Abstract
The main methods for producing genetically engineered cells use viral vectors for which safety issues and manufacturing costs remain a concern. In addition, selection of desired cells typically relies on the use of cytotoxic drugs with long culture times. Here, we introduce an efficient non-viral approach combining the Sleeping Beauty (SB) Transposon System with selective proliferation of engineered cells by chemically induced dimerization (CID) of growth factor receptors. Minicircles carrying a SB transposon cassette containing a reporter transgene and a gene for the F36VFGFR1 fusion protein were delivered to the hematopoietic cell line Ba/F3. Stably-transduced Ba/F3 cell populations with >98% purity were obtained within 1 week using this positive selection strategy. Copy number analysis by quantitative PCR (qPCR) revealed that CID-selected cells contain on average higher copy numbers of transgenes than flow cytometry-selected cells, demonstrating selective advantage for cells with multiple transposon insertions. A diverse population of cells is present both before and after culture in CID media, although site-specific qPCR of transposon junctions show that population diversity is significantly reduced after selection due to preferential expansion of clones with multiple integration events. This non-viral, positive selection approach is an attractive alternative for producing engineered cells.
INTRODUCTION
Genetically engineered cells are essential elements of biotechnologies ranging from recombinant protein production to cell therapy (1,2). However, several limitations and inefficiencies still remain in generating engineered cells. The preparation of stably transformed cell lines with high protein production efficiency is a current limitation in the manufacture of recombinant proteins (3). As a result, hundreds of clonal cell lines are typically screened to identify high production cell lines. For cell therapy applications, integrating viral vectors such as retrovirus are commonly used to stably transduce cells, and this approach raises safety concerns due to the possibility of insertional mutagenesis mediated by the non-random integration pattern of most integrating viruses (4). Clinical grade viral vectors are also costly to manufacture. The desired cells then need to be selected and/or amplified to necessary quantities and purities (5,6). Technologies that improve and simplify the manufacturing process for cell therapies are critical in facilitating clinical translation and adoption of these approaches.
The Sleeping Beauty (SB) transposon system is an attractive, non-viral alternative to viral delivery systems (7). This DNA transposon system was revived by molecular reconstruction of inactive sequences from fish by Ivics and coworkers (8). Since this discovery, the SB transposon system has become one of the most widely used transposition systems for non-viral gene therapy and has been approved for human clinical trials in a T-cell therapy application (9). Although insertional mutagenesis caused by transposition is still a possibility, SB transposition has safety advantages over most of the commonly-used integrating vector systems [retrovirus, lentivirus and adeno-associated virus (AAV)] because of weaker preference for integrating in transcriptional units and their upstream regulatory sequences (10,11). The SB transposase enzyme, that can be given in trans as DNA or RNA, inserts the transposon expression cassette into mammalian cells through a ‘cut and paste’ mechanism. The transposase binds to terminal inverted repeats (IR) that contain SB-binding elements (DR), excises the sequence flanked by the repeats, and integrates the transposon into thymine–adenine (TA) sites in the genome (8). However, both non-viral transfection and transposon integration efficiencies are low in primary cell types and therefore prolonged culture time is typically required to propagate the desired cells.
In this work, we present a non-viral approach for expedited and enriched generation of engineered cells by combining SB integration technology with chemically responsive amplification mechanisms. Specific signaling pathways can be chemically controlled using fusion proteins of small molecule-responsive dimerization domains with signaling domains that are activated upon dimerization (12,13). A F36VFGFR-1 fusion protein constructed by Whitney et al. (14) contains the modified FK506 protein (F36V) that binds to the chemical inducer of dimerization (CID) AP20187 fused to the cytoplasmic domain of the fibroblast growth factor receptor-1 (FGFR-1). When dimerized, FGFR-1 initiates several signaling pathways that include cell proliferation (15). CID-dependent signaling through the F36VFGFR-1 construct provides both specificity and control. While FGFR-1 is activated through different fibroblast growth factors and requires heparin, F36VFGFR-1 is activated specifically by the presence of AP20187 (15). Removal of AP20187 also terminates activation of F36VFGFR-1 (16,17).
A SB transposon cassette containing two genes, encoding enhanced green fluorescent protein (EGFP) reporter and F36VFGFR-1, was constructed. We hypothesized that cells with successful transposition could be selectively amplified through CID-stimulated proliferation as an approach to overcome the limitations of poor non-viral transfection and low integration frequency. We report that using this non-viral gene transfer and amplification approach, nearly-pure populations (>98% containing integrated transposon cassettes) of a murine hematopoietic cell line (Ba/F3) could be obtained within 7 days of CID selection. Integration analysis suggested that multiple insertion sites were being preferentially selected through this process.
MATERIALS AND METHODS
Plasmids
The pT3/eGIF plasmid that carries the T3 SB transposon cassette containing an EF1α promoter, EGFP gene, IRES and F36VFGFR1 gene was constructed using standard molecular biology cloning techniques from pT3-MSCV-U3-GFP (generously provided by Richard A. Morgan, National Cancer Institute) and pMGIF36Vfgf (generously provided by Charles Murry, University of Washington) (14,18). To create pMCT3/eGIF, the plasmid for producing minicircles, the NdeI/SbfI fragment of pT3/eGIF containing only the transposon cassette was blunt-ended and cloned into EcoRV site of pMC.BESPX (generously provided by Mark A. Kay, Stanford University) (19,20). Minicircles were produced from this plasmid and purified according to System Biosciences user manual for minicircle DNA vector technology. The CMV(CAT)T7-SB100X plasmid containing the hyperactive variant of Sleeping Beauty transposase SB100X was a generous gift from Zsuzsanna Izsvak (Max Delbruck Center for Molecular Medicine, Berlin, Germany). All plasmids were amplified under endotoxin-free conditions using Qiagen Endofree Plasmid Kit.
Ba/F3 culture and transfection
Ba/F3 cells were cultured in RPMI with 10% FBS and 1% antibiotic/antimicotic supplemented with mrIL-3 5 ng/ml (BD Bioscience). The optimized nucleofection protocol for Ba/F3 cells using Nucleofector (Lonza) was followed (program X-001, Nucleofector Kit V). Two million cells were used per nucleofection with varying amounts of plasmid DNA. Cells were grown in IL-3 containing media for a week after nucleofection to achieve stable transfection. For CID selection, cells were cultured in RPMI with 10% FBS and 1% antibiotic/antimicotic with 100 nM AP20187 (ARIAD Pharmaceuticals, Inc).
Flow cytometric analysis
For flow cytometry analysis, live cells were selected based on propidium iodide exclusion by adding propidium iodide in the flow cytometry buffer to 4 µg/ml. To stain for the HA tag on F36VFGFR1, cells were fixed on ice in 2% paraformaldehyde, resuspended in ice-cold methanol, washed with permeabilization staining buffer (PBS, 0.5% BSA, 0.1% saponin), resuspended in PBS with anti-HA-PE antibodies (Miltenyi Biotec Inc) at a 1:10 dilution and incubated for 30 min in the dark at room temperature. Cells were then washed and resuspended in PBS with 0.3% BSA for flow cytometry. Flow cytometric analysis was carried out on Becton Dickinson FACScan using analysis using FlowJo software. Appropriate negative controls (untransfected Ba/F3 cells with and without propidium iodide staining) were used for compensation and gating. A Becton Dickinson FACSAria II was used for cell sorting. All flow cytometry work was conducted at the UW Immunology Flow Cytometry Facility.
Growth analysis
Ba/F3 cells were washed with PBS and diluted to the concentration of 300 000 cells/ml and grown in RPMI with 10% FBS and 1% antibiotic/antimicotic with, or without factors (IL-3 or AP20187). Concentration of live cells was measured by trypan blue exclusion in hemocytometer every 2 days.
Determination of transposon copy number
Genomic DNA was extracted using Puregene Kit A according to manufacturer’s instructions (Qiagen) and qPCR was performed in Applied Biosystems 7300 instrument (Real-Time PCR System) using PowerSYBR Green Master Mix (Applied Biosystems). Primers used for qPCR were designed using Primer3 software (http://simgene.com/Primer3): EGFP forward primer: 5′-ACGTAAACGGCCACAAGTTC, EGFP reverse primer: 5′-AAGTCGTGCTGCTTCATGTG, albumin forward primer: 5′-CTCAGGTGTCAACCCCAACT, albumin reverse primer: 5′-CCACACAAGGCAGTCTCTGA. EGFP primers are specific for the EGFP gene in the transposon, and ALBm primers are specific for the Albumin gene, that is represented at a single copy in mouse haploid genome. Standard curves were generated using genomic DNA of a BaF3 clone with a single insertion of transposon. This clone was obtained by diluting the culture to single cell suspension and growing colonies in 96-well plate. Several colonies were then screened for single insertions by using qPCR to determine the absolute copy number of GFP gene. An amount of 10 ng of genomic DNA was used in qPCR for determination of transposon copy number. Copy number was calculated using CT method (21). To determine the representation of identified integration sites in cell populations, primer pairs were designed for individual transposon junctions (Supplementary Table S2). PCR products were used to generate standard curves, and copy number determined using an absolute quantitation method (22).
Integration site analysis
Integration site analysis was done by Linear Amplification Mediated Polymerase Chain Reaction (LAM-PCR) (23), as modified by Harkey et al. (24) and adapted here for SB vectors. Briefly, a single-stranded copy of the vector-host junction was generated by linear extension from a biotinylated vector primer. This single-stranded junction fragment was trapped and isolated on Streptavidin-coated magnetic beads, and converted to double-stranded DNA by random primed polymerization. Three aliquots of junction DNA were digested with three separate four-base restriction endonucleases (Tsp509I, RsaI or HaeIII) chosen to cut at the nearest host site. An anchor primer was ligated to the newly exposed ends and the junction region was amplified by nested PCR. The complexity of the population of integration sites was initially observed by the complexity of PCR products seen by 4–20% polyacrylamide gel electrophoresis. Individual insertion sites were identified by cloning fragments eluted from the gel and sequencing individual clones. Unique primers for SB were as follows: biotinylated extension primer: 5′-biotin-TGTGGTGTAGTTGAAAAACGAG, nested primer #1: TTTTAATGACTCCAACTTAAGTG, and nested primer #2: TATGTAAACTTCCGACTTCAAC. Additional primers were as described previously (24).
RESULTS
pT3/eGIF plasmid and minicircle construction
The pT3/eGIF plasmid that carries the hyperactive transposon (T3) cassette containing both the EGFP reporter gene and the F36VFGFR-1 gene expressed under the EF1α (elongation factor-1α) promoter through an internal ribosome entry sequence (IRES) was constructed (Figure 1). The hyperactive transposon, developed by Yant et al., contains a duplication of the left IR/DR structure in the transposon along with additional flanking TA dinucleotides and has been shown to be three times more active than the original transposon sequences (25). The analogous plasmid (pT3/mGIF, 8.3 kb) with the murine stem cell virus (MSCV) promoter was also constructed. These two promoter sequences were selected because they have both been shown to drive strong expression across different types of cells, including stem cells (26,27) The EF1α promoter drove more robust transgene expression (4–5 times) than the MSCV promoter in Ba/F3 cells (data not shown) so the EF1α promoter construct was used for all remaining studies.
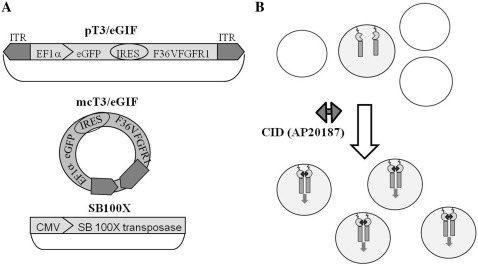
Figure 1.
The SB transposon system combined with chemical-induced dimerization (CID). (A) Constructs used in this study. pT3/eGIF plasmid carries the SB transposon containing of the EF1α promoter, eGFP gene, Internal Ribosome Entry Site (IRES) and a fusion gene of synthetic dimerization domain F36V and a Fibroblast Growth Factor Receptor 1 (FGFR1), positioned between Inverted Terminal Repeats (ITRs, black arrows); mcT3/eGIF is the minicircle derived from pT3/eGIF and SB100X plasmid contains the hyperactive SB transposase SB100X under the CMV promoter. (B) CID (AP20187) selection for cells with integrated transposon. Transfected cells expressing the F36VFGFR1 protein undergo selective proliferation through CID-induced cell signaling. After selection, a nearly homogenous population of stably transduced cells is obtained.
In our initial studies, we noted that cell viability after nucleofection was inversely related to the amount of DNA present during nucleofection. To optimize the delivery of our transgene, we also prepared minicircles of the T3/eGIF transposon (mcT3/eGIF) that lack the bacterial plasmid backbone (19,20). Transposon cassettes were cloned into a minicircle production plasmid and mini-circles were generated by recombination of flanking attB and attP sites followed by degradation of the plasmid DNA backbone.
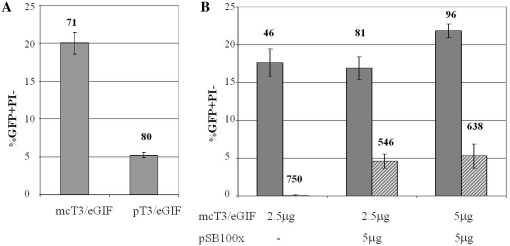
Transfection studies were performed in the IL-3 dependent, FGFR-1 responsive pro-B cell line Ba/F3 using nucleofection for gene delivery. The efficiency of nucleofection was 4 times higher for minicircles carrying transposon, than the pT3/eGIF plasmid, when equal amount of the DNA was used (Figure 2A). The mean GFP fluorescence of the transfected population is similar for both minicircle- and plasmid-transfected cells, indicating that expression efficiency is similar between the two constructs. No difference in cell viability after nucleofection was observed as well (data not shown). Next, the transposon integration efficiency was tested by codelivery of mcT3/eGIF with the SB100X transposase plasmid by flow cytometry analysis for EGFP-positive cells 24 h and 7 days post-nucleofection to assess for efficiency of transient and stable expression, respectively. By Day 7 post-nucleofection, delivered transposon in the absence of transposase was eliminated, resulting in 0.1% EGFP+ cells. In contrast, ∼5% of live cells (or 25% of transiently transfected population) carried integrated transposons due to codelivery of transposase (Figure 2B). Increasing the transposon to transposase plasmids ratio from 1:2 to 1:1 (by mass) did not significantly affect transposition efficiency.
Figure 2.
Transient and stable transfection of Ba/F3 cells using SB transposon system. (A) Comparison of transient transfection efficiency by nucleofection delivery of plasmid and minicircle bearing T3/eGIF transposon. Ba/F3 cells were nucleofected with 5 µg of either plasmid pT3/eGIF or minicircle mcT3/eGIF. Flow cytometry was performed 24 h after nucleofection. (B) Integration of T3/eGIF transposon. Ba/F3 cells were nucleofected with varying DNA amounts of transposon minicircle mcT3/eGIF and transposase plasmid pSB100x. Flow cytometry was performed 24 h (gray bars) and 7 days (striped bars) after nucleofection to assess transient and stable transfection efficiency, respectively. Bars represent SD of three independent transfections. Averages of mean fluorescence intensity of GFP+/PI− populations are indicated above the graphs in RFU.
Transformed Ba/F3 cells are selectively amplified by CID selection and show CID-dependent proliferation
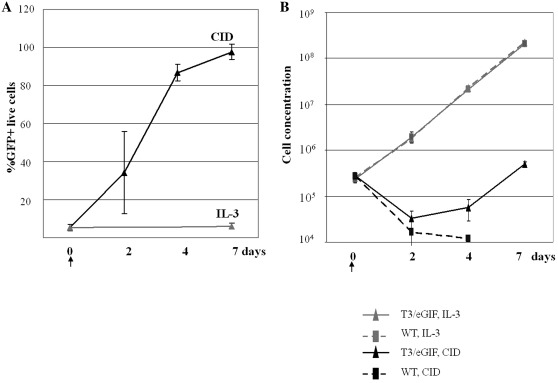
Pools of Ba/F3 cells with a starting integrated transposon frequency of ∼5% were cultured in the presence of 100 nM AP20187 in IL3-free conditions to allow for CID selection. Ba/F3 cells are normally IL-3 dependent. The population of transformed cells was monitored by flow cytometry at 2, 4 and 7 days post-selection. The percentage of EGFP+ cells in the live cell population (negative for propidium iodide staining) increased steadily, reaching >98% with 7 days of CID selection (Figure 3A). Transformed cells maintained in IL-3 kept a stable population of 5.6 ± 1.8% of EGFP+ cells (Figure 3A). The growth kinetics of transformed Ba/F3 cells grown in IL3 and CID was also determined (Figure 3B). Control (untransfected) Ba/F3 and transfected Ba/F3 cells showed similar growth curves when cultured in IL-3 containing media. Transfected Ba/F3 cells decreased in population 2 days after transitioning to CID-containing, IL-3-deficient media due to death of untransformed cells. However, cell numbers began to increase at 4 days post-selection and continued to expand with CID-induced proliferation. The doubling time of transformed Ba/F3 cells under CID expansion was similar as control cells growing in IL-3 media (doubling time of 19–23 h, data not shown). As expected, control Ba/F3 cells were unable to survive in the absence of IL-3.
Figure 3.
Proliferation of transfected Ba/F3 cells cultured in IL-3 or CID-containing media. (A) Flow cytometry analysis of stably transfected populations of BaF3 cells with T3/eGIF transposon, grown in the presence of CID (AP20187) or IL-3 at different time points. (B) Growth curve of untransfected (WT) or SD transfected (T3/eGIF) Ba/F3 cells cultured in either IL-3 or CID (AP20187) containing media. An arrow indicates time (Day 0) when CID selection commenced. Bars represent SDs of three independent cultures.
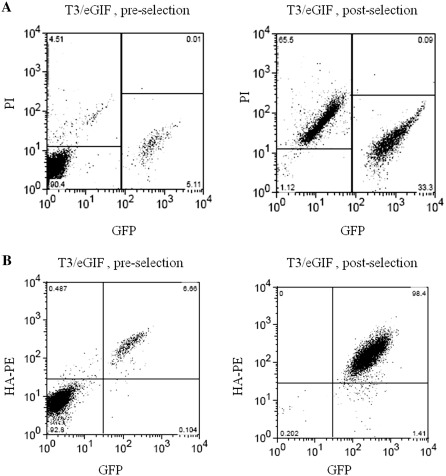
Expression of EGFP and F36VFGFR1 was monitored by EGFP fluorescence and an antibody against a hemagglutinin (HA) tag on the F36VFGFR1 both pre- and post-selection (representative plots shown in Figure 4B). The dual positive cell population increased from 6.4 ± 2.1% pre-selection to 96.6 ± 2.0% post-selection. Another 1.4% of cells were EGFP positive but did not stain positive for F36VFGFR1. The mean fluorescence of the EGFP+ population also increased from 565 ± 25 relative fluorescence units (RFU) to 1065 ± 91 RFU post-selection. Cell viability after selection was assessed by propidium iodide (PI) exclusion staining (Figure 4A). Before selection, cells that have recovered from nucleofection are healthy (<5% PI+). After CID selection, two major populations of cells exist: live, EGFP+ cells (33%) and dead cells (65%). The dead cells are the untransfected cells (EGFP−) that do not survive after IL-3 withdrawal.
Figure 4.
Flow cytometry scattergrams of Ba/F3 cells stably transfected with T3/eGIF transposon, pre- and post-selection with CID. (A) GFP versus Propidium Iodine (PI). (B) GFP versus HA-PE (F36VFGFR1-HA expression). Pre-selection, cells were cultured in IL-3 media. Post-selection, cells were cultured in media containing 100 nM CID (AP20187) for 7 days.
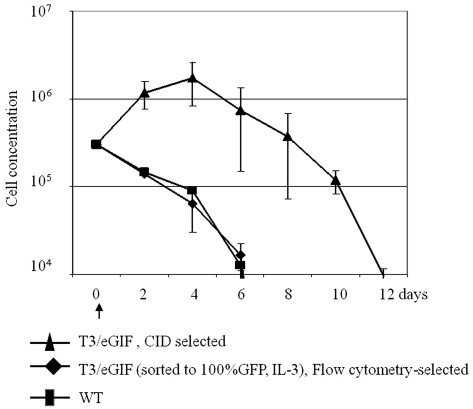
Because SB integration is random, it is theoretically possible that transformed cells could proliferate independently of IL-3 or CID due to an insertional mutagenesis event. To exclude this low probability event, a cell survival study after IL-3 and CID withdrawal was conducted with three Ba/F3 populations: untransfected Ba/F3 (control WT), stably transfected Ba/F3 selected by flow cytometry, and stably transfected Ba/F3 selected by CID culture for 2 weeks (Figure 5). The first two populations were cultured in IL-3 containing media prior to factor withdrawal. As reported previously, removal of IL-3 from control Ba/F3 cells results in loss of cell viability that begins within 24 h after factor withdrawal. (28) Removal of IL-3 from stably transfected cells also resulted in cell death with similar kinetics as control cells. CID withdrawal from CID-selected cells reversed proliferation and resulted in cell death. The population decline rate for this group was similar to the control, except for the 4 days offset.
Figure 5.
Cell survival after withdrawal of IL-3 or CID. Three different Ba/F3 cell populations were investigated for survival: wild-type BaF3 (WT) after IL-3 withdrawal, Ba/F3 cells stably transduced with T3/eGIF transposon, selected by flow cytometry, after IL-3 withdrawal and stably transduced Ba/F3cells with T3/eGIF transposon selected by CID, after CID withdrawal. An arrow indicates the time of factors withdrawal (Day 0). Bars represent SDs of three independent cultures.
Growth of stably transduced Ba/F3 cells in the presence of CID leads to selection of a subpopulation with multiple transposon insertions
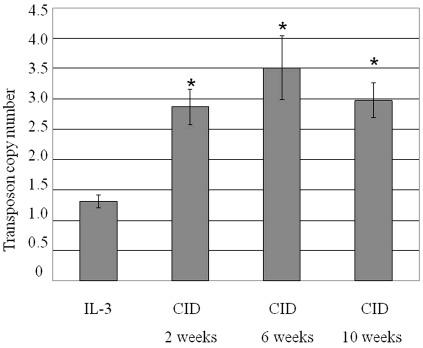
The average copy number of integrated transposons before and at various durations of CID selection (2, 6 and 10 weeks) was determined by real-time quantitative polymerase chain reaction (qPCR) with primers specific for the Egfp gene (transposon detection) and Album (albumin, a single copy gene) (Figure 6). A purified population of stably transduced Ba/F3 cells growing in IL-3 conditions was selected by flow cytometry 3 weeks post-transfection. The average transposon copy number in this stably transfected cell pool was 1.3 ± 0.1 per haploid genome. This number was increased to 2.9 ± 0.3 after 2 weeks of CID selection and did not increase further with longer selection times. These data suggest that cells with multiple insertion sites have an advantage when cultured with dimerizer.
Figure 6.
Transposon copy number per mouse haploid genome. Genomic DNA was isolated from population of Ba/F3 cells stably transfected with T3/eGIF transposon, before and after CID selection at different time points. The average transposon copy number was determined by quantitative PCR. Bars represent SDs of three independent experiments. *P < 0.01 compared to IL-3-treated cells by t-test analysis.
It is possible that cells with F36VFGFR1 integration at certain sites show preferential growth characteristics due to either insertional mutagenesis or chromosomal positional effects and as a result may be over-represented in CID-amplified populations. CID withdrawal experiments showed that Ba/F3 cell growth remained dependent on either IL-3 or CID (Figure 5). To assess the possibility of preferential site integration, integration sites in Ba/F3 cells before and after CID amplification were determined by linear amplification-mediated PCR (LAM–PCR) Multiarm LAM–PCR has been shown to improve detection capacity compared to single-arm LAM–PCR; thus, three-arm LAM–PCR with restriction enzymes HaeIII, Tsp509I and RsaI was conducted (24). Each of the three independently-derived populations of Ba/F3 cells transfected with the SB100X and mcT3/eGIF constructs were cultured in IL-3 for 2 weeks and then split into two groups. One group was cultured in IL-3 conditions (‘IL-3’ group) and the second group (‘CID’ group) was grown with AP20187 for 2 weeks before LAM–PCR analysis. The electrophoretic banding profiles generated by the three-arm LAM–PCR analyses are shown in Supplementary Figure S1. The non-selected samples show complex but distinct patterns, indicative of multiple integration events. The three CID-selected samples also show complex patterns but the distribution of bands changes after CID-selection and remain unique for each sample. We also took one sample and separated it into three groups for CID selection. The distributions of bands from these three CID-selected samples were similar, showing that the initial population does determine the integration patterns after CID selection (Supplementary Figure S1, lanes B1, B2, B3).
Finally, at least 10 integration sites from independent CID-selected populations were determined by sequencing (Supplementary Table S1). In random samplings of insertion site sequences, 80–100% of the reactions yielded unique sites for each cell population tested. These results suggest that a heterogeneous population of clones remains after 2 weeks of CID selection. To ascertain relative representation of specific clones after CID selection, the copy number of specific integration sites was determined by absolute quantitation of qPCR results. Three randomly selected individual transposon junctions were detected in CID-selected populations from which they were identified at all time points (2, 6 and 10 weeks of selection) at anywhere between 0.7% and 25% of cells. These junctions were not detected in either the parent-unselected population nor the other CID selected pools. These results indicate that there is a substantial variation in the representation of individual clones, and that there is no detectable overlap of integration sites between individual experiments.
DISCUSSION
In this work, we present a positive-selection approach for generating engineered mammalian cells. By combining SB transposon technology and CID technology, desired cells are generated by non-viral transfection followed by selective proliferation of desired cells responsive to a small molecule trigger. This method provides safety, cost, and time advantages over the commonly used protocols of viral transduction followed by drug selection. In comparison with integrating viruses, such as human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) (29,30), SB shows lower integration preference for intragenic regions (10,25,31). For clinical cell therapy applications, GMP-grade virus must also be produced.
Positive selection of cells addresses the main disadvantages of drug selection, which is the predominant approach for isolating engineered cells (6,32). Cytocidal culture conditions reduce the growth rate of cells, exacerbating the already limiting parameter of prolonged culture times required for cell production (33). In contrast, CID-induced proliferation by FGFR activation has been shown to increase the proliferation rate in several cell types compared to control cells (34,35). Also, the method of selecting stably transduced cells has been shown to affect the heterogeneity of the resulting population, with drug selection yielding cells with mosaic expression patterns due to gene silencing (36). We show in this work that CID-dependent cell amplification selects for high expression of transgene. Because growth is dependent on continuous CID selection over several days, a bias toward engineered cells with more temporally homogeneous expression patterns is also expected.
CID technology has been used to affect cell signaling via several protein partners including T cell receptor domains, erythropoietin receptor (EpoR) and mpl receptor. For this work, the F36VFGFR1 construct was chosen because it has been used to trigger expansion of both short-term repopulating hematopoietic stem cells (HSCs) and skeletal myoblasts, and is therefore relevant for cell therapy applications (14,16,17). IL-3 dependent, proB Ba/F3 cells were selected as a model hematopoietic cell line because the constitutively active BCR–FGFR1 fusion protein responsible for cases of chronic myeloid leukemia has been shown to render this cell line IL-3 independent (37). In addition, Blau et al. (13) showed that CID-based signaling through the EpoR can also remove IL-3 dependency in these cells. Transfection of Ba/F3 cells has been used extensively as an approach to identify and/or validate potential oncogenes (38–40).
Lipid, cationic polymer and electroporation delivery approaches were tested to optimize transfection of Ba/F3 cells. Of these, nucleofection was found to give relatively high transient transfection of plasmids. The latest generation of hyperactive SB transposon and transposase were both used. The T3 transposon contains extra flanking TA dinucleotides, important for transposon excision, and a duplication of the left IR/DR sequence, important for transposase binding and multimerization (25,41,42). SB100X is a hyperactive transposase developed through a PCR-based DNA-shuffling strategy followed by screening of activity in mammalian cells (43). SB100X was shown to be 100-fold more potent in transposition than the original protein.
Minicircle vectors were used to carry the transposon cassette. The cell survival after nucleofection has been reported to increase with decreasing amount of delivered DNA (44), and we also observed this for Ba/F3 cells. The amount and sequence of DNA between the transposon ends has also been shown to affect transposition; shorter DNA segments result in increased transposition efficiency (42). This observation was attributed to the transposase complex binding to one end of the transposon and then searching bidirectionally for the second end. Minicircle delivery of transposon cassettes is therefore an attractive alternative to plasmid delivery. Transfection efficiency by nucleofection of Ba/F3 cells is four times higher when minicircles are used (Figure 2A). In addition to improved delivery and transposition efficiencies, minicricles may offer reduced inflammation from CpG responses (20).
CID selection for 1 week allowed us to produce a nearly pure (98%) population of cells carrying transposon due to preferential survival and proliferation of cells expressing F36VFGFR1 when grown in CID media (Figure 3A). Dual expression of both transgenes, EGFP and F36VFGFR1, was confirmed in >96% of surviving cells by flow cytometry (Figure 4). The growth rate of cells after CID selection in the presence of AP20187 is similar to that of control Ba/F3 cells grown in Il-3 conditions, with a doubling time of 19–23 h (data not shown). While FGFR1 dimerization initiates a complicated signaling cascade, there are common proteins involved with IL-3 activation. The MAP kinase signaling pathway is activated by CID-induced FGFR1 dimerization and also by IL-3 stimulation of Ba/F3 cells (14,34,45,46). In addition, inhibition of FGFR activity has been shown to lead to downregulation of cyclin D proteins, a MAP kinase-regulated protein and IL-3 withdrawal from IL-3 cells also reduces cyclin D levels in Ba/F3 (47,48).
CID-dependent proliferation of engineered cells was confirmed by cell death upon withdrawal of CID (Figure 5). The time lag observed between CID withdrawal and cell death may be attributed to remaining intracellular CID and the positive feedback downstream of FGFR1 signaling. Indeed, Xian et al. note that phosphorylated MAP kinase ERK1 (pERK1) levels are reduced after dimerizer withdrawal from mammary epithelial cells but remain elevated compared to controls (49). The same effect was also observed after CID withdrawal from F36VFGFR1 transduced myoblasts (14). Finally, it is interesting to note that withdrawal of dimerizer from Ba/F3 cells induced to proliferate due to EpoR1 activation results in a 5-day time lag before decline in cell numbers is observed (13).
Integration site analysis revealed the presence of a complex population of clones bearing stable SB integrants whether the cells were grown in IL-3 or selected with CID treatment (Supplementary Figure S1). The altered integration site patterns caused by CID suggest selection of a subpopulation of clones. This supports our conclusion from qPCR that clones with higher copy numbers are selected (Figure 6). Selection of clones with higher copy numbers may be advantageous for biotechnology applications due to the potential of increased protein production efficiency. For cell therapy applications, higher number of integration events increases the possibility of insertional mutagenesis. Copy number analysis of identified individual integration sites reveal that representation of clones is uneven, with some being represented in 25% of the CID-selected population. Therefore, the diversity of clones is substantially reduced after CID selection. The pattern of CID selected sites is determined by the original pool of transposition events, since specific transposon junctions were detected only in the population in which they were identified.
In summary, we demonstrate here that a nearly pure population of stably transduced cells can be generated by non-viral delivery of desired transgenes through a combination of SB transposon-mediated integration and selective amplification using chemically induced dimerizer. We anticipate that this approach may be used to produce engineered cells for biotechnology and clinical cell therapy applications. Dimerizer-induced growth provides cost and reproducibility advantages to natural ligand stimulation in ex vivo cell culture, and also provides a means to control engineered cell behavior in vivo (17). While the FGFR1-signaling pathway was used in this work to stimulate proliferation, the choice of CID-responsive fusion protein can be tailored for the application of interest. Future work will include application of this described approach to primary HSCs. F36VFGFR1-induced proliferation of murine HSCs has been shown to expand short term HSCs that eventually differentiate to granulocytes and monocytes (17), and may be a viable approach for generating engineered monocytes or monocyte-derived cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1 and S2, Supplementary Figure S1.
FUNDING
Core Center of Excellence in Molecular Hematology (CCEMH) (Grant P30 DK56465) for performing the LAM–PCR analyses; Life Sciences Discovery Fund [grant number 2361524]; NIH R01DK74522. Funding for open access charge: Life Science Discoveries fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Maja Zavaljevski and David Emery (University of Washington) for helpful discussions. The authors are grateful to Mark A. Kay (Stanford University) for providing the minicircle expression system (pMC.BESPX plasmid and ZYCY10P3S2T E. coli producer strain), Richard A. Morgan (National Cancer Institute) for providing pT3-MSCV-U3-GFP plasmid, Charles Murry (University of Washington) for providing the pMGIF36Vfgf plasmid and Zsusanna Izsvak (Max Delbruck Center for Molecular Medicine, Berlin, Germany) for providing the CMV(CAT)T7-SB100X plasmid. The authors thank ARIAD Pharmaceuticals for providing AP20187.
REFERENCES
- 1.Birch JR, Racher AJ. Antibody production. Adv. Drug Deliv. Rev. 2006;58:671–685. doi: 10.1016/j.addr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Trounson A, Thakar RG, Lomax G, Gibbons D. Clinical trials for stem cell therapies. BMC Med. 2011;9:52. doi: 10.1186/1741-7015-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - Where do we go from here? Biotechnol. Adv. 2009;27:1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCcormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 5.Ahrlund-Richter L, De Luca M, Marshak DR, Munsie M, Veiga A, Rao M. Isolation and Production of Cells Suitable for Human Therapy: Challenges Ahead. Cell Stem Cell. 2009;4:20–26. doi: 10.1016/j.stem.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Cooper LJN, Ausubel L, Gutierrez M, Stephan S, Shakeley R, Olivares S, Serrano LM, Burton L, Jensen MCV, Forman SJ, et al. Manufacturing of gene-modified cytotoxic T lymphocytes for autologous cellular therapy for lymphoma. Cytotherapy. 2006;8:105–117. doi: 10.1080/14653240600620176. [DOI] [PubMed] [Google Scholar]
- 7.Izsvak Z, Hackett PB, Cooper LJN, Ivics Z. Translating Sleeping Beauty transposition into cellular therapies: victories and challenges. Bioessays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 9.Aronovich EL, McIvor RS, Hackett PB. The Sleeping Beauty transposon system: a non-viral vector for gene therapy. Hum. Mol. Genet. 2011;20:R14–R20. doi: 10.1093/hmg/ddr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 11.Yant SR, Wu XL, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal-transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 13.Blau CA, Peterson KR, Drachman JG, Spencer DM. A proliferation switch for genetically modified cells. Proc. Natl Acad. Sci. USA. 1997;94:3076–3081. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitney ML, Otto KG, Blau CA, Reinecke H, Murry CE. Control of myoblast proliferation with a synthetic ligand. J. Biol. Chem. 2001;276:41191–41196. doi: 10.1074/jbc.M103191200. [DOI] [PubMed] [Google Scholar]
- 15.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Stevens KR, Rolle MW, Minami E, Ueno S, Nourse MB, Virag JI, Reinecke H, Murry CE. Chemical dimerization of fibroblast growth factor receptor-1 induces myoblast proliferation, increases intracardiac graft size, and reduces ventricular dilation in infarcted hearts. Hum. Gene Ther. 2007;18:401–412. doi: 10.1089/hum.2006.161. [DOI] [PubMed] [Google Scholar]
- 17.Weinreich MA, Lintmaer I, Wang LL, Liggitt HD, Harkey MA, Blau CA. Growth factor receptors as regulators of hematopoiesis. Blood. 2006;108:3713–3721. doi: 10.1182/blood-2006-01-012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol. Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 20.Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 2010;28:1287–U96. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat. Protocol. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Whelan JA, Russell NB, Whelan MA. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Meth. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H, Williams DA, Ragg S, Hesemann CU, von Kalle C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- 24.Harkey MA, Kaul R, Jacobs MA, Kurre P, Bovee D, Levy R, Blau CA. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cell. Dev. 2007;16:381–392. doi: 10.1089/scd.2007.0015. [DOI] [PubMed] [Google Scholar]
- 25.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong S, Hwang DY, Yoon S, Isacson O, Ramezani A, Hawley RG, Kim KS. Functional analysis of various promoters in lentiviral vectors at different stages of in vitro differentiation of mouse embryonic stem cells. Mol. Ther. 2007;15:1630–1639. doi: 10.1038/sj.mt.6300251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol. Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 28.Collins MKL, Marvel J, Malde P, Lopezrivas A. Interleukin-3 protects murine bone-marrow cells from apoptosis induced by dna damaging agents. J. Exp. Med. 1992;176:1043–1051. doi: 10.1084/jem.176.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 30.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Guo HF, Tammana S, Jung YC, Mellgren E, Bassi P, Cao Q, Tu ZJ, Kim YC, Ekker SC, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and PiggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahara M, Ueda H, Morita S, Tsumoto K, Kumagai I, Nagamune T. Bypassing antibiotic selection: positive screening of genetically modified cells with an antigen-dependent proliferation switch. Nucleic Acids Res. 2003;31:e32. doi: 10.1093/nar/gng032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu MB, Kern JA, Todd P, Kompala DS. Effect of amplification of dhfr and lac z genes on growth and beta-galactosidase expression in suspension-cultures of recombinant cho cells. Cytotechnolol. 1992;9:237–245. doi: 10.1007/BF02521751. [DOI] [PubMed] [Google Scholar]
- 34.Nourse MB, Rolle MW, Pabon LM, Murry CE. Selective control of endothelial cell proliferation with a synthetic dimerizer of FGF receptor-1. Laboratory Investigation. 2007;87:828–835. doi: 10.1038/labinvest.3700597. [DOI] [PubMed] [Google Scholar]
- 35.Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J. Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufman WL, Kocman , Agrawal V, Rahn HP, Besser D, Gossen M. Homogeneity and persistence of transgene expression by omitting antibiotic selection in cell line isolation. Nucleic Acids Res. 2008;36:e111. doi: 10.1093/nar/gkn508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demiroglu A, Steer EJ, Heath C, Taylor K, Bentley M, Allen SL, Koduru P, Brody JP, Hawson G, Rodwell R, et al. The t(8;22) in chronic myeloid leukemia fuses BCR to FGFR1: transforming activity and specific inhibition of FGFR1 fusion proteins. Blood. 2001;98:3778–3783. doi: 10.1182/blood.v98.13.3778. [DOI] [PubMed] [Google Scholar]
- 38.Jiang JR, Paez JG, Lee JC, Bo RH, Stone RM, DeAngelo DJ, Galinsky I, Wolpin BM, Jonasova A, Herman P, et al. Identifying and characterizing a novel activating mutation of the FLT3 tyrosine kinase in AML. Blood. 2004;104:1855–1858. doi: 10.1182/blood-2004-02-0712. [DOI] [PubMed] [Google Scholar]
- 39.Lierman E, Van Miegroet H, Beullens E, Cools J. Identification of protein tyrosine kinases with oncogenic potential using a retroviral insertion mutagenesis screen. Haematologica Hematol. J. 2009;94:1440–1444. doi: 10.3324/haematol.2009.007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata-Minoshima F, Oki T, Doki N, Nakahara F, Kageyama SI, Kitaura J, Fukuoka J, Kitamura T. Identification of RHOXF2 (PEPP2) as a cancer-promoting gene by expression cloning. Int. J. Oncol. 2012;40:93–98. doi: 10.3892/ijo.2011.1173. [DOI] [PubMed] [Google Scholar]
- 41.Cui ZB, Geurts AM, Liu GY, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 42.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 43.Mates L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 44.Iversen N, Birkenes B, Torsdalen K, Djurovic S. Electroporation by nucleofector is the best nonviral transfection technique in human endothelila and smooth muscle cells. Genet Vaccines Ther. 2005;3:2. doi: 10.1186/1479-0556-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freeman KW, Gangula RD, Welm BE, Ozen M, Foster BA, Rosen JM, Ittmann M, Greenberg NM, Spencer DM. Conditional activation of fibroblast growth factor receptor (FGFR) 1, but not FGFR2, in prostate cancer cells leads to increased osteopontin induction, extracellular signal-regulated kinase activation, and in vivo proliferation. Cancer Res. 2003;63:6237–6243. [PubMed] [Google Scholar]
- 46.Terada K, Kaziro Y, Satoh T. Ras-dependent activation of c-Jun N-terminal kinase/stress-activated protein kinase in response to interleukin-3 stimulation in hematopoietic BaF3 cells. J. Biol. Chem. 1997;272:4544–4548. doi: 10.1074/jbc.272.7.4544. [DOI] [PubMed] [Google Scholar]
- 47.Koziczak M, Holbro T, Hynes NE. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene. 2004;23:3501–3508. doi: 10.1038/sj.onc.1207331. [DOI] [PubMed] [Google Scholar]
- 48.Parada Y, Banerji L, Glassford J, Lea NC, Collado M, Rivas C, Lewis JL, Gordon MY, Thomas NSB, Lam EWF. BCR-ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27(Kip1) expression. J. Biol. Chem. 2001;276:23572–23580. doi: 10.1074/jbc.M101885200. [DOI] [PubMed] [Google Scholar]
- 49.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J. Cell Biol. 2005;171:663–673. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.