NPM3 is a negative regulator of cell growth, possibly through the inhibition of NPM1 function. In contrast, endogenous NPM3 is highly expressed in mouse ES cells, where the expression level is decreased after differentiation, and NPM3 overexpression in ES cells increases cell growth (25). We also found that NPM3 expression in normal human fibroblast cells is much lower than that found in cancer-derived cell lines, including HeLa and 293T cells (data not shown). Therefore, it seems likely that NPM3 expression is associated with cell growth rate. As shown in Figure 8, NPM1 mobility appeared to be increased by complex formation with NPM3. Nucleo-cytoplasmic shuttling of NPM1 was shown to contribute to the export of ribosome subunits (38–40), thereby decreasing cell growth. These results suggest that the nucleo-cytoplasmic shuttling of NPM1 is important for positive cell growth. In this regard, NPM3 may have a growth-associated function.
Finally, it should be mentioned that the NPM1–NPM3 complex functions as an active histone chaperone (Figure 6). NPM3 incorporation into the NPM1 oligomer increases NPM1 retention time in the nucleoplasm, and it is possible that the NPM1–NPM3 complex is involved in the regulation of chromatin structure at the specific gene loci driven by RNA polymerase II. NPM1 is associated with a variety of RNA polymerase II-regulated genes (41). Therefore, it would be interesting to test whether NPM1 and NPM3 cooperatively regulate the gene expression profile in somatic growing cells.
FUNDING
PRESTO from Japan Science and Technology Agency (JST); Special Coordination Funds for Promoting Science and Technology from JST; Grants–in–aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Special Coordination Funds for Promoting Science and Technology from JST.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Drs Alain Verreault (University of Montreal) and Martin Matzuk (Baylor College of Medicine) for reagents, and Keijiro Kato for his technical assistance. The authors also thank Dr Kensaku Murano (University of Tsukuba) for his help to prepare mouse sperms and MII oocytes.
REFERENCES
- 1.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 2.Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- 3.Tamada H, Van Thuan N, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, Wakayama T, Kikyo N. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sylvestre EL, Pennetier S, Bureau M, Robert C, Sirard MA. Investigating the potential of genes preferentially expressed in oocyte to induce chromatin remodeling in somatic cells. Cell Reprogram. 2010;12:519–528. doi: 10.1089/cell.2010.0012. [DOI] [PubMed] [Google Scholar]
- 5.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 6.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 7.McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 9.Platonova O, Akey IV, Head JF, Akey CW. Crystal structure and function of human nucleoplasmin (npm2): a histone chaperone in oocytes and embryos. Biochemistry. 2011;50:8078–8089. doi: 10.1021/bi2006652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 11.Namboodiri VM, Akey IV, Schmidt-Zachmann MS, Head JF, Akey CW. The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus. Structure. 2004;12:2149–2160. doi: 10.1016/j.str.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Namboodiri VM, Dutta S, Akey IV, Head JF, Akey CW. The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding. Structure. 2003;11:175–186. doi: 10.1016/s0969-2126(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 13.Lee HH, Kim HS, Kang JY, Lee BI, Ha JY, Yoon HJ, Lim SO, Jung G, Suh SW. Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface. Proteins. 2007;69:672–678. doi: 10.1002/prot.21504. [DOI] [PubMed] [Google Scholar]
- 14.Frehlick LJ, Eirín-López JM, Ausió J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Umekawa H, Olson MO. Expression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cells. Cell Mol. Biol. Res. 1993;39:33–42. [PubMed] [Google Scholar]
- 16.Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 2006;80:794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto K, Nagata K, Ui M, Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 1993;268:10582–10587. [PubMed] [Google Scholar]
- 18.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 19.Samad MA, Okuwaki M, Haruki H, Nagata K. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007;581:3283–3288. doi: 10.1016/j.febslet.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Sung MT, Cao TM, Coleman RT, Budelier KA. Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc. Natl Acad. Sci. USA. 1983;80:2902–2906. doi: 10.1073/pnas.80.10.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto K, Nagata K, Miyaji-Yamaguchi M, Kikuchi A, Tsujimoto M. Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins. Mol. Cell Biol. 1999;19:6940–6952. doi: 10.1128/mcb.19.10.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hisaoka M, Ueshima S, Murano K, Nagata K, Okuwaki M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol. Cell Biol. 2010;30:4952–4964. doi: 10.1128/MCB.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuwaki M, Kato K, Shimahara H, Tate S, Nagata K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motoi N, Suzuki K, Hirota R, Johnson P, Oofusa K, Kikuchi Y, Yoshizato K. Identification and characterization of nucleoplasmin 3 as a histone-binding protein in embryonic stem cells. Dev. Growth Differ. 2008;50:307–320. doi: 10.1111/j.1440-169X.2008.01034.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang N, Negi S, Szebeni A, Olson MO. Protein NPM3 interacts with the multifunctional nucleolar protein B23/nucleophosmin and inhibits ribosome biogenesis. J. Biol. Chem. 2005;280:5496–5502. doi: 10.1074/jbc.M407856200. [DOI] [PubMed] [Google Scholar]
- 27.Inoue A, Ogushi S, Saitou M, Suzuki MG, Aoki F. Involvement of mouse nucleoplasmin 2 in the decondensation of sperm chromatin after fertilization. Biol. Reprod. 2011;85:70–77. doi: 10.1095/biolreprod.110.089342. [DOI] [PubMed] [Google Scholar]
- 28.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 29.Gadad SS, Shandilya J, Kishore AH, Kundu TK. NPM3, a member of the nucleophosmin/nucleoplasmin family, enhances activator-dependent transcription. Biochemistry. 2010;49:1355–1357. doi: 10.1021/bi9021632. [DOI] [PubMed] [Google Scholar]
- 30.Bañuelos S, Omaetxebarria MJ, Ramos I, Larsen MR, Arregi I, Jensen ON, Arizmendi JM, Prado A, Muga A. Phosphorylation of both nucleoplasmin domains is required for activation of its chromatin decondensation activity. J. Biol. Chem. 2007;282:21213–21221. doi: 10.1074/jbc.M702842200. [DOI] [PubMed] [Google Scholar]
- 31.Vitale AM, Calvert ME, Mallavarapu M, Yurttas P, Perlin J, Herr J, Coonrod S. Proteomic profiling of murine oocyte maturation. Mol. Reprod. Dev. 2007;74:608–616. doi: 10.1002/mrd.20648. [DOI] [PubMed] [Google Scholar]
- 32.Negi SS, Olson MO. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J. Cell Sci. 2006;119:3676–3685. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- 33.Salvany L, Chiva M, Arnan C, Ausió J, Subirana JA, Saperas N. Mutation of the small acidic tract A1 drastically reduces nucleoplasmin activity. FEBS Lett. 2004;576:353–357. doi: 10.1016/j.febslet.2004.07.095. [DOI] [PubMed] [Google Scholar]
- 34.Leno GH, Mills AD, Philpott A, Laskey RA. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J. Biol. Chem. 1996;271:7253–7256. doi: 10.1074/jbc.271.13.7253. [DOI] [PubMed] [Google Scholar]
- 35.Ito T, Tyler JK, Bulger M, Kobayashi R, Kadonaga JT. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J. Biol. Chem. 1996;271:25041–25048. doi: 10.1074/jbc.271.40.25041. [DOI] [PubMed] [Google Scholar]
- 36.Loppin B, Bonnefoy E, Anselme C, Laurençon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 37.Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol. Cell Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, Weber JD. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol. Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shandilya J, Swaminathan V, Gadad SS, Choudhari R, Kodaganur GS, Kundu TK. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol. Cell Biol. 2009;29:5115–5127. doi: 10.1128/MCB.01969-08. [DOI] [PMC free article] [PubMed] [Google Scholar]