Abstract
Most restriction endonucleases, including FokI, interact with two copies of their recognition sequence before cutting DNA. On DNA with two sites they act in cis looping out the intervening DNA. While many restriction enzymes operate symmetrically at palindromic sites, FokI acts asymmetrically at a non-palindromic site. The directionality of its sequence means that two FokI sites can be bridged in either parallel or anti-parallel alignments. Here we show by biochemical and single-molecule biophysical methods that FokI aligns two recognition sites on separate DNA molecules in parallel and that the parallel arrangement holds for sites in the same DNA regardless of whether they are in inverted or repeated orientations. The parallel arrangement dictates the topology of the loop trapped between sites in cis: the loop from inverted sites has a simple 180° bend, while that with repeated sites has a convoluted 360° turn. The ability of FokI to act at asymmetric sites thus enabled us to identify the synapse geometry for sites in trans and in cis, which in turn revealed the relationship between synapse geometry and loop topology.
INTRODUCTION
Genetic processes often rely on DNA-binding proteins that interact with multiple target sites on DNA (1). These processes include DNA replication and repair (2,3), site-specific recombination (4,5), the initiation and regulation of transcription (6–9) and DNA cleavage by restriction enzymes (10–13). Most restriction endonucleases [REase(s)] cleave DNA substrates containing two or more copies of their recognition site more rapidly than DNA with a single site. The Type I and the Type III REases need two sites because they operate by 1D tracking mechanisms from one site to another along the DNA (14). The Type II enzymes, on the other hand, act at fixed loci relative to their recognition sites (15) and those that need two sites for full activity bind both sites at the same time, spanning the distant loci through 3D space. A protein that binds two DNA sites does so more readily with sites in cis on the same molecule of DNA, than with sites in trans on two unconnected molecules, simply because sites on the same molecule will almost always be in closer proximity than sites on different molecules (6,12). On the two-site DNA, the intervening DNA is held in a loop, while interactions in trans require the separate DNA molecules to be held together in a synaptic complex. The Type II nucleases that need two sites for optimal activity thus make excellent test systems to study protein-mediated DNA looping (12,13,16–19).
Many Type II REases are homodimeric proteins that recognize palindromic DNA sequences and cut both strands at equivalent positions within the site (20). Such enzymes interact symmetrically with their sites so that the contacts from one subunit of the protein to one half of the recognition site are duplicated by the second subunit with the other half (21,22). The Type II enzymes that require two copies of a palindromic sequence commonly act as tetramers with two DNA-binding clefts, each formed from two subunits like a dimeric enzyme at an individual site (23–28). On account of the symmetry within the recognition sequence, the DNA-binding surfaces can accommodate the segments of DNA carrying the sequence in either possible orientation (29). Nevertheless, a large number of the Type II REases recognize asymmetric rather than palindromic sequences and cleave the DNA at fixed loci outside of the site (20). FokI is perhaps the best known example (30). Almost all of the enzymes that act at asymmetric sites, including FokI (31,32), cleave two-site substrates more rapidly than DNA with a single site (33,34).
The FokI REase recognizes the non-palindromic sequence, 5′–GGATG(9/13)–3′, and in the presence of Mg2+ cleaves both strands at non-equivalent positions downstream of the site, 9 nt away in the strand shown (the ‘top’ strand) and 13 nt away in the complementary (‘bottom’) strand (30). No cleavage is observed with Ca2+, though Ca2+ promotes the assembly of the cognate complex at the recognition site (31). FokI is a monomer of 66 kDa in free solution (35) and when bound to DNA in the absence of metal ions (31). It contains two domains connected by a flexible linker: an N-terminal DNA recognition domain that binds but cannot cleave the cognate site; and a C-terminal catalytic domain that can act as a non-specific nuclease (36,37). Mutational studies indicated that the monomer harbours a single active site for phosphodiester hydrolysis (38). A crystal structure of FokI bound to specific DNA in the absence of metal ions showed it did indeed possess two domains but that its catalytic domain, containing the solitary active site, was located beside the recognition sequence rather than the downstream cleavage loci (39).
To cut both DNA strands, the catalytic domain of the monomer bound to the recognition site associates with the catalytic domain of a second monomer to form a dimer with two active sites (40,41). The monomer bound directly to the specific site via its DNA recognition domain cuts the scissile bond in the bottom strand, 13 nt away from the site, while the second monomer held at that site via protein–protein interactions cuts the top strand (42). However, the catalytic domain of the DNA-bound protein moves from its initial position next to the recognition site to its target phosphodiester bond before capturing the second monomer: the dimerization event thus occurs not at the recognition site itself but at the downstream loci for DNA cleavage (43). On DNA molecules with one FokI site, the recruited monomer is supplied in trans, from either free solution (41) or from a second DNA (31). On account of the small surface area of the dimer interface (40), the interaction in trans is weak so the DNA-bound protein is seldom in its active dimeric state (32). In contrast, on DNA molecules with two or more FokI sites, two monomers bound in cis are tethered together and associate readily to the dimer, trapping the intervening DNA in a loop (32,44). The two proteins bound to a two-site substrate are thus often in the active dimeric state and so cleave that DNA more rapidly, at least at one site, than a DNA that has only one site.
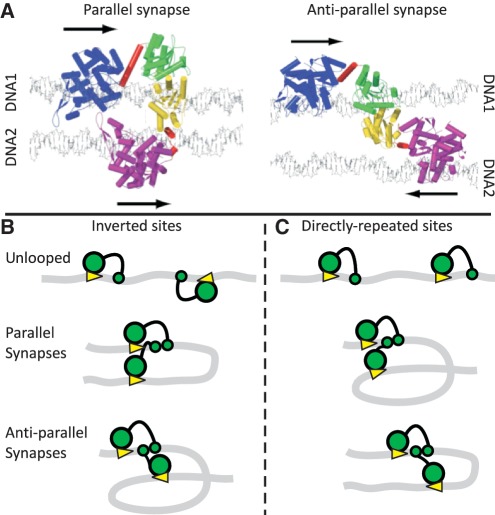
Most studies of systems spanning two DNA sites have employed proteins that recognize rotationally symmetric sequences (6–9,16–19,23–29), which are functionally identical in left-to-right and right-to-left orientations. In contrast, FokI recognizes an asymmetric sequence that has an inherent directionality. Thus, unlike SfiI or Lac repressor sites for example, two FokI sites in trans can be juxtaposed against each other in two distinctly different arrangements, either parallel or anti-parallel (Figure 1). Models have been constructed for the synaptic complex for FokI containing two monomers of the protein and two cognate DNA duplexes, with the catalytic domains from both monomers positioned to make a double-strand break on one of the two duplexes [(31) Figure 1A]. By reconfiguring the flexible linker between the catalytic and recognition domains, both parallel and anti-parallel arrangements are possible. In the anti-parallel model, the two protein subunits are in similar conformations and only minimal rearrangements are needed to move the two catalytic domains from one duplex to the other. In contrast, the parallel model requires radically different conformations for the linker regions in the two monomers and thus extensive conformational changes within both monomers to move the catalytic domains from one site to the other. The anti-parallel model might thus seem more plausible but, while both arrangements retain the dimer interface between the catalytic domains (40), the parallel scheme also features a protein–protein interaction between the two recognition domains (31).
Figure 1.
Synaptic complexes and loop topologies. (A) Shown here (with permission from Elsevier Press) are the models of the FokI synapse from Vanamee et al. (31). The complexes feature two monomers of the FokI endonuclease holding together two DNA duplexes (1 and 2) in either parallel or anti-parallel arrangements, with the catalytic domains from the two monomers each positioned to cut one strand of DNA1: the arrows denote the 5′→3′ direction of the recognition sequences (5′-GGATG-3′). The DNA recognition and catalytic domains of the protein bound to DNA1 are in blue and green, respectively, while the equivalent domains of the protein bound to DNA2 are in purple and yellow: α-helical segments are marked as cylinders. In both monomers, the linker region is in red. Note that in the model of the parallel synapse, the linker region of the protein bound to DNA1 contains a long α-helix, which is snapped into two for the protein bound to DNA2. (B and C) The DNA molecules (grey ribbons) carry two recognition sites for FokI (yellow arrowheads, pointing towards the downstream sites for DNA cleavage), in either inverted (IF) or directly repeated (DF) orientations along the DNA. The recognition and catalytic domains of the FokI monomers bound to each site are shown as large and small green circles, respectively. (B) On an IF DNA, juxtaposition of the sites in the parallel manner must bend the intervening DNA through 180° while juxtaposition in the anti-parallel alignment requires it to be bent through 360°. (C) The converse applies to a DF DNA: the parallel synapse traps a convoluted 360° loop and the anti-parallel a simple 180° loop.
For the synapsis of two FokI sites in trans, the sites will become aligned in whichever arrangement from the parallel and anti-parallel options has the lower free energy. Cryo-EM studies of FokI bound to short oligoduplexes revealed a croissant-shaped object that could be reconciled more readily to the parallel rather than the anti-parallel alignment (45). However, the synapsis of sites in cis may be governed by other factors. On a DNA with two FokI sites, different outcomes can arise depending on whether the sites are present in inverted (head-to-head) or directly repeated (head-to-tail) orientations along the DNA (Figure 1B and C): substrates with two inverted FokI sites are noted as IF and those with two directly repeated FokI sites as DF. On an IF substrate, a parallel synapse traps a simple 180° loop, while an anti-parallel synapse requires the intervening DNA to be bent through 360° (Figure 1B). The exact opposite applies to a DF substrate: the parallel alignment of FokI sites gives the convoluted 360° loop and the anti-parallel arrangement the simple 180° bend (Figure 1C). Consequently, if FokI can form only one particular synapse, either the parallel or the anti-parallel but not both, it must trap loops of different topologies on the IF and the DF DNA. On the other hand, if one loop topology possesses a substantially lower free energy than the other, then FokI will need to be able to synapse sites in either parallel or anti-parallel alignments in order to act both on inverted and on directly repeated sites. Yet, FokI cleaves both IF and DF substrates readily (32,33). The question is thus whether a fixed-synapse geometry dictates alternate loop topologies or whether a fixed-loop topology requires alternate synapse geometries.
This study sought to find out first if the FokI endonuclease can form alternate synaptic complexes, one with the sites held in parallel and another with anti-parallel sites. Or is it limited to a unique complex with the sites held in one particular arrangement? If the latter applies, the requisite alignment needs to be identified. These questions were addressed by using DNA substrates tagged at one or both ends with fluorescent labels, so that the synapsis of two FokI sites places one label in close proximity to the other and gives rise to fluorescence resonance energy transfer (FRET). FRET had been used previously to record DNA looping by the Lac repressor (46). In this study, synapsis in trans was detected by attaching different labels to samples of DNA with one FokI site, while synapsis in cis was examined on DNA with two sites with a different label at each end. A further objective was to see if the loops trapped between IF or DF sites have the same or different topologies. This was achieved by the single-molecule technique, tethered particle motion (TPM). In TPM, a DNA molecule is used to tether a polystyrene bead to a glass slide: the position of the bead is then monitored over time with an inverted microscope (7,9,18,19,28). The range of positions that the bead can reach due to Brownian motion is governed by the end-to-end length of the DNA tether, so any reduction in this length—from, for example, sequestering a segment of the DNA in a loop—causes a corresponding reduction in the range of motion of the bead. The TPM method was applied here to look for differences between the loops formed on IF and DF substrates. In the following article (47), the TPM studies were extended to analyse the kinetics and the thermodynamics of loop capture and release.
MATERIALS AND METHODS
Proteins and buffers
The FokI endonuclease was purified from an over-producing strain of Escherichia coli (from Dr W.E. Jack, New England Biolabs) and stored at −20°C as described previously (32). Protein concentrations were evaluated from A280 readings using a molar extinction coefficient calculated for the monomeric form of FokI. Immediately prior to each experiment, it was diluted to the requisite concentration in the buffer for that experiment.
DNA cleavage reactions were done in M-buffer [20 mM Tris–acetate (pH 7.9), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT], while DNA binding was studied in C-buffer (the same as M-buffer but with 2 mM CaCl2 in place of the magnesium acetate). CC-buffer is C-buffer supplemented with 100 μgċml−1 α-casein.
DNA
The plasmid pIF190 was described previously (32): it carries two FokI sites in head-to-head orientation 190 bp apart. A derivative of pIF190 with one FokI site, pF1, was constructed by using standard recombinant DNA methods (48) to replace the segment of DNA carrying its second FokI site with an identical segment that differed only at the recognition sequence: GTGCG in place of GGATG. Standard methods were also used to construct a plasmid, pDF190, with two FokI sites 190 bp apart but in directly repeated orientation. The FokI sites on all of these plasmids (and the mutated site on pF1) were flanked by identical sequences for at least 5 bp upstream of the site and for all the downstream sequence to 6 bp beyond the cleavage site: changes to the intervening sequence outside of the segment with the FokI site were kept to a minimum. Lengths between sites were counted from the base pair immediately following one site to the base pair immediately preceding the next site.
Oligonucelotides tagged at their 5′-termini with an Alexa Fluor dye (Invitrogen), either Alexa Fluor 546 (Ax546) or Alexa Fluor 647 (Ax647), were obtained from Purimex (Grebenstein, Germany) as samples that had been HPLC purified both before and after conjugation with the dye. Other oligonucleotides, including those 5′-tagged with biotin or digoxigenin (DIG) were from Eurofins MWG Operon.
To generate linear substrates, the region of each of the above plasmids encompassing the FokI site(s) was amplified by PCR using forward and reverse primers tagged at their 5′-ends with an appropriate label. The constructs carrying one recognition site were derived from pF1, and those with two sites 190 bp apart in either inverted or directly repeated orientations from pIF190 and pDF190, respectively. For the FRET experiments, either one or both primers carried at their 5′-ends the requisite fluorophore, either Ax546 or Ax647. In some instances, the DNA was labelled with one dye at one end only and mixed with a second DNA labelled with a different dye, again at one end only. In other cases, the DNA was labelled with the two different dyes, one at each end. The priming sites for the FRET substrates yielded PCR products of 260 bp, with 30 bp of DNA between each FokI site and its proximal terminus. For the TPM experiments, one primer carried a biotin label and the other a DIG, to give a DNA with biotin at the 5′-end of one strand and DIG at the opposite 5′-end on the complementary strand. The priming sites for the TPM substrates yielded PCR products of 536 bp, with the two FokI sites almost equidistant from the mid-point, 166 and 170 bp from their proximal ends. PCR products were purified using a QIAquick PCR purification kit (Qiagen) and their concentrations determined by A260 measurements.
FRET
Equal volumes of FokI and DNA, both in C- or in M-buffer, were mixed in a Hi-Tech Scientific SF61-DX2 stopped-flow fluorimeter (TgK Scientific Ltd., UK) at 20°C. After mixing, the total DNA concentration was 25 nM, while the enzyme was kept equal to the concentration of FokI recognition sites: i.e. 25 nM enzyme for reactions on DNA with one site and 50 nM enzyme for reactions on two-site DNA. The DNA was appropriately labelled with Ax546 and/or Ax647. Excitation of Ax546 was at 545 nm and emission observed through a Schott RG645 long-pass filter, which cuts off wavelengths <645 nm and so excludes most of the emission from Ax546, while transmitting emission from Ax647. Any signal from this set-up is thus due primarily to FRET between the Ax546 and Ax647 dyes. Data were recorded for 60 s but the reactions generally reached their end-points within 10 s. Each record shown is the average of at least five transients.
TPM
TPM experiments were performed by attaching the DIG end of a 536 bp PCR product to a glass cover slip that had been coated with Anti-DIG (Roche) and the biotin end to a 440 nm streptavidin-coated polystyrene bead (18). The cover slips were mounted in flow cells held at 20°C (47) through which CC-buffer was flushed followed by the FokI enzyme at 5 nM. The motion of up to 50 beads were tracked simultaneously at a frame rate of 50 Hz and converted in real time to bead positions (x, y). The data were analysed by converting the positions to a root mean square (RMS) motion of the bead {√[(x − xm)2 + (y − ym)2], where xm and ym are the mean values averaged over 100 frames}. The histogram of this motion was fitted to either a single or a double Gaussian to find the RMS values of the looped and the unlooped states (19).
RESULTS
FRET from synapsis in trans
Two fluorophores can give rise to FRET provided the emission spectrum of one, the donor, overlaps the excitation spectrum of the other, the acceptor and that the two dyes are in close physical proximity, typically <100Å apart (49). To see if FRET could be used to detect the synapsis of DNA sites in trans, pairs of DNA molecules carrying a single FokI site were generated: in each pair, one was labelled with Ax546 and the other with Ax647 (Figure 2). These two dyes have overlapping spectra and constitute a FRET pair with a Förster radius (R0) of 74 Å (50). If the synapsis of the FokI sites brings the Ax546 moiety on one DNA into close proximity of the Ax647 on the second DNA, to within a distance approaching the R0, the excitation of the donor will lead directly to emission from the acceptor due to FRET without any direct excitation of the latter. This possibility was examined by mixing rapidly in a stopped-flow fluorimeter one solution of FokI endonuclease with another containing the pair of DNA molecules labelled with their respective fluorophores. The buffer contained Ca2+ rather than Mg2+, to permit the formation of synaptic complexes while preventing DNA cleavage (31). Following excitation of the Ax546 dye, emission from Ax647 was recorded over time (Figure 2).
Figure 2.
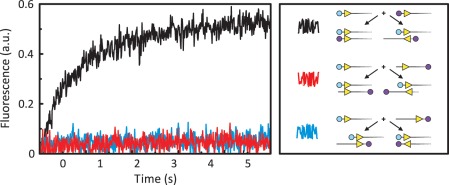
FRET from synapsis in trans. Equal volumes of FokI endonuclease and a solution containing two DNA molecules were mixed in the stopped-flow fluorimeter to give reactions in C-buffer at 20°C with FokI at 25 nM and both DNA at 12.5 nM. Excitation was at 545 nm and the change in emission (in arbitrary units: a.u.) at >645 nm recorded over time. The black, red and blue traces each came from a reaction with a particular pair of DNA molecules, as indicated in the right-hand panel. The DNA molecules were 260 bp long and contained a single recognition site for FokI (yellow arrowhead) 30 bp from the nearest end of the DNA: the arrowheads mark the orientation of the recognition sequence by pointing towards the downstream sites for DNA cleavage. For each experiment, one of the DNA molecules was labelled at one end with Ax546 (cyan circles) whereas the other DNA was labelled with Ax647 (mauve circles) at either the same or the opposite end. The pairs of the substrates that gave rise to the black, the red and the blue trace are depicted next to the squiggle of the same colour. The schemes below each pair show the structures formed by parallel and anti-parallel synapses of the FokI sites on both substrates within that pair: the relative locations of the Ax546 and Ax647 dyes are indicated.
In each pair, both DNA molecules were 260 bp long and contained one copy of the recognition sequence for FokI, located 30 bp from one end of the chain (as noted in Figure 2). The Ax546 fluorophore on one DNA was at the 5′-end of its top strand (the left-hand end) while the Ax647 label on the second DNA was at either the left- or the right-hand end. The relative positions of the recognition sites and the fluorescent labels on each DNA are shown in Figure 2 (right-hand panel).
In the first pair (Figure 2, black record), both substrates carried the FokI site near the left-hand end and were labelled at that end, with Ax546 and Ax647, respectively. If FokI can align these two DNA molecules in parallel, the Ax546 on one DNA might lie close enough to the Ax647 on the other to allow for FRET. On the other hand, if FokI forms exclusively anti-parallel synapses, the two fluorescent labels will be held physically distant from each other, on opposite sides of the synaptic complex, too far apart for FRET. All of the DNA constructed here contain ∼45 bp between the location of the synapse (the cleavage loci) and the nearest DNA end, so termini held on opposite sides of the complex must be the equivalent of at least 90 bp (>300 Å) apart. On adding FokI to this mixture of DNA molecules and exciting the Ax546 dye, the emission from the Ax647 label increased over a 0–5 s time scale, with first-order kinetics. But when a mutant of FokI incapable of dimerization was added to these two DNA species, no increase in Ax647 emission was observed (data not shown). The FRET thus depends on the association of two monomers of FokI, each bound to a DNA, to form a synaptic complex containing the dimeric protein holding together two copies of its target site in trans. Moreover, the FRET signal with this pair can only come from a parallel synapse. It has, however, yet to be established whether it can only form parallel synapses, since this particular pair would not generate a FRET signal from an anti-parallel synapse.
The second pair (Figure 2, red trace) differed from the first only in the relocation of the Ax647 dye from the terminus proximal to the FokI site to the distal end. However, in this case, both the parallel and the anti-parallel synapsis of FokI sites leave the labels on the two DNA molecules too distant from each other for FRET. This combination thus constitutes a negative control for the other pairs. As expected, no increase in Ax647 emission was seen.
In the third pair (Figure 2, blue trace), one DNA molecule had both the FokI site and the fluorescent label close to (or at) the left-hand end of the DNA, whereas the other had both recognition site and label towards the right-hand end. In this case, a parallel synapse of the FokI sites on the two separate DNA molecules ought not to yield a FRET signal as the two dyes would be located on opposite sides of the complex. Conversely, an anti-parallel synapse from this pair should place the Ax546 dye attached to one DNA on the same side of the complex as the Ax647 dye on the other DNA and so might generate a FRET signal. This arrangement is thus the opposite of the first pair, which had the potential to give rise to FRET from a parallel but not an anti-parallel synapse. While an increase in FRET was observed with the first pair (Figure 2, black trace), no such increase was seen with the latter pair (Figure 2, blue trace). Hence, while FokI can hold together two copies of its recognition site in trans in the parallel arrangement, the data provide no support for the view that it might also be able to hold sites in the anti-parallel arrangement.
FRET from synapsis in cis
The synapsis of two DNA sites in cis, on the same molecule of DNA, traps the intervening DNA in a loop, with different outcomes depending on whether the sites are aligned in parallel or anti-parallel and whether they are in directly repeated or inverted orientations (Figure 1). Two substrates were constructed to examine the synapsis of FokI sites in cis (Figure 3). Both were 260 bp long with two FokI sites 30 bp from each end; i.e. 190 bp apart. Both were labelled at one end with the donor fluorophore Ax546 and at the other with the acceptor Ax647. The only significant difference between them was the relative orientation of the two FokI sites: in one case, inverted (head-to-head); in the other, directly repeated (head-to-tail). On the IF DNA (Figure 3, black record), the formation of a parallel synapse should leave the donor at one end of the DNA close to the acceptor at the other end and thus permit FRET, while an anti-parallel synapse would hold the two labels too far apart for FRET. The reverse applies to the DF DNA (Figure 3, red record). In this case, a parallel alignment should leave the labels out of range for FRET, while the anti-parallel scheme could lead to FRET.
Figure 3.
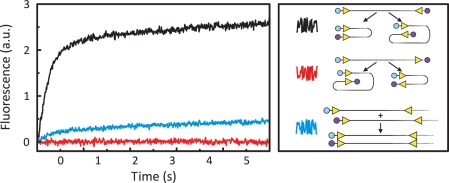
FRET from synapsis in cis. Equal volumes of FokI endonuclease and a solution of the requisite DNA(s) were mixed by stopped-flow to give reactions in C-buffer at 20°C containing DNA at a total concentration of 25 nM and FokI at 50 nM. Excitation was at 545 nm and the change in emission (a.u.) at >645 nm recorded over time. The black, red and blue traces each come from reactions with the DNA molecule(s) indicated in the right-hand panel. The DNA constructs were 260 bp long and contained two FokI sites (yellow arrowheads, marking their directionality) 30 bp from each end, in inverted or in directly repeated orientations. The DNA was labelled with Ax546 (cyan circle) at one end and with Ax647 (mauve circles) at either the opposite or the same end. The DNA substrate(s) that gave rise to the black, red and blue traces are depicted next to the squiggle in the same colour. The schemes below each set of DNA substrate(s) show the structures formed by parallel and anti-parallel synapses of the FokI sites within that set: the relative locations of the Ax546 and Ax647 dyes are indicated. (For the set of substrates in the blue record, only one of the many possible synapses is shown, a parallel assembly).
When FokI was mixed with each two-site substrate in the presence of Ca2+, a large increase in acceptor emission was observed with the IF construct but no increase was detected with the DF construct (Figure 3, black and red traces, respectively). Hence, FokI can hold together two copies of its recognition site from separate locations in the same DNA in a parallel assembly. The IF DNA labelled at both ends (Figure 3, black) delivered a much larger increase in Ax647 emission than that from a parallel synapse on the pair of one-site substrates each labelled at one end (Figure 2, black). The larger amplitude may reflect the fact that two sites in cis can be bridged more readily than sites in trans. However, the concentrations of the fluorescent dyes in the reactions on the two-site constructs were higher than those on the one-site species, even though both sets employed the same overall concentration of DNA. On the two-site substrates, both the Ax546 and Ax647 labels—one at each end—are present at the same concentration as the DNA. In contrast, for the reactions on pairs of one-site substrates, one half of the DNA molecules carry the Ax546 label and half the Ax647 dye, so the one-site mixtures should yield at most half the fluorescence from Ax647 as the two-site constructs.
The increase in FRET on the IF substrate (Figure 3, black trace) occurred into at least two kinetically distinct phases; a fast phase lasting for about 1 s (k1 = 4 s−1) followed by a slower phase that took >5 s to approach its end-point (k2 = 0.2 s−1). One explanation for the biphasic response is that the fast phase originates from the intramolecular DNA looping reaction bridging sites in cis, while the slow phase is due to intermolecular interactions spanning two labelled DNA molecules in trans. To see if in trans interactions make any contribution to the signal obtained with the IF DNA, one sample of this DNA was labelled at its left-hand end with the Ax546 donor and another sample labelled at the same end with the Ax647 acceptor (Figure 3, blue record). Since both of these two-site substrates were labelled at one end only, interactions spanning the sites in cis cannot give a FRET signal but FRET can come from interactions bridging sites in trans, as seen before with the one-site substrates (Figure 2, black record). Upon adding FokI to a solution containing equal concentrations of the donor- and the acceptor-labelled IF DNA, the emission from the Ax647 dye increased, albeit to a lower extent than that observed with the construct labelled at both ends (Figure 3, blue trace). The two-site substrates can thus form complexes in trans as well as in cis, and the former must contribute to the overall signal. The rate of the FRET change from the reaction in trans was, however, much slower than the fast phase seen with the construct labelled at both ends and was instead similar to that with the one-site substrates (Figure 2). Hence, the fast phase seen with the two-site DNA labelled at both ends can be assigned to FokI looping sites in cis and the slow phase to bridging interactions in trans. Nevertheless, the loop capture rate measured here by FRET (k1 = 4 s−1) is faster than that obtained in the following paper (47) by TPM on a DNA with the same 190 bp spacing between the FokI sites (1 s−1). The difference may be due to the fact that the FRET experiments were carried out on an unconstrained DNA, dynamically mobile in free solution, while the TPM studies employed DNA molecules immobilized at both ends, by attaching one to the glass surface and the other to a 440 nm bead.
No increase in Ax647 emission was observed with the DF substrate (Figure 3, red trace). However, if FokI had formed a parallel synapse with directly repeated sites, as it had with sites in inverted orientation, no increase in FRET would be observed since this would leave the two fluorophores far apart across the complex (Figure 3, red record). Nevertheless, these experiments essentially exclude the possibility that FokI sites can be held together in an anti-parallel alignment since this would have placed the two labels on the DF DNA sufficiently close together for FRET. The anti-parallel synapse with directly repeated sites and the parallel synapse with inverted sites should both trap 180° loops (Figure 1) and leave the two dyes a similar distance apart, yet only the IF construct yielded a FRET signal. The possibility that the lack of FRET from the DF DNA was due to one or both labels being defective was excluded: the Ax546 dye at the left-hand end of the DF DNA gave a FRET signal when paired in trans with a second DNA tagged with Ax647, and likewise the Ax647 label at the other end when paired with a DNA carrying Ax546 (Supplementary Figure S1).
DNA cleavage from synapsis in trans and in cis
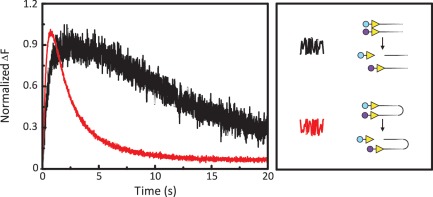
The FRET signals from adding FokI to DNA carrying the appropriate fluorescent labels in the presence of Ca2+ show that, at equilibrium, stable synaptic complexes are formed that contain two recognition sites in cis or in trans, aligned in parallel. But the FokI endonuclease has no activity in Ca2+. To see if similar complexes are formed as transient intermediates during DNA cleavage, the substrates that had given a FRET signal upon mixing with FokI and Ca2+ (black records in Figure 2 and 3) were re-examined in reactions containing Mg2+. The test for the transient synapsis of sites in trans employed a pair of DNA molecules, both with one FokI site close to the left-hand end, tagged at that end with either Ax546 or Ax647. To look for short-lived looping interactions in cis, the experiments employed the IF construct with two FokI sites in head-to-head orientation, with a different label at each end. In both cases, the ratio of the concentration of FokI monomer to DNA sites was fixed at 1:1.
The Mg2+-dependent action of FokI on these constructs was monitored from the changes in acceptor emission (Figure 4). For both the one- and two-site substrates, the emission from the Ax647 moiety first rose but then decayed back to the level seen before adding the enzyme. The rates of the initial increases were similar to those seen with the same substrates in Ca2+ buffer. The rise in emission thus denotes the formation of the equivalent synaptic complex, in trans with the pair of one-site substrates and in cis with the two-site DNA. As in Ca2+, the initial increase was faster on the DNA with two sites than on the one-site constructs. The subsequent decline seen in the presence of Mg2+, but not Ca2+, can be assigned to the release of at least one of the fluorophores on the DNA into bulk solution after the cleavage reaction. The rate of the decrease, and thus the overall rate of the reaction, was much faster with the two-site construct than with the one-site DNA. To observe directly DNA cleavage on the one- and the two-site substrates constructed here, the same DNA species but lacking the fluorescent label(s) were mixed with FokI endonuclease in a quench-flow apparatus and the reactions stopped at various times after mixing: the quenched samples were then analysed by gel electrophoresis (Supplementary Figure S2). The one-site construct was indeed cleaved more slowly than the constructs containing a second FokI site in either inverted or repeated orientations. The synaptic complexes detected by FRET thus appear to be catalytically competent species that can proceed to cleave DNA in the presence of Mg2+.
Figure 4.
FRET from cutting one-site and two-site DNA. Equal volumes of FokI endonuclease and DNA labelled with Alexa Fluor dye(s) were mixed by stopped-flow to give the following reactions in M-buffer at 20°C. One reaction (black trace), contained 25 nM FokI and two DNA molecules with one FokI site, both at 12.5 nM: the DNA was labelled at the end proximal to the FokI site, one with Ax546 and the other with Ax647. The second reaction (red trace) contained FokI (50 nM) and one DNA molecule (25 nM) with two inverted FokI sites, labelled at one end with Ax546 and at the other with Ax647. [All DNA were 260 bp long, with 30 bp from the FokI site(s) to the proximal end(s). The locations and orientations of the recognition sites (yellow arrowheads) and the fluorescent labels (Ax546, in cyan; Ax647 in mauve) on the substrates are indicated in the right-hand panel, as are also representative cleavage products.] Excitation was at 545 nm and the change in emission at >645 nm recorded over time. Since the emission from the reaction on the one-site substrates was lower than that from the two-site substrate, the fluorescence signal (ΔF) is shown normalized to a value of 1 for the maximal signal from each reaction.
TPM on one and two-site DNA
The FRET experiments in both Ca2+ and Mg2+ show that the synaptic complex that FokI forms on a DNA with two FokI sites in inverted orientation contains the two sites aligned in parallel, and thus sequesters the intervening DNA into a simple 180° loop (Figure 1). On a DF DNA with directly repeated FokI sites, a 180° loop would be formed only if the sites are arranged in the anti-parallel manner: a parallel synapse would result in a convoluted 360° loop. Although the DF construct appears not to produce an anti-parallel synapse with concomitant trapping of a 180° loop (Figure 3, red record), the FRET strategy used here cannot demonstrate that the directly repeated sites are held together in the parallel manner to give a 360° loop, as the parallel structure would leave the two ends of the DNA too far apart for a FRET signal.
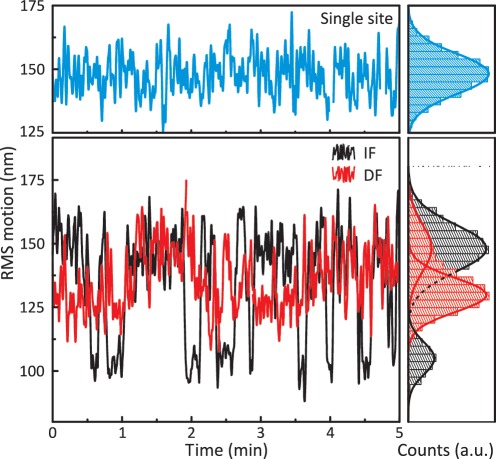
TPM was therefore employed to reveal the nature of the DNA loops trapped between FokI sites in either IF or DF configurations. In these experiments, linear DNA molecules were generated by using PCR to amplify the segments of DNA spanning the FokI site(s) on the same one- and two-site plasmids that had been used to make the substrates for the FRET experiments. The two-site substrates for TPM retained the same inter-site distance as before but were longer overall, with increased distances from the FokI sites to the proximal ends of the DNA (166 and 170 bp). However, this time, the constructs carried a DIG moiety at one 5′-end and a biotin at the other. The DIG-labelled end was fixed to a glass surface coated with an anti-DIG antibody, to immobilize the DNA, and the biotin-tagged end was attached to a streptavidin-coated polystyrene bead. The bead tethered by this procedure remains subject to Brownian motion but the extent of motion is governed by the length of the tether, as any reduction in this length restricts the motion of the bead. An inverted microscope was used to follow the motion of up to 50 beads concurrently, both before and after adding FokI endonuclease: the buffer for these experiments contained Ca2+ to prevent DNA cleavage as this would have released the bead from the tether. As a measure of the effective end-to-end length (the distance between the two ends of the DNA), the RMS motion of the bead over ∼0.5 s intervals was logged.
The trace for the one-site construct in the presence of FokI exhibited a single defined state (Figure 5, blue trace). The histogram for the distribution of its RMS values fitted readily to a single Gaussian, centred at about 150 nm. The same distribution was observed on the one-site DNA in the absence of FokI (data not shown). In the absence of FokI, the data from the DNA with two sites, in either inverted or repeated orientations, was also indistinguishable from that with the one-site DNA. In contrast, the traces for both two-site substrates showed transitions between two well-defined states in the presence of FokI (Figure 5, red and black traces). For both traces, a double Gaussian was needed to fit the histogram. For both IF and DF DNA, the larger of the two RMS motions (∼150 nm) was identical to the RMS measured for the one-site DNA and for the two-site DNA in the absence of protein, so this corresponds to the unlooped state of the tether. Yet the two substrates clearly gave different values for the smaller RMS motion, that for the looped state; ∼105 nm for the IF construct and ∼130 nm for the DF construct. The difference in the RMS motions of the beads held by these two tethers cannot be attributed to the length of DNA between the sites, since the IF and DF substrates have identical inter-site spacing. They must instead be due to different loop topologies.
Figure 5.
TPM experiments. Upper panel: the RMS motion for an individual bead tethered by a DNA containing one FokI site, in the presence of 5 nM FokI in CC-buffer, is plotted as a function of time (blue trace). Lower panel: as above except that the DNA contained two FokI sites 190 bp apart in either inverted (IF: black trace) or directly repeated (DF: red trace) orientation. In all three cases, the distribution of the RMS values is plotted as a histogram in the right-hand panels, in corresponding colours. The histogram for the single-site DNA was fitted to a single Gaussian and those for the two-site constructs to double Gaussians.
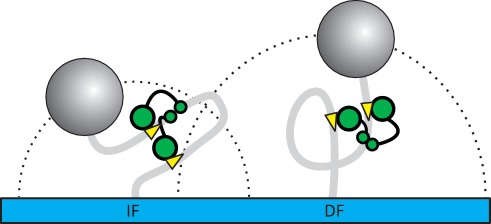
A 360° loop has no net effect on the direction of the helical axis of double-stranded DNA, while a 180° loop bends the DNA back onto itself. Hence, in a TPM set-up, the tethered bead is sent closer to the glass surface by trapping a 180° loop than a 360° loop (Figure 6). However, the bead is repelled from the surface by an entropic volume-exclusion effect (51). Consequently, a DNA in which one section is held in a 180° loop must also be bent through a total of 180° in the section(s) outside of the trapped loop, as the two ends of the DNA, one anchored to the surface and the other to the bead, are forced in opposite directions. Nevertheless, the mean 3D distance between the ends will still be shorter with a 180° loop than with the 360° topology, on account of the stiffness of the DNA (Figure 6). The 180° loop thus results in a lower RMS value than the 360° loop. The TPM experiments show that FokI traps loops of different topologies on the IF and DF substrates. The FRET experiments showed that FokI forms a parallel synapse on a DNA with two recognition sites in inverted orientation, which would trap a 180° loop. Hence, the different loop topology on a DNA with two directly repeated sites indicates that FokI retains the same parallel synapse geometry regardless of the orientation of the sites along the DNA, so trapping a 180° loop from inverted sites and a 360° loop from repeated sites.
Figure 6.
RMS motions for different loop topologies. The 180° and 360° loop topologies trapped by FokI on the IF and DF construct result in a lower RMS motion on the IF species compared to the DF species. This is due to the additional 180° bend outside of the loop trapped on the IF DNA that is not present with the 360° loop topology generated on the DF DNA. The net result is that the average end-to-end length of the tether is shorter with the IF DNA than with the DF DNA, thus positioning its bead closer to the glass slide. Dotted lines reflect RMS motion.
DISCUSSION
To cleave DNA on one side of its asymmetric recognition sequence, two monomers of the FokI restriction endonuclease, each bound to its cognate sequence, associate with each other to create a synaptic complex that contains the dimeric form of the protein bridging two copies of the DNA sequence (32,41). Models for the dimeric form of FokI bound to two sites can be constructed with the sites in either parallel or anti-parallel alignments (31; Figure 1). To place the two active sites of the dimer against the two target phosphodiester bonds at one recognition site, the parallel synapse calls for one protein monomer to be in a significantly different conformation from the other while the anti-parallel assembly retains essentially the same spatial relationship between catalytic and recognition domains in both subunits. The relocation of the two catalytic domains from one site to the other, therefore, entails a wholesale rearrangement of the parallel complex but relatively minor adjustments to the anti-parallel complex. Nevertheless, single-particle EM studies of the complexes formed by FokI with two cognate duplexes in trans revealed compact shapes consistent with the model for the parallel synapse, but not the elongated contour expected for the anti-parallel scheme (45).
The FRET experiments described here confirm the above suggestion for the synapsis of FokI sites in trans. They showed conclusively that the FokI protein holds together two separate DNA molecules, each carrying one FokI site in a parallel arrangement (Figure 2). FRET was also used here to examine the nature of the synaptic complex trapped by FokI across two sites in cis, on DNA substrates with two cognate sites in inverted or directly repeated orientations. Sites in inverted orientation were clearly shown by FRET to be synapsed together in the parallel arrangement. Moreover, while the FRET strategy employed here cannot reveal a parallel synapse with directly repeated FokI sites, it excluded the possibility of an anti-parallel synapse on the DF construct (Figure 3). Hence, it appears that FokI is limited to a unique geometry in its synaptic complexes, invariably aligning the sites in the parallel rather than the anti-parallel pattern, regardless of whether the sites are present in trans or in cis and, in the latter case, whether present in inverted or repeated orientations. While stable synaptic complexes were detected using a buffer containing Ca2+ to prevent DNA cleavage, the FRET experiments in Mg2+ revealed equivalent complexes which then proceeded to cleave the DNA (Figure 4).
For sites in cis, the demand for a parallel alignment of recognition sites dictates the topology of the looped-out DNA (Figure 1): a simple 180° loop for sites in inverted orientation along the DNA and a convoluted 360° turn for directly repeated sites (Figure 1). The TPM studies reported here show that the loop trapped by FokI on an IF construct does indeed have a different topology from the loop trapped on a DF construct (Figure 5). The loop between the inverted sites has a greater effect on the 3D distance between the two ends of the DNA than that with directly repeated sites, indicating that the former possesses a 180° bend and the latter a 360° turn (Figure 6). Strikingly, the DNA with directly repeated sites retains the parallel synapse even though it entails a greater degree of DNA bending than would be the case if it had switched to the anti-parallel arrangement. The additional bending energy trapped in the DF DNA must, therefore, be insufficient to block the formation of the parallel synapse though we show in the following article that this energy affects the dynamics of DNA loop capture by FokI (47). The parallel synapse may be favoured over the anti-parallel on account of protein-protein interactions between the two DNA recognition domains present in the model for the parallel but not the anti-parallel scheme (45).
The principal interaction between the protein monomers occurs, however, between the catalytic domains (40,41). But this interaction is formed only after the catalytic domain of the monomer at the cognate site relocates from its initial position adjacent to the recognition sequence to its downstream position for DNA cleavage, 13 nt away (43). The DNA substrates constructed here thus carry about 45 bp between the synapse junction and the fluorescent label on the 5′-end of the DNA, a distance of about 150 Å. The R0 for the FRET pair used here, Ax546 and Ax647, is 74 Å (50). The detection of a FRET signal from a parallel alignment of sites, one tagged with the donor and the other with the acceptor, implies that the angle subtended between the two DNA segments exiting the complex must be comparatively small. If it had been close to a right angle, the distance between the two fluorescent dyes would have been >200 Å and thus out of range for FRET. A crystal structure for dimeric FokI bound to two DNA segments has yet to be obtained but structures have been solved for several other restriction enzymes bound to two duplexes (25,27,52). One of these, Ecl18kI, involves, like FokI, the association of two DNA–protein complexes (28) and in this case, the segments subtend an angle of 30° (52).
Many proteins that bind two DNA loci at the same time recognize rotationally symmetric DNA sequences: well-studied examples include the Lac repressor (7,53), the Cre recombinase (4,5) and the SfiI restriction endonuclease (16,19,27). In these cases, no distinction can be made between the parallel and the anti-parallel alignment of two such sites in trans nor, on DNA with two palindromic sequences in cis, can any distinction be made between inverted and directly repeated orientations along the DNA. Nevertheless, DNA loops trapped between two functionally symmetric sites in cis can possess alternate topologies encompassing either a 180° or a 360° loop (8,53). Moreover, these loops possess a handedness depending on whether the DNA between the sites needs to be under- or over-wound to align the sites against the DNA binding surfaces of the protein (12). SfiI for example traps a right-hand loop akin to a negative supercoil when the DNA needs to be under-wound and a left-hand positively coiled loop from over-wound DNA (29). But in all of these cases, the geometry of the synapse can only be deduced indirectly from the topology of the loop. Only with asymmetric sites, such as the recognition sequence for FokI, is it possible to distinguish unambiguously parallel from anti-parallel synapses.
Proteins that bridge two copies of an asymmetric DNA sequence are often able to act only on sites present in cis in a particular orientation (4,12). Some such as the Type III restriction endonucleases do not act by DNA looping but instead follow the DNA path from one site to the other (10,14). Others trap loops but demand a unique synapse geometry than can only arise on a supercoiled DNA with sites in a specified orientation: for example, the resolvases from the Tn3-like transposons require supercoiled substrates with two target sites in direct repeat (4,5). Of the many situations that can lead to the juxtaposition of two sites in a supercoiled DNA, the most common is when the sites are located on interwound DNA segments directly opposite each other across the superhelical axis (54,55). When juxtaposed across the superhelical axis, two directly repeated sites run in anti-parallel directions while inverted sites lie parallel to each other. Only the latter would appear to be aligned appropriately for FokI. Nevertheless, unlike resolvase, FokI can cleave supercoiled DNA with two recognition sites regardless of whether the sites are present in directly repeated or inverted orientations, and does so with similar turnover rates (32,33). Turnover rates of FokI are, however, limited by the release of the cleaved DNA at the end of the reaction, and not by the DNA loop capture step prior to phosphodiester hydrolysis. Hence, it is not yet known if the orientation of the sites on a supercoiled DNA has any effect on the ability of FokI to loop out the intervening DNA, or even whether it retains the parallel synapse for both site orientations on supercoiled substrates.
In the following article (47), the ability of FokI to trap DNA loops with predictable topologies, given its invariant synapse geometry, is exploited to reveal the impact of DNA bending and twisting rigidity on the dynamics of protein-induced DNA looping.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1 and S2.
FUNDING
Wellcome Trust (grant 078794 to D.A.R. and S.E.H.); the Marie Curie Research Training Network on ‘DNA Enzymes’ (to C.P.); the Fundamenteel Onderzoek der Materie (FOM) research program ‘Physics of the Genome’ and by the ‘Nederlandse Organisatie voor Wetenschappelijk’ (NWO; to N.L. and G.J.L.W.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lucy Catto for her valued input into the early stages of this study and Rachel Smith for advice on the article.
REFERENCES
- 1.Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J. Biol. Chem. 1990;265:14697–14700. [PubMed] [Google Scholar]
- 2.Singh SK, Sabatinos S, Forsburg S, Bastia D. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell. 2010;142:868–878. doi: 10.1016/j.cell.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 4.Hallet B, Sherratt DJ. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 5.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 6.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 7.Finzi L, Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 8.Semsey S, Virnik K, Adhya S. A gamut of loops: meandering DNA. Trends Biochem. Sci. 2005;30:334–341. doi: 10.1016/j.tibs.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Finzi L, Dunlap DD. Single-molecule approaches to probe the structure, kinetics, and thermodynamics of nucleoprotein complexes that regulate transcription. J. Biol. Chem. 2010;285:18973–18978. doi: 10.1074/jbc.R109.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dryden DTF, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mucke M, Kruger DH, Reuter M. Diversity of type II restriction endonucleases that require two DNA recognition sites. Nucleic Acids Res. 2003;31:6079–6084. doi: 10.1093/nar/gkg836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 13.Gemmen GJ, Millin R, Smith DE. Tension-dependent DNA cleavage by restriction endonucleases: two-site enzymes are "switched off" at low force. Proc. Natl Acad. Sci. USA. 2006;103:11555–11560. doi: 10.1073/pnas.0604463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczelkun MD, Friedhoff P, Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem. Soc. Trans. 2010;38:404–409. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DTF, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wentzell LM, Halford SE. DNA looping by the SfiI restriction endonuclease. J. Mol. Biol. 1998;281:433–444. doi: 10.1006/jmbi.1998.1967. [DOI] [PubMed] [Google Scholar]
- 17.Welsh AJ, Halford SE, Scott DJ. Analysis of type II restriction endonucleases that interact with two recognition sites. In: Pingoud A, editor. Nucleic Acids and Molecular Biology, Restriction Endonucleases. Vol. 14. Berlin: Springer; 2004. pp. 297–317. [Google Scholar]
- 18.van den Broek B, Vanzi F, Normanno D, Pavone FS, Wuite GJL. Real-time observation of DNA looping dynamics of Type IIE restriction enzymes NaeI and NarI. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurens N, Bellamy SRW, Harms AF, Kovacheva YS, Halford SE, Wuite GJL. Dissecting protein-induced DNA looping dynamics in real time. Nucleic Acids Res. 2009;37:5454–5464. doi: 10.1093/nar/gkp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal AK. Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 1995;5:11–19. doi: 10.1016/0959-440x(95)80004-k. [DOI] [PubMed] [Google Scholar]
- 22.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wentzell LM, Nobbs TJ, Halford SE. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 24.Daniels LE, Wood KM, Scott DJ, Halford SE. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J. Mol. Biol. 2003;327:579–591. doi: 10.1016/s0022-2836(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 25.Siksnys V, Grazulis S, Huber R. Structure and function of the tetrameric restriction enzymes. In: Pingoud A, editor. Nucleic Acids and Molecular Biology, Restriction Endonucleases. Vol. 14. Berlin: Springer; 2004. pp. 237–259. [Google Scholar]
- 26.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 27.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. EMBO J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaremba M, Owsicka A, Tamulaitis G, Sasnauskas G, Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL, Laurens N, van den Broek B, Wuite GJL, et al. DNA synapsis through transient tetramerization triggers cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 2010;38:7142–7154. doi: 10.1093/nar/gkq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson MA, Gowers DM, Halford SE. Alternative geometries of DNA looping: an analysis using the SfiI endonuclease. J. Mol. Biol. 2000;298:461–475. doi: 10.1006/jmbi.2000.3676. [DOI] [PubMed] [Google Scholar]
- 30.Sugisaki H, Kanazawa S. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI) Gene. 1981;16:73–78. doi: 10.1016/0378-1119(81)90062-7. [DOI] [PubMed] [Google Scholar]
- 31.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 32.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 34.Marshall JJT, Gowers DM, Halford SE. Restriction endonucleases that bridge and excise two recognition sites from DNA. J. Mol. Biol. 2007;367:419–431. doi: 10.1016/j.jmb.2006.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaczorowski T, Skowron P, Podhajska AJ. Purification and characterization of the FokI restriction endonuclease. Gene. 1989;80:209–216. doi: 10.1016/0378-1119(89)90285-0. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. Proc. Natl Acad. Sci. USA. 1992;89:4275–4279. doi: 10.1073/pnas.89.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YG, Li L, Chandrasegaran S. Insertion and deletion mutants of FokI restriction endonuclease. J. Biol. Chem. 1994;269:31978–31982. [PubMed] [Google Scholar]
- 38.Waugh DS, Sauer RT. Single amino acid substitutions uncouple the DNA binding and strand scission activities of FokI endonuclease. Proc. Natl Acad. Sci. USA. 1993;90:9596–9600. doi: 10.1073/pnas.90.20.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 40.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders KL, Catto LE, Bellamy SRW, Halford SE. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 2009;37:2105–2115. doi: 10.1093/nar/gkp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pernstich C, Halford SE. Illuminating the reaction pathway of the FokI restriction endonuclease by fluorescence resonance energy transfer. Nucleic Acids Res. 2012;40:1203–1213. doi: 10.1093/nar/gkr809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catto LE, Bellamy SRW, Retter SE, Halford SE. Dynamics and consequences of DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2008;36:5122–5122. doi: 10.1093/nar/gkn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanamee ES, Berriman J, Aggarwal AK. An EM view of the FokI synaptic complex by single particle analysis. J. Mol. Biol. 2007;370:207–212. doi: 10.1016/j.jmb.2007.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelman LM, Cheong R, Kahn JD. Fluorescence resonance energy transfer over ∼130 basepairs in hyperstable Lac repressor-DNA loops. Biophys. J. 2003;84:1131–1145. doi: 10.1016/S0006-3495(03)74929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurens N, Rusling DA, Pernstich C, Brouwer I, Halford SE, Wuite GJL. DNA looping by FokI: the impact of twisting and bending rigidity on protein-induced looping dynamics. Nucleic Acids Res. 2012;40:4988–4997. doi: 10.1093/nar/gks184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 49.Lakowizc JR. Principles in Fluorescence Spectroscopy. 3 rd edn. New York: Kulwer Academic/Plenum Press; 2006. [Google Scholar]
- 50.Johnson I, Spence MTZ. Molecular Probes Handbook, a Guide to Fluorescent Probes and Labeling Technologies. 11th edn. Carlsbad, California: Invitrogen; 2010. [Google Scholar]
- 51.Segall DE, Nelson PC, Phillips R. Volume-exclusion effects in tethered-particle experiments: bead size matters. Phys. Rev. Lett. 2006;96:088306. doi: 10.1103/PhysRevLett.96.088306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: the influence of DNA looping on Lac repressor conformation. Proc. Natl Acad. Sci. USA. 2006;103:9879–9884. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker CN, Halford SE. Dynamics of long-range interactions in DNA: the speed of synapsis during site-specific recombination by resolvase. Cell. 1991;66:781–791. doi: 10.1016/0092-8674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, Schlick T, Vologodskii A. Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl Acad. Sci. USA. 2001;98:968–973. doi: 10.1073/pnas.98.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.