Abstract
In eukaryotic cells, ribosomal DNA (rDNA) forms the basis of the nucleolus. In Saccharomyces cerevisiae, 100–200 copies of a 9.1-kb rDNA repeat exist as a tandem array on chromosome XII. The stability of this highly repetitive array is maintained through silencing. However, the precise mechanisms that regulate rDNA silencing are poorly understood. Here, we report that S. cerevisiae Ydr026c, which we name NTS1 silencing protein 1 (Nsi1), plays a significant role in rDNA silencing. By studying the subcellular localization of 159 nucleolar proteins, we identified 11 proteins whose localization pattern is similar to that of Net1, a well-established rDNA silencing factor. Among these proteins is Nsi1, which is associated with the NTS1 region of rDNA and is required for rDNA silencing at NTS1. In addition, Nsi1 physically interacts with the known rDNA silencing factors Net1, Sir2 and Fob1. The loss of Nsi1 decreases the association of Sir2 with NTS1 and increases histone acetylation at NTS1. Furthermore, Nsi1 contributes to the longevity of yeast cells. Taken together, our findings suggest that Nsi1 is a new rDNA silencing factor that contributes to rDNA stability and lifespan extension in S. cerevisiae.
INTRODUCTION
The synthesis and processing of rRNAs and ribosome assembly occur in the nucleolus, and ribosomal DNA (rDNA) constitutes the basis of this organelle. In Saccharomyces cerevisiae, 100–200 copies of a 9.1-kb rDNA repeat exist as a tandem array on chromosome XII (1). Each repeat contains a Pol I-transcribed 35S rRNA gene and a non-transcribed spacer (NTS) that is divided by a Pol III-transcribed 5S rRNA gene into NTS1 and NTS2 (Figure 2A). Due to its highly repetitive nature, an rDNA array is an easy target for homologous recombination events, and recombination between rDNA repeats, which leads to the formation of extrachromosomal rDNA circles (ERCs) that accumulate to toxic levels in mother cells, is a primary cause of aging in S. cerevisiae (2). Cells have therefore evolved mechanisms to protect rDNA arrays, as their stability is critical for growth and survival. Under normal conditions, rDNA repeats remain relatively stable because the homologous recombination between them is negatively regulated through a mechanism referred to as rDNA silencing.
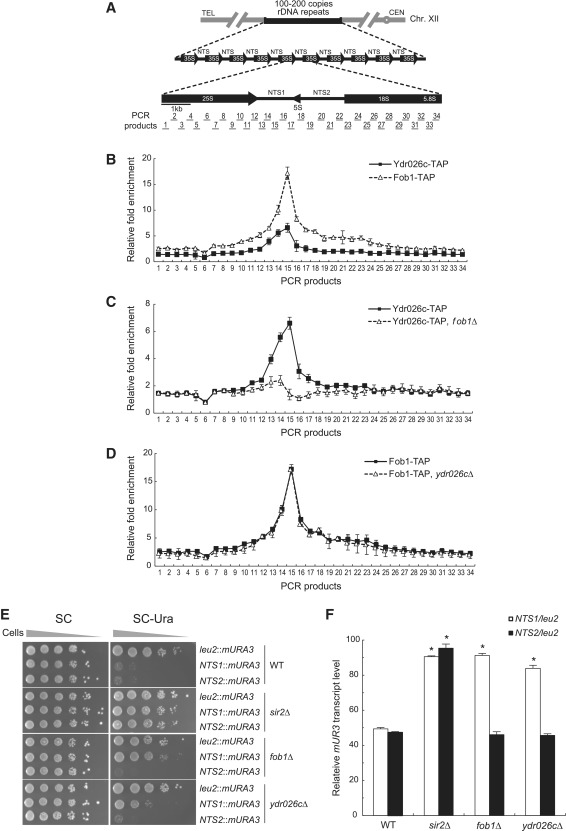
Figure 2.
Ydr026c is associated with the NTS1 region of rDNA and is required for NTS1-specific rDNA silencing. (A) The structure of the tandemly repeating rDNA of S. cerevisiae is shown above, and a single 9.1-kb rDNA unit is shown expanded below. PCR amplicons used in ChIP assays are indicated below the rDNA unit. (B) Ydr026c and Fob1 are associated with the NTS1 region of rDNA. Shown is the degree of association of Ydr026c (solid line) and Fob1 (dashed line) with rDNA. The degree of association with rDNA was measured using ChIP assay. Relative fold enrichment refers to the relative ratio of PCR products amplified from immunoprecipitated DNA to products from input DNA. Values represent the average of three independent experiments, and error bars indicate the SEM. (C) Fob1 is required for the recruitment of Ydr026c to NTS1. Shown is the degree of association of Ydr026c with rDNA in the presence (solid line) or absence (dashed line) of Fob1. (D) Ydr026c is not required for the association of Fob1 with NTS1. Shown is the degree of association of Fob1 with rDNA in the presence (solid line) or absence (dashed line) of Ydr026c. (E) Ydr026c contributes to rDNA silencing at NTS1 but not at NTS2. Silencing within rDNA was assessed by monitoring the growth of cells (10-fold serial dilutions) plated on SC medium without uracil. SC medium was used as a plating control. (F) Ydr026c contributes to transcriptional silencing of the mURA3 reporter gene at NTS1 but not at NTS2. Total RNA was extracted from wild-type (WT), sir2Δ, fob1Δ and ydr026cΔ cells. Quantitative real-time reverse transcription–PCR analysis was performed to measure the transcript levels of the mURA3 gene inserted inside (RDN1-NTS1::mURA3 and RDN1-NTS2::mURA3) or outside the rDNA array (leu2::mURA3). Amplification efficiencies were validated and normalized against ACT1. Relative mURA3 transcript levels were calculated as the ratio of the normalized transcript level of the mURA3 reporter gene inside the NTS1 or NTS2 region to that outside the rDNA array. Primers used for the amplification of mURA3 were 5′-CTGTTGACATTGCGAAGAGC-3′ and 5′-TCTCCCTTGTCATCTAAACC-3′, and those for ACT1 were 5′-TGACTGACTACTTGATGAAG-3′ and 5′-TGCATTTCTTGTTCGAAGTC-3′. All reactions were carried out in triplicate and error bars indicate the SEM. Asterisks indicate P < 0.05, compared with wild-type cells (Student’s t-test).
Sir2 is a subunit of regulator of nucleolar silencing and telophase exit (RENT), an rDNA silencing complex which represses Pol II-dependent transcription at rDNA loci (3). Sir2 is an NAD+-dependent histone deacetylase (4–6), and its activity is required for the spreading of silencing complexes along chromatin via interactions with the N-terminal tails of histones (7–9). It has been reported that Sir2 extends replicative lifespan by stabilizing rDNA loci (2,10,11). Net1, another subunit of RENT, recruits Sir2 to rDNA and is also required for rDNA silencing (12). Additionally, Net1 interacts with Pol I and regulates the structure of the nucleolus (12). It has been shown that Net1 and Sir2 are associated with two regions of rDNA, the NTS1 region and the NTS2/Pol I promoter (3). Fob1 physically interacts with Net1 and Sir2 and is required for rDNA silencing by recruiting Net1 and Sir2 to the NTS1 region (3). In addition to RENT complex proteins, several other proteins, including Tof2, Lrs4, Csm1, Heh1 and Nur1, have been shown to participate in rDNA silencing (13–15).
Recent studies suggest that the nucleolus not only coordinates the synthesis and assembly of ribosomal subunits but also plays important roles in cell-cycle regulation, senescence and stress responses (16–21). The nucleolus has been shown to contain several hundred proteins, which are required for the diverse roles that it plays (22–24). However, the precise functions for a significant number of nucleolar proteins remain unknown. To gain new insights into nucleolar architecture and nucleolar protein function, we performed a systematic analysis of nucleolar protein localization under various environmental conditions. We identified 11 proteins whose localization pattern was similar to that of Net1, including an uncharacterized nucleolar protein, Ydr026c, which we named NTS1 silencing protein 1 (Nsi1). Nsi1 was associated with the NTS1 region of rDNA and was required for rDNA silencing at NTS1. We found that Nsi1 is involved in the association of Sir2 with NTS1 and histone deacetylation at the NTS1 region. We also found that the loss of Nsi1 reduces the longevity of yeast cells. Collectively, our results suggest that Nsi1 is a new rDNA silencing factor that contributes to rDNA stability and lifespan extension in S. cerevisiae.
MATERIALS AND METHODS
Yeast strains and growth conditions
The yeast strains used in this study are listed in Supplementary Table S1. Yeast strains were genetically manipulated according to the one-step PCR-mediated gene targeting procedure as previously described (25,26). Yeast transformation was performed using the lithium acetate method (27), and proper integration was confirmed by PCR. In all cases, deletion strains were constructed by the replacement of the corresponding open reading frames with a selectable marker. Rich medium (yeast extract, peptone, glucose; YPD) and synthetic complete (SC) medium lacking the appropriate amino acids for selection were prepared as previously described (28). Unless otherwise noted, cells were grown in YPD medium at 30°C.
Fluorescence microscopy
Fluorescence microscopy was performed on a Zeiss Axiovert 200M inverted microscope, as previously described (26). Strains expressing GFP-tagged nucleolar proteins have been described previously (24). Nuclei in live cells were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were grown in SC medium at 30°C.
Environmental conditions
Environmental conditions were applied to cells as previously described (29) with some modifications. Cells were grown to log phase (OD600 = 1.0) in SC medium at 30°C and were subjected to the following conditions: stationary phase, incubation for 3days in SC medium; carbon source depletion, incubation for 2 h in SC medium lacking glucose; nitrogen source depletion, incubation for 2 h in SC medium lacking amino acids and ammonium sulfate; amino acid starvation, incubation for 2 h in SC medium lacking amino acids; actinomycin D, incubation for 2 h in SC medium containing a final concentration of 1 mg/ml actinomycin D; rapamycin, incubation for 2 h in SC medium containing a final concentration of 2 ng/ml rapamycin; heat shock, incubation in SC medium at 37°C for 2 h; dithiothreitol, incubation for 2 h in SC medium containing a final concentration of 2.5 mM dithiothreitol; menadione, incubation for 30 min in SC medium containing a final concentration of 1 mM menadione; diamide, incubation for 30 min in SC medium containing a final concentration of 1.5 mM diamide; hydrogen peroxide, incubation for 30 min in SC medium containing a final concentration of 0.3 mM hydrogen peroxide; methyl methanesulfonate, incubation for 6 h in SC medium containing a final concentration of 0.02% methyl methanesulfonate; ultraviolet light, exposure to 1400 mJ/cm2 for 10 s.
Chromatin immunoprecipitation assay and quantitative real-time PCR analysis
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (30). For TAP ChIP experiments, pre-washed IgG Sepharose beads (17-0969-01, GE Healthcare) were used. For total or acetylated histone H3 ChIP experiments, anti-histone H3 (ab1791, Abcam) and anti-acetyl-histone H3 (Lys9/18) (07-593, Millipore) were used. ChIP samples were analyzed by quantitative real-time PCR using SYBR Green and the Applied Biosystems 7300 real-time PCR system. Relative fold enrichment was determined by calculating the ratio of rDNA to CUP1, an internal control, as follows: [rDNA(IP)/CUP1(IP)]/[rDNA(input)/CUP1(input)]. To determine the enrichment of Sir2 at telomeres and the silent mating type loci, the subtelomeric region on the right arm of chromosome VI (TEL-VI) and the E silencer region of the mating type locus HMR on chromosome III (HMR-E) were analyzed by ChIP assay as previously described (7), using CUP1 as an internal control. The sequences of PCR primers used in ChIP experiments are shown in Supplementary Table S2. Each set of experiments was performed at least three times. Statistical analysis was performed using Student’s t-test.
Silencing assay
Silencing at heterochromatic regions (rDNA, telomeres and the silent mating type loci) was assayed as previously described (13,31). Yeast cells were grown to OD600 = 2.0 in SC medium, and 3 µl of 10-fold serial dilutions of the cell suspensions were spotted on the appropriate media. Plates were incubated at 30°C for 2days.
Bimolecular fluorescence complementation assay
Bimolecular fluorescence complementation (BiFC) assays to analyze protein–protein interaction were performed using the yeast BiFC vector system as previously described (26). Net1, Sir2 and Fob1 were N-terminally fused to the N-terminal fragment of Venus (VN) under the control of the RPL7B promoter in MATa cells. Nsi1 was N-terminally fused to the C-terminal fragment of Venus (VC) under the control of the RPL7B promoter in MATα cells. Diploid cells expressing both the VN fusion protein and the VC fusion protein were generated by mating. Fluorescence was monitored on a Zeiss Axiovert 200 M inverted microscope.
Coimmunoprecipitation experiment
Coimmunoprecipitation (CoIP) experiments were performed as described previously (12) with some modifications. Mouse anti-Myc antibody (sc-40, Santa Cruz Biotechnology) was added to cell extracts prepared from yeast strains expressing HA-Nsi1 together with Net1-Myc, Sir2-Myc or Fob1-Myc and incubated for 2 hr. Protein A-agarose (P3476, Sigma) was added to the immunoprecipitation reaction and incubated for 1 h. Proteins were detected with an HRP-conjugated anti-HA antibody (sc-7392, Santa Cruz Biotechnology) and a rabbit anti-Myc antibody (06-549, Millipore). Hexokinase was used as a loading control and detected by an anti-hexokinase antibody (H2035-02, United States Biological).
Purification of glutathione S-transferase fusion proteins
Glutathione S-transferase (GST)-Sir2 fusion protein was expressed from pHB0315, which was constructed by ligation of a 1.7-kb EcoRI-XhoI PCR product containing the Sir2 coding sequence with pGEX-4T-1 (28-9545-49, GE Healthcare). GST-Fob1 protein was expressed from pHB0316, which was constructed by ligation of a 1.7-kb BamHI-XhoI PCR product containing the Fob1 coding sequence with pGEX-4T-1. GST-Nsi1 fusion protein was expressed from pHB0330, which was constructed by ligation of a 1.7-kb EcoRI-XhoI PCR product containing the Nsi1 coding sequence with pGEX-4T-1. Rosetta cells containing expression vectors were grown in 2 l of Luria–Bertani medium to an OD600 of 0.4 and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at 30°C for 4 h. Cells were washed with cold phosphate-buffered saline and stored at −80°C. The cell pellet was resuspended in lysis buffer (phosphate-buffered saline with 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonylfluoride, 1 mM benzamidine, 1 µg/ml leupeptin and 1 µg/ml pepstatin), sonicated and centrifuged at 4°C for 15 min at 12 000 g in an SS-34 rotor (Sorvall). The supernatant was incubated with glutathione agarose (70541-3, Novagen) at 4°C for 1.5 h. The resin was loaded on a column and washed with lysis buffer and 50 mM Tris–HCl, pH 8.0. Column was eluted with 50 mM Tris–HCl, pH 8.0 and 10 mM glutathione.
GST pull-down assay
Purified GST fusion proteins (5 µg) were bound to 50 µl of glutathione agarose (70541-3, Novagen) at 4°C for 1 h in 200 µl of yeast lysis buffer (50 mM HEPES–NaOH, pH 7.6, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40, 1 mM phenylmethylsulfonylfluoride, 1 mM benzamidine, 1 µg/ml leupeptin and 1 µg/ml pepstatin). Ten microliters of beads were washed three times with yeast lysis buffer and incubated with 90 µl of whole yeast cell extract at 4°C for 2 h. Beads were washed three times with yeast lysis buffer and resuspended in SDS sample buffer. Proteins were detected with an HRP-conjugated anti-HA antibody (sc-7392, Santa Cruz Biotechnology), an HRP-conjugated anti-GFP antibody (600-103-215, Rockland) or a rabbit anti-Myc antibody (06-549, Millipore). Hexokinase was used as a loading control and detected by an anti-hexokinase antibody (H2035-02, United States Biological).
Analysis of ERCs
Analysis of ERCs was performed using Southern blots as previously described (32) with a digoxigenin (DIG)-labeled 25 S rDNA probe. Cells were spheroplasted by incubation in 1 ml of sorbitol buffer [0.9 M sorbitol, 0.1 M Tris–HCl, pH 8.0, 0.1 M EDTA, 150 µg/ml zymolyase and 1% (v/v) 2-mercaptoethanol] at 30°C with gentle shaking for 1 h. Of 10% SDS, 100 µl was added and incubated at 65°C for 30 min, followed by incubation with 333 µl of 5 M potassium acetate on ice for 1 h. After centrifugation for 3 min at 16 000×g, the supernatant was removed and the DNA was precipitated with 1 ml of ice-cold absolute ethanol. The DNA pellet was resuspended in 200 µl TE containing 0.1 mg/ml RNase and incubated at 37°C for 30 min. The purified DNA was heated with loading buffer at 55°C for 10 min before being loaded onto a 0.6% TBE-buffered agarose gel. Southern blot hybridization was carried out with a DIG-labeled 25 S rDNA probe. ERC levels were quantitated with a LAS-3000 image analyzer (Fujifilm). Statistical analysis was performed using Student’s t-test.
rDNA recombination assay
The rDNA recombination rate was determined by measuring the frequency of loss of ADE2 integrated at the rDNA locus of strain DMY3010 as previously described (10). Exponentially growing cells (OD600 = 1.0) in SC medium were sonicated briefly to prevent aggregation and were spread on SC plates. Colonies were allowed to grow for 2days at 30°C and then placed at 4°C for 3days to enhance color development. The rDNA recombination rate was calculated by dividing the number of half-red/half-white colonies by the total number of colonies. Entirely red colonies were excluded from all calculations. Three independent experiments were performed, and more than 20 000 colonies were examined for each assay. Statistical analysis was performed using Student’s t-test.
Analysis of the replicative lifespan
Analysis of the replicative lifespan was carried out by micromanipulation as previously described (33) using a Zeiss Tetrad Microscope. Lifespan analysis was performed on YPD plates. Cells that never budded were excluded from the calculation. For statistical analysis, lifespan data sets were compared by a two-tailed Wilcoxon rank-sum test. Lifespan was determined for 40 cells in each experiment.
RESULTS
Nucleolar protein localization study under various environmental conditions
The yeast nucleolus normally appears as a crescent shape localized at the periphery of the nucleus. The shape of the nucleolus can be dynamically altered, and some proteins move out of the nucleolus in response to environmental stresses. For example, when the target of rapamycin complex 1 (TORC1) signaling is inhibited, Pol I components are released from the nucleolus, and nucleolar size is rapidly reduced (34). In contrast, some nucleolar proteins, such as Net1 and Sir2, are retained in the nucleolus and are stably associated with rDNA under TORC1 inhibition. The stable association of Net1 and Sir2 with rDNA under TORC1 inhibition leads to increased rDNA stability through the reduction of rDNA recombination (30). It is well established that alterations in the subcellular localization of proteins can be important for regulating their function. Thus, a change in a protein’s subcellular localization in response to a specific condition may suggest a functional change in the protein under that condition. Based on this correlation, we performed a systematic analysis of nucleolar protein localization under various environmental conditions to better understand nucleolar architecture and nucleolar protein function.
To examine the subcellular localization of nucleolar proteins, we analyzed 159 strains expressing chromosomally GFP-tagged nucleolar proteins (24) under the following environmental conditions: stationary phase, carbon source depletion, nitrogen source depletion, amino acid starvation, exposure to ultraviolet light, heat shock, treatment with actinomycin D (an RNA synthesis inhibitor), rapamycin (a TORC1 inhibitor), dithiothreitol (a disulfide-reducing agent), menadione (a superoxide-generating drug), diamide (a sulfhydryl-oxidizing agent), hydrogen peroxide and methyl methanesulfonate (an alkylating agent) (29). Consistent with the dynamic nature of the nucleolus, many proteins exhibited dynamic changes in localization under various environmental conditions (Supplementary Figure S1). However, out of 159 proteins, 32 remained localized to the nucleolus under all conditions.
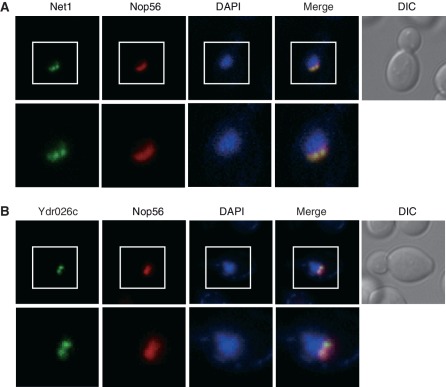
Net1, a well-known nucleolar protein, is stably associated with rDNA and is required for rDNA silencing (3,30). Interestingly, the Net1-GFP signal did not appear homogeneously distributed in the nucleolus (Figure 1A). Among 32 proteins identified above, we observed that the localization pattern of 10 proteins, Cdc14, Csm1, Fob1, Hmo1, Rrn5, Rrn7, Rrn9, Rrn10, Tof2 and Uaf30, was similar to that of Net1 (Supplementary Figure S2). Results from previous studies suggest a tight association of these proteins with rDNA. Fob1, Csm1 and Tof2 are recruited to the NTS1 region and are specifically required for silencing at the rDNA region (3,13). Cdc14 associates with the NTS1 region (35). Hmo1 associates throughout the entire 35 S rDNA region and is required for the repression of Pol II-dependent transcription at the rDNA region (36,37). Rrn5, Rrn9, Rrn10 and Uaf30 are components of the Pol I-specific transcription factor complex UAF, which functions as both an activator of Pol I and a silencer of Pol II for rDNA transcription (38). Rrn7, a component of the rDNA transcription factor complex CF, is involved in the Pol I-dependent transcription of rDNA (39,40). In addition to these proteins, we observed that an uncharacterized nucleolar protein Ydr026c showed a similar localization pattern to Net1 (Figure 1B).
Figure 1.
Subcellular localization analysis of Net1 and Ydr026c. Cells with chromosomally GFP-tagged Net1 or Ydr026c were grown to logarithmic phase in SC medium and analyzed by fluorescence microscopy. RFP-tagged Nop56 was used as a nucleolar marker. Shown are representative images of Net1-GFP (A) and Ydr026c-GFP (B). Boxed areas are enlarged at the bottom for better visualization. DAPI staining for visualization of the nucleus (blue) and differential interference contrast (DIC) images are also shown.
Ydr026c is associated with the NTS1 region of rDNA and is an NTS1-specific rDNA silencing factor
There is some data to suggest that Ydr026c may associate with ribosomes (41) and physically interact with Fob1 (42,43), but how Ydr026c is linked to nucleolar function is not known. Given that Ydr026c shows a localization pattern similar to Net1, it is plausible that Ydr026c may be tightly associated with rDNA. To check whether Ydr026c is associated with rDNA, we constructed a yeast strain in which the endogenous gene encoding Ydr026c was modified to express a C-terminal TAP fusion protein, and performed a ChIP experiment followed by quantitative real-time PCR assay using a panel of 68 primers, which were designed to tile across an entire 9.1-kb rDNA unit in fragments of ∼0.25 kb each (3). The structure of an rDNA repeat unit and the PCR products analyzed in ChIP assay are shown in Figure 2A. Consistent with the above result, Ydr026c was primarily associated with the region of rDNA that overlaps with the NTS1 region (Figure 2B). Furthermore, the overall association profile of Ydr026c with rDNA closely resembled that of Fob1 (3), suggesting a functional relationship between Ydr026c and Fob1. To better understand the recruitment of Ydr026c and Fob1 to the NTS1 region, we examined the association of Ydr026c with rDNA in the absence of Fob1 and vice versa. In fob1Δ cells, the association of Ydr026c with NTS1 was almost completely abolished (Figure 2C), indicating that Fob1 is required for the association of Ydr026c with NTS1. However, the association of Fob1 with rDNA was not affected by the absence of Ydr026c (Figure 2D). These results suggest that Ydr026c is recruited to NTS1 via Fob1, while Fob1 is associated with NTS1 in a Ydr026c-independent manner. To rule out the possibility that the loss of association of Ydr026c with NTS1 in fob1Δ cells was due to reduced expression of Ydr026c, we examined the expression level of Ydr026c in this strain. Western blot analysis of whole cell extracts showed that the protein level of Ydr026c was not altered by the absence of Fob1 (Supplementary Figure S3). Therefore, we conclude that Fob1 has no effect on the protein level of Ydr026c, but it is essential for the association of Ydr026c with rDNA.
The colocalization of Ydr026c with known rDNA silencing factors and its Fob1-dependent association with NTS1 suggest that Ydr026c may play a role in rDNA silencing. To investigate this possibility, we performed an rDNA silencing assay using the mURA3 silencing reporter gene integrated at the rDNA locus. We deleted SIR2, FOB1 and YDR026C in strains carrying the mURA3 silencing reporter gene integrated inside (RDN1-NTS1::mURA3 and RDN1-NTS2::mURA3) or outside the rDNA array (leu2::mURA3) (13). Cells were spotted in 10-fold serial dilutions on SC medium as a plating control and on medium without uracil to monitor the silencing of the mURA3 reporter gene. In wild-type cells, the reporter gene was efficiently silenced at both the NTS1 and NTS2 regions, as indicated by poor growth on medium lacking uracil (Figure 2E). The loss of SIR2 abolished the silencing of mURA3 at both the NTS1 and NTS2 regions, while the deletion of FOB1 abolished the silencing of mURA3 only at NTS1 but not at NTS2. Similar to the deletion of FOB1, the loss of YDR026C compromised the silencing of mURA3 at the NTS1 region but had no effect on this process at the NTS2 region. To more directly examine rDNA silencing, we performed a real-time reverse transcription–PCR analysis to measure the transcript levels of the Pol II-transcribed mURA3 gene integrated inside or outside the rDNA array. In wild-type cells, the reporter mURA3 gene was efficiently silenced at both NTS1 and NTS2 (>50%) compared to outside rDNA (Figure 2F). As expected, the silencing of the reporter gene at NTS1 and NTS2 was almost completely abolished in sir2Δ cells. In fob1Δ cells, the silencing of mURA3 was compromised at NTS1 but not at NTS2. The NTS1-specific defect in the silencing of mURA3 was also observed in ydr026cΔ cells. This result is consistent with the specific association of Ydr026c with NTS1 and indicates that Ydr026c contributes to rDNA silencing at NTS1. Based on the NTS1-specific silencing activity of Ydr026c, we decided to name this uncharacterized 66-kDa nucleolar protein Nsi1.
We next tested whether Nsi1 is required for silencing at other heterochromatic regions, telomeres and the silent mating type loci. We deleted NSI1 in strains carrying the URA3 reporter gene integrated at the right telomere of chromosome V or the TRP1 reporter gene integrated at a modified allele of the HMR locus, hmrΔE (31). The reporter genes at the telomere and the silent mating type locus were both silenced in wild-type cells, as indicated by the poor growth on medium lacking uracil or tryptophan (Supplementary Figure S4). Consistent with previous reports, the loss of SIR2 abolished the silencing of reporter genes at both the telomere and the silent mating type locus. However, the loss of NSI1 had no effect on the silencing of reporter genes at the right telomere of chromosome V or at the hmrΔE locus. This result suggests that Nsi1 plays a role in silencing specifically at the NTS1 region of rDNA, but not at other heterochromatic regions.
Nsi1 interacts with Net1, Sir2 and Fob1 and plays a positive role in the interaction between Fob1 and Sir2
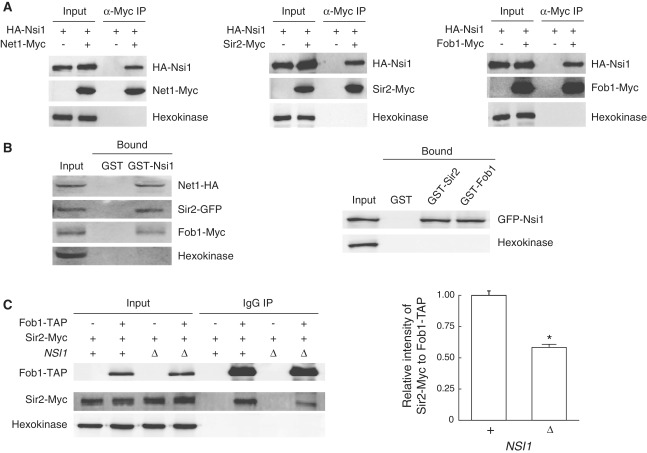
The association of Nsi1 with NTS1 and its involvement in NTS1 silencing raise the possibility that Nsi1 may physically interact with known rDNA silencing factors, such as Net1, Sir2 and Fob1. To determine whether Nsi1 interacts with Net1, Sir2 and Fob1, we performed a BiFC assay (26). We tagged NET1, SIR2 and FOB1 with VN in MATa cells and NSI1 with VC in MATα cells, and then generated diploid strains co-expressing VC-tagged Nsi1 and VN-tagged Net1, Sir2 or Fob1. When analyzed by fluorescence microscopy, all cells showed clear BiFC signals that overlapped exactly with the signal of Nop56-RFP, a well-characterized nucleolar marker (Supplementary Figure S5A). To confirm the interaction of Nsi1 with Net1, Sir2 and Fob1, we carried out CoIP experiments using cell extracts prepared from yeast strains co-expressing HA-Nsi1 and Net1-Myc, Sir2-Myc or Fob1-Myc. Immunoprecipitation of Net1-Myc, Sir2-Myc and Fob1-Myc all resulted in the coprecipitation of HA-Nsi1 (Figure 3A). We also tested the interaction of Nsi1 with Net1, Sir2 and Fob1 using a GST pull-down assay. We purified bacterially expressed GST and GST-Nsi1, incubated them with whole yeast cell extracts, and analyzed the bound fractions by western blotting. We found that GST-Nsi1 associated with Net1-HA, Sir2-GFP and Fob1-Myc, whereas the GST control did not (Figure 3B, left panel). We also observed that both GST-Sir2 and GST-Fob1 associated with GFP-Nsi1 (Figure 3B, right panel). Taken together, these results suggest that Nsi1 physically interacts with the Net1 and Sir2 subunits of the RENT complex and Fob1.
Figure 3.
Nsi1 physically interacts with Net1, Sir2 and Fob1, and the loss of Nsi1 decreases the interaction between Fob1 and Sir2. (A) Western blots showing that HA-Nsi1 coprecipitates with Net1-Myc (left panel), Sir2-Myc (middle panel) and Fob1-Myc (right panel) from whole cell extracts. (B) GST pull-down assay showing direct association of Nsi1 with Net1, Sir2 and Fob1. GST-Nsi1 associates with Net1, Sir2 and Fob1 from whole cell extracts (left panel). GST-Sir2 and GST-Fob1 associate with Nsi1 from whole cell extracts (right panel). (C) Western blots showing that the loss of Nsi1 decreases the amount of Sir2-Myc that coprecipitates with Fob1-TAP. Hexokinase was used as a loading control in all blots. The level of Sir2-Myc coprecipitated with Fob1-TAP in the presence or absence of Nsi1 was quantified and is shown to the right of the blots. Values represent the average of three independent experiments, and error bars indicate the SEM. Asterisk indicates P < 0.05, compared with wild-type cells (Student’s t-test).
Fob1 has been shown to play a role in rDNA silencing by recruiting the RENT complex to the replication fork block region of NTS1 (3). To better understand the role of Nsi1 in rDNA silencing, we performed CoIP experiments to monitor the interaction between Net1, Sir2 and Fob1 in the absence of Nsi1. The deletion of NSI1 had little effect on the amount of Net1 and Sir2 that precipitated together (Supplementary Figure S5B). An insignificant difference was also observed in Fob1-Net1 interaction in the absence of Nsi1 (Supplementary Figure S5C). These observations indicate that the Net1-Sir2 and Fob1-Net1 interactions occur independently of Nsi1. Interestingly, however, the amount of Sir2 that precipitated together with Fob1 was significantly decreased in the absence of Nsi1 (Figure 3C and Supplementary Figure S5D). This result suggests that Nsi1 plays a positive role in the interaction between Fob1 and Sir2.
Loss of Nsi1 decreases the association of Sir2 with NTS1, decreases rDNA stability and shortens replicative lifespan
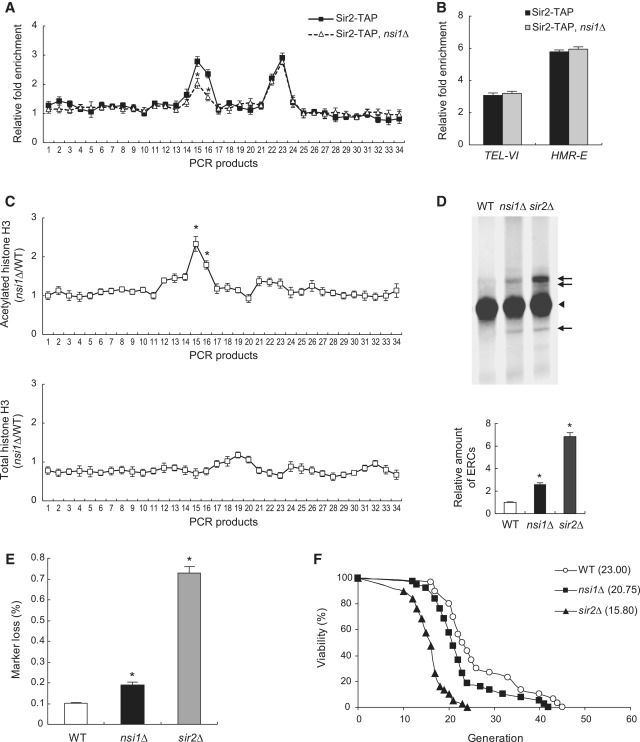
A previous study showed that Fob1 recruits Sir2 to the NTS1 region (3). Based on our finding that the association between Fob1 and Sir2 does not occur at the same level in the absence of Nsi1, we predicted that the loss of Nsi1 might reduce the association of Sir2 with NTS1. To test this, we performed ChIP experiments to examine the association of Sir2 with the rDNA region in the presence or absence of Nsi1. In the absence of Nsi1, the association of Sir2 with NTS1 was significantly reduced (Figure 4A), indicating that the loss of Nsi1 leads to a partial loss of Sir2 from the NTS1 region. This result suggests that Nsi1 may act as an adaptor that helps localize Sir2 to the NTS1 region. In contrast, deleting NSI1 had no effect on the association of Fob1 and Net1 with the NTS1 region (Figure 2D and Supplementary Figure S6A). We also examined whether the loss of Nsi1 affects the association of Sir2 with heterochromatic regions other than rDNA. However, there was little change in the association of Sir2 with the subtelomeric region on the right arm of chromosome VI (TEL-VI) or the E silencer region of the mating type locus HMR (HMR-E) whether Nsi1 was present or not (Figure 4B). Given that Sir2 is an NAD+-dependent histone deacetylase (4–6), we anticipated that the decreased association of Sir2 with NTS1 in the absence of Nsi1 would lead to increased acetylation levels of histones at this region. To determine whether the absence of Nsi1 affects histone acetylation at NTS1, we measured the acetylation level of histone H3 at rDNA by ChIP using an antibody that recognizes acetylated histone H3. As expected, the acetylation level of histone H3 at the NTS1 region was increased in nsi1Δ cells compared with that in wild-type cells (Figure 4C, upper panel). It has been shown that the level of total histone H3 is significantly lower at several positions in the rDNA repeat in sir2Δ cells than in wild-type cells (44). To determine whether Nsi1 affects the level of total histone H3 at rDNA, we performed ChIP experiments with antisera that recognize the C-terminal tail of histone H3. We observed that the level of total histone H3 was slightly decreased at several positions in the rDNA region in nsi1Δ cells compared with wild-type cells (Figure 4C, lower panel), indicating that the increase in histone H3 acetylation at NTS1 in nsi1Δ cells is at least not due to the increase in the level of total histone H3. Taken together, these results suggest that Nsi1 contributes to the association of Sir2 with NTS1 and to the increase in histone acetylation at NTS1.
Figure 4.
The loss of Nsi1 decreases the association of Sir2 with NTS1, decreases rDNA stability and shortens replicative lifespan. (A) The loss of Nsi1 reduces the association of Sir2 with the NTS1 region of rDNA. Shown is the degree of association of Sir2 with rDNA measured by ChIP in the presence (solid line) or absence (dashed line) of Nsi1. PCR products analyzed in ChIP assay are as indicated in Figure 2A. Relative fold enrichment refers to the relative ratio of PCR products amplified from immunoprecipitated DNA to products from whole cell extract DNA. Values represent the average of three independent experiments, and error bars indicate the SEM. Asterisks indicate the sites of rDNA with significantly lower association of Sir2 in nsi1Δ cells than in wild-type cells, as determined by Student’s t-test (P < 0.05). (B) Nsi1 does not influence the association of Sir2 with heterochromatic regions other than rDNA. Shown is the degree of association of Sir2 with the subtelomeric region on the right arm of chromosome VI (TEL-VI) and the E silencer region of the mating type locus HMR (HMR-E) in the presence (black bar) or absence (gray bar) of Nsi1. Relative fold enrichment refers to the relative ratio of PCR products amplified from immunoprecipitated DNA to products from whole cell extract DNA. Values represent the average of three independent experiments, and error bars indicate the SEM. (C) The loss of Nsi1 increases histone H3 acetylation at the NTS1 region of rDNA. The levels of acetylated (upper panel) or total histone H3 (lower panel) associated with the rDNA region were measured by ChIP in wild-type and nsi1Δ cells. Values represent the average of three independent experiments, and error bars indicate the SEM. Asterisks indicate the sites of rDNA with significantly increased association of acetylated histone H3 in nsi1Δ cells compared with wild-type cells, as determined by Student’s t-test (P < 0.05). (D) The loss of Nsi1 causes the accumulation of ERCs. Total DNA from wild-type (WT), nsi1Δ or sir2Δ cells was isolated, separated by agarose DNA gel electrophoresis and analyzed by Southern blot with a DIG-labeled 25 S rDNA probe. Chromosomal DNA is denoted by an arrowhead and ERC species by arrows. sir2Δ cells were used as a control. Asterisks indicate P < 0.05, compared with wild-type cells (Student’s t-test). (E) The loss of Nsi1 increases rDNA recombination. rDNA recombination is represented by the rate of loss of the ADE2 marker gene integrated at the rDNA locus (see ‘Materials and methods’ section). Values represent the average of three independent experiments, and error bars indicate the SEM. Asterisks indicate P < 0.05, compared with wild-type cells (Student’s t-test). (F) The loss of Nsi1 decreases the replicative lifespan. Replicative lifespan was determined by scoring the number of daughter cells produced by each mother cell. Mean lifespans are shown in parentheses. The P values for sir2Δ and nsi1Δ cells versus wild-type cells (WT) are 4.4 × 10−10 and 2.7 × 10−2, respectively.
Aging in yeast cells is associated with the progressive instability of rDNA and the accumulation of ERCs (2). We tested whether Nsi1 is required for maintaining rDNA integrity. To confirm that the loss of Nsi1 leads to the accumulation of ERCs, we isolated total DNA from wild-type, nsi1Δ and sir2Δ cells and performed Southern blot analysis with a DIG-labeled 25 S rDNA probe. Different ERCs migrate slower or faster than chromosome XII DNA during gel electrophoresis (2,32). As expected, exponentially growing wild-type cells contained only a few ERCs, whereas sir2Δ cells exhibited a 6.8-fold increase in ERCs relative to wild-type cells (Figure 4D). Although the accumulation of ERCs in nsi1Δ cells was not as strong as in sir2Δ cells, nsi1Δ cells showed a 2.6-fold increase in ERCs relative to wild-type cells, suggesting that Nsi1 contributes to rDNA stability. Based on this result, we next examined whether Nsi1 reduces rDNA recombination by promoting rDNA stability. To measure the rDNA recombination rate, the frequency of loss of the ADE2 marker gene, which was integrated at the rDNA locus, was monitored (10). In agreement with the observation that the loss of Nsi1 causes the accumulation of ERCs, nsi1Δ cells showed about a 2-fold increase in the rate of marker loss relative to wild-type cells (Figure 4E). Taken together, these results suggest that Nsi1 contributes to repressing rDNA recombination and to rDNA stability.
It has been reported that Sir2 mediates lifespan extension in yeast by stabilizing the rDNA locus (2,10,11). Given that Nsi1 contributes to NTS1-specific rDNA silencing, promotes the association of Sir2 with NTS1 and maintains rDNA stability, it is plausible that Nsi1 may play a positive role in lifespan extension in yeast. To confirm the positive role of Nsi1 in longevity, we measured the replicative lifespan of nsi1Δ cells and compared it with that of wild-type and sir2Δ cells. Consistent with previous reports, the deletion of SIR2 caused a dramatic decrease in lifespan (Figure 4F). The lifespan of nsi1Δ cells was also shortened compared with wild-type cells, although it was longer than that of sir2Δ cells. Collectively, these results indicate that Nsi1 plays a positive role in the longevity of yeast cells by promoting rDNA stability.
DISCUSSION
In this study, we demonstrated that Nsi1, encoded by the YDR026C gene, is a new NTS1-specific rDNA silencing factor in S. cerevisiae. Nsi1 associates with the NTS1 region of rDNA and interacts with known rDNA silencing factors such as Net1, Sir2 and Fob1. In addition, Nsi1 contributes to the association of Sir2 with Fob1 and plays a positive role in rDNA stability. Furthermore, the absence of Nsi1 shortens the replicative lifespan of yeast cells. These data suggest that Nsi1 plays a role in the recruitment of Sir2 to rDNA, thus regulating rDNA silencing and maintaining rDNA integrity.
Nsi1 is a 66-kDa nucleolar protein that has a Myb-like DNA-binding domain, which is composed of tandem repeats of a helix-turn-helix motif that fit into the major groove of the DNA double helix. Previous two-hybrid analyses have shown that Nsi1 physically interacts with Fob1 (42,43). In this study, we found that Nsi1 also interacts with Net1 and Sir2, subunits of the RENT complex, in addition to Fob1. Large-scale protein interaction studies have identified nine more proteins—Cdc23, Mif2, Nip100, Prp9, Rpa34, Rpa49, Rpc19, Smt3 and Spc25—that physically interact with Nsi1 (45–48). Among the 12 Nsi1-interacting proteins identified so far, six of them—Fob1, Net1, Rpa34, Rpa49, Rpc19, Sir2—are localized to the nucleolus (24). Fob1, Net1 and Sir2 are well-known rDNA silencing factors, which makes sense given the functional role of Nsi1 in maintaining rDNA integrity. Rpa34, Rpa49 and Rpc19 are subunits of Pol I, however, whether Nsi1 has a functional relationship with Pol I is not yet clear. In addition to the NTS1 region, Net1 and Sir2 are associated with the NTS2/18 S region of the rDNA locus, which overlaps with the Pol I transcription initiation region (3,13,30). A previous CoIP analysis revealed that both Net1 and Sir2 interact with Pol I, suggesting that the association of Net1 and Sir2 to the NTS2/18 S region may result from their physical interaction with Pol I (3). Given that some Pol I subunits interact with Nsi1, it is possible that Nsi1 may also be recruited to the NTS2/18 S region by Pol I. However, we could not detect any significant association of Nsi1 with the NTS2/18 S region by conventional ChIP analysis (Figure 2B and C). Further studies with more sensitive methods are needed to determine whether Pol I recruits Nsi1 to the NTS2/18 S region of rDNA.
Other rDNA silencing factors have been reported to bind to the NTS1 region of rDNA. For example, Tof2 was shown to be primarily associated with the NTS1 region of rDNA in a Fob1-dependent manner (13). In addition, Csm1 and Lrs4, subunits of the monopolin complex, are also recruited to the NTS1 region by Fob1 and Tof2. Tof2, Csm1 and Lrs4 are specifically required for silencing at NTS1. We wondered whether the association of these rDNA silencing factors with rDNA is influenced by Nsi1. However, the association of Tof2, Csm1 and Lrs4 with rDNA was not altered in the absence of Nsi1 (Supplementary Figure S6B–D). Previous findings suggest that there are Sir2-dependent rDNA silencing pathway, which involves Net1 and Sir2, and Sir2-independent rDNA silencing pathway, relying on Tof2, Csm1 and Lrs4 (3,13,30). In the Sir2-dependent pathway, Net1 is associated with NTS1 by Fob1, and they recruit Sir2 to NTS1. In the Sir2-independent pathway, Fob1 recruits Tof2 to NTS1, and Csm1 and Lrs4 are associated with NTS1 via interaction with Tof2. Our data suggest that Nsi1 plays a role in the Sir2-dependent pathway and acts as an adaptor that helps localize Sir2 to the NTS1 region. However, given that the deletion of NSI1 leads to a partial loss of Sir2 from the NTS1 region (Figure 4A), the role of Nsi1 in the recruitment of Sir2 to NTS1 seems to be somewhat limited. A significant but small effect of deletion of NSI1 on the ADE2 marker loss and the cell longevity is likely to be due to the limited role of Nsi1 in the recruitment of Sir2 to the NTS1 region of rDNA.
The chromosomal regions containing the tandem arrays of rDNA genes constitute the nucleolar-organizing regions and are the basis of the structural organization of the nucleolus (49). There is growing evidence to suggest that the nucleolus is involved in additional cellular functions that are not directly related to ribosomal biogenesis (17,19–21). The non-ribosomal functions of the nucleolus include cell cycle control, apoptosis, viral infection, DNA replication, DNA repair and stress signaling. Given these diverse roles, it is unsurprising that the nucleolus contains several hundred proteins, and that the protein content of the nucleolus dynamically changes under stress conditions (22–24,50,51). Despite extensive investigation, however, the precise functions for a significant number of nucleolar proteins remain unknown. In this study, we successfully identified a new rDNA silencing factor, Nsi1, by systematically analyzing the dynamic localization of nucleolar proteins in response to a variety of environmental stresses. As evidenced by our study, this functional proteomic approach provides new insights into nucleolar architecture and nucleolar protein function. Combined with conventional biochemical and cell biological analyses, functional proteomic approaches will be useful in elucidating novel functions of uncharacterized proteins in other organelles as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–6.
FUNDING
Basic Science Research Program of the National Research Foundation of Korea (2010-0013678); 21C Frontier Functional Proteomics Project (FPR08A1-060) and 21C Frontier Microbial Genomics and Application Center Program (MG-11-2008-09-004-00), Republic of Korea. Funding for open access charge: The Brain Korea 21 Project from the Ministry of Education, Science and Technology, Republic of Korea.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Danesh Moazed and Dr Kurt W. Runge for generously providing the yeast strains. We also thank the members of our laboratory for helpful discussions.
REFERENCES
- 1.Johnston M, Hillier L, Riles L, Albermann K, Andre B, Ansorge W, Benes V, Bruckner M, Delius H, Dubois E, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature. 1997;387:87–90. [PMC free article] [PubMed] [Google Scholar]
- 2.Sinclair DA, Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 5.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11:2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- 17.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 18.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 19.Visintin R, Amon A. The nucleolus: the magician’s hat for cell cycle tricks. Curr. Opin. Cell Biol. 2000;12:752. doi: 10.1016/s0955-0674(00)00165-4. [DOI] [PubMed] [Google Scholar]
- 20.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J. Cell. Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 23.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 24.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 25.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 27.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 28.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 29.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray A, Hector RE, Roy N, Song JH, Berkner KL, Runge KW. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 2003;33:522–526. doi: 10.1038/ng1132. [DOI] [PubMed] [Google Scholar]
- 32.Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park PU, McVey M, Guarente L. Separation of mother and daughter cells. Methods Enzymol. 2002;351:468–477. doi: 10.1016/s0076-6879(02)51865-6. [DOI] [PubMed] [Google Scholar]
- 34.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegmeier F, Huang J, Rahal R, Zmolik J, Moazed D, Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqi IN, Dodd JA, Vu L, Eliason K, Oakes ML, Keener J, Moore R, Young MK, Nomura M. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 2001;20:4512–4521. doi: 10.1093/emboj/20.16.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keys DA, Vu L, Steffan JS, Dodd JA, Yamamoto RT, Nogi Y, Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- 40.Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 41.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohanty BK, Bastia D. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 2004;279:1932–1941. doi: 10.1074/jbc.M309078200. [DOI] [PubMed] [Google Scholar]
- 43.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 44.Li C, Mueller JE, Bryk M. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol. Biol. Cell. 2006;17:3848–3859. doi: 10.1091/mbc.E06-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 46.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 47.Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, et al. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheer U, Hock R. Structure and function of the nucleolus. Curr. Opin. Cell. Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 50.Boisvert FM, Lamond AI. p53-Dependent subcellular proteome localization following DNA damage. Proteomics. 2010;10:4087–4097. doi: 10.1002/pmic.201000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell Proteomics. 2010;9:117–130. doi: 10.1074/mcp.M900338-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.