Abstract
Non-coding RNAs (ncRNA) account for a large portion of the transcribed genomic output. This diverse family of untranslated RNA molecules play a crucial role in cellular function. The use of ‘deep sequencing’ technology (also known as ‘next generation sequencing’) to infer transcript expression levels in general, and ncRNA specifically, is becoming increasingly common in molecular and clinical laboratories. We developed a software termed ‘RandA’ (which stands for ncRNA Read-and-Analyze) that performs comprehensive ncRNA profiling and differential expression analysis on deep sequencing generated data through a graphical user interface running on a local personal computer. Using RandA, we reveal the complexity of the ncRNA repertoire in a given cell population. We further demonstrate the relevance of such an extensive ncRNA analysis by elucidating a multitude of characterizing features in pathogen infected mammalian cells. RandA is available for download at http://ibis.tau.ac.il/RandA.
INTRODUCTION
Non-coding RNAs (ncRNA) account for a large portion of the transcribed genomic output (1). They are a diverse family of untranslated transcripts that have crucial roles in cellular function. It has been shown, for example, that ncRNAs modulate the biogenesis and activity of ribosomes [small nucleolar RNA, (snoRNA)] (2), repress gene expression [via microRNAs (miRNA)] (3), facilitate mRNA splicing and regulate transcription factors [small nuclear RNA (snRNA)] (4,5), alter cellular proliferation and apoptosis (small interfering RNA) (6) and play a role in infrastructural functions (tRNA and rRNA). Not surprisingly, ncRNA have been implicated in human health and disease (7).
NcRNA expression profile can serve as an initial step for multiple sequence alignment-based phylogeny, homology and conservation studies (8). It can also be used for detecting RNA library preparation biases such as failure in tRNA and rRNA filtration, or undesirable abundance of transcript degradation products. Differential expression of several types of ncRNAs can be of great value in a number of scientific fields such as: assessment of viral infection (9), diagnosis and prognosis of different tumor types (10), analysis of neurological disorders (11) and directing personalized medicine (12).
Up till recently, the most common tool for ncRNA expression analysis was either custom-designed microarrays (13) or tiling microarrays (14). The use of ‘deep sequencing’ technology (15) (also known as ‘next generation sequencing’) to infer transcript expression levels is becoming increasingly common in molecular and clinical laboratories. For the purpose of exploring the diverse world of ncRNAs, deep sequencing has many advantages. Deep sequencing improves the sensitivity and specificity above microarray techniques (16,17) and allows the identification of novel ncRNA transcripts (18). Moreover, sequencing does not require any prior knowledge of the actual transcript sequence, and any relevant database can be utilized in order to compare and characterize the sequence population (19).
The massive amount of data produced by deep sequencing requires several computational analysis procedures. These stages can be performed by employing a variety of tools that process and analyze the data. However, these tools necessitate the user to be familiar with Linux command lines and programming data manipulation. There are currently several available tools that reduce the need for computer savvy expertise when looking at ncRNAs. Tools such as DSAP (20) and miRTools (21) utilize Rfam, an open access database containing information about all known ncRNA families (22) in order to characterize the sample. However, differential expression analysis is performed only on miRNA and the amount of uploaded data is limited. Tools like miRExpress (23) and miRNAkey (19) allow data analysis on a local computer, but these are dedicated for miRNA analysis, skipping the potential information encompassed in the entire spectrum of ncRNA transcripts.
We developed a software termed RandA (ncRNA Read-and-Analyze), which performs comprehensive ncRNA expression profiling and differential expression analysis on deep sequencing data while running on a local computer (operated by a Linux operating system). RandA has a user-friendly graphical interface (avoiding the requirement for command lines) which allows the user to analyze and compare several samples using a pre-defined set of ncRNAs from Rfam. We demonstrate the fidelity of RandA by comparing its analysis to published data and by experimentally verifying its differentially expressed ncRNAs. We then use RandA to reveal the complexity of the ncRNA repertoire in a given pathogen infected cell population. RandA is available for download at http://ibis.tau.ac.il/RandA.
MATERIALS AND METHODS
RandA software pipeline
RandA is free-access software with a graphical user interface that carries through the essential steps in ncRNA analysis after the acquisition of deep sequencing data. The workflow of the tool is divided into three main sections:
Input and Output: the user should choose one or more deep sequencing read files for analysis. For each sequenced sample, the user assigns a condition name, allowing several samples to be assigned under the same condition (technical/biological replicates). Reads deriving from short RNA transcripts (e.g. miRNA) will usually include the adapter sequence in addition to the transcript sequence. By utilizing ea-utils' fastq-mcf tool (http://code.google.com/p/ea-utils/wiki/FastqMcf) RandA allows the user to clip this adapter sequence from the 3′ ends of the reads. RandA can also trim bases from the 3′ end of each sequence read if their quality is below a user defined threshold. Finally, RandA will filter out reads that were reduced to less than a set number of bases by either the clipping or the quality trimming.
Database preparation: for the purpose of ncRNA alignment and subsequent annotation, we utilize Rfam (v10.1), an extensive database of known ncRNA. The database contains almost 200 000 different organisms, and >1 million unique sequences for a variety of RNA species such as rRNA, tRNA, miRNA, cis-regulatory elements, snRNA, snoRNA, ribozymes and other documented non-coding transcripts. Due to the high level of sequence diversity and magnitude of Rfam, RandA allows the user to perform a variety of manipulations, creating a novel ad hoc database, specifically tailored according to the relevant experimental needs. Further refining of the database can be carried out by specifying an organism(s) and RNA families, then collapsing (joining) transcripts that are either identical in sequence, or share the same description.
Analysis: RandA maps the reads against the newly formed database (from Step 2, above) using a Burrows–Wheeler transform based alignment tool (24) summing the number of reads that mapped uniquely to each of the annotated ncRNA sequence. Due to the short read length produced by deep sequencing platforms, and the sequence similarity between the same family ncRNAs, a significant amount of reads align to several different reference transcripts and their respective isoforms. Although these multiple aligned reads may add up to >50% of the total amount of mapped reads (25), they are usually excluded from the analysis, possibly leading to a biased and misleading expression profile. RandA introduces multiple hits handling (reads mapped to more than one unique location or RNA sequence on the reference library) by implementing an expectation maximization-based algorithm called SEQ-EM (26). SEQ-EM enables the inclusion of these multiple hits in the transcripts' final expression assessment, resulting in increased accuracy and power.
The user may choose whether to continue and perform differential expression between the given samples, or to simply perform a transcript expression (read count) profiling. If the user selects to perform only transcript expression, RandA standardizes the number of reads mapped to each transcript according to its length and the initial total number of mapped reads in the sample based on the ‘reads per kilo-base per million’ (RPKM) method (27). The RandA will then output a count table with the read count and RPKM for each of the given samples. For the purpose of differential expression analysis, RandA employs DESeq (28), an ‘R’-based tool that performs differential expression analysis on deep sequencing data, and utilizes a negative binomial distribution model for variance estimation. Prior to the analysis, RandA reviews the number of samples assigned to each condition and sets the appropriate input parameters to the DESeq tool. Once finished, RandA outputs the differential expression analysis results combined with additional transcript-specific links.
All through the workflow, RandA generates a comprehensive summary including clipping, alignment and differential expression (summaries and plots), depicting multiple alignments and post-clipping read length distribution.
Sample preparation for deep sequencing
SupT1 cells (human Caucasian lymphoma T cells) were infected with human immunodeficiency virus (HIV1, HXB2 strain) and Mycoplasma hyorhinis. Eight days post-infection cells were harvested, total RNA was extracted using TRIzol (Invitrogen) and 10 µg of each sample were prepared for deep sequencing following Illumina’s Small RNA sample preparation protocol (v1.5). During this process, samples were ligated with 3′ and 5′ adapters, reverse-transcribed and then amplified using a PCR. Libraries of cDNA were prepared from ∼100 bp PCR products (representing ∼25 nt RNA molecules) and sequenced in separate lanes on an Illumina Genome Analyzer IIx instrument at the Tel Aviv University Genome High-Throughput Sequencing Laboratory.
Real time PCR
Real time PCR was carried out on the same RNA samples. One microgram of RNA was used to generate cDNA using the TaqMan MicroRNA Reverse Transcription Kit and Megaplex RT Pools (Applied Biosystems) in a final volume of 7.5 μL, according to the manufacturer's instructions. Six microliters of the reverse transcription reaction were used for real time PCR in a volume of 900 μL containing TaqMan Universal PCR Master Mix and TaqMan Low Density Arrays (TLDA) arrays (Applied Biosystems). U6 RNA was used as reference. Reactions were run on an Applied Biosystems 7900HT Fast Real-Time PCR System. Normalization of the results was done by reducing the cycle threshold (Ct) of each miRNA from the average Cts of four U6-snRNA replicates in each of the TLDAs. For each miRNA, the normalized Ct in HIV-infected TLDA was reduced from the normalized Ct in the non-infected one. Relative quantity (RQ) is calculated by 2 exponent the remainder from the last step.
RESULTS AND DISCUSSION
RandA produces a table comprising of all the mapped ncRNAs in a given sample. The output includes either expression profiles sorted from most to least expressed or differential expression between selected sample pairs. This enables the user to prioritize the massive amount of data and to focus on the most relevant ones. The output includes the: Rfam accession; RNA type; number of mapped reads for each condition; normalized count (RPKM); fold change; derived P-value (chi-square test) and the corrected P-value. For runs with multiple samples where DESeq (28) is implemented a normalized base mean count is given for each condition instead of the actual read count.
We demonstrate the applicability of RandA using small RNA samples taken from two human T cell cultures, one co-infected by both Mycoplasma and human immunodeficiency virus (HIV) and one uninfected [henceforth referred to as the ‘infected’ and ‘uninfected’ samples respectively; see Methods and (29)]. Each sample (condition) was run twice on the deep sequencer. The database was set to include all non-coding transcripts derived from either Homo sapiens or bacteria. Other organisms were excluded. The newly generated database entailed 793 118 transcripts out of the original 2 756 313 registered in Rfam. Since Rfam transcript annotation is highly extensive, identical sequences might appear under different accession numbers. Therefore, RandA enables the user to reduce possible redundancy by collapsing transcripts either based on sequence or description identity (each might fit a different experimental question). Collapsing the combined human and bacteria database by sequence identity resulted in 394 240 unique sequences, a 50% reduction.
The sequence reads were clipped and aligned against the ad hoc novel database. The alignment resulted in 88% and 54% of the mapped reads' unique alignment to the database in the uninfected and infected samples, respectively. The remaining reads that mapped to multiple locations were distributed using the inherent SEQ-EM algorithm (26) to produce a new read count for each transcript. The counts in each deep sequencing run for both conditions were analyzed using the DESeq tool, to produce a table describing the difference in expression between mapped transcripts (Table 1).
Table 1.
Ten most differently expressed RNA transcripts in our experiment
| RNA accession | RNA description | Organism | Base mean 1 | Base mean 2 | Fold change | Adjusted P-value |
|---|---|---|---|---|---|---|
| AK292330.1/1-191 | U2 spliceosomal RNA | H. sapiens (human) | 1.4 | 10384.93 | 7418.416 | 1.46 E-30 |
| AE017243.1/178458-178387 | tRNA | Mycoplasma hyopneumoniae J | 4.55 | 8283.43 | 1820.685 | 1.62 E-29 |
| ABBA01175726.1/642-527 | microRNA mir-689 | H. sapiens (human) | 5.375 | 3934.62 | 732.047 | 7.13 E-28 |
| ABSL01060990.1/9336-9469 | U11 spliceosomal RNA | H. sapiens (human) | 1.584 | 4291.48 | 2709.088 | 5.27 E-27 |
| AE017332.1/337236-337309 | tRNA | M. hyopneumoniae 232 | 0.508 | 2095.19 | 4121.295 | 2.12 E-24 |
| AADD01000927.1/23641-23760 | U5 spliceosomal RNA | H. sapiens (human) | 24.439 | 1749.02 | 71.566 | 1.60 E-22 |
| AE017332.1/830793-830880 | tRNA | M. hyopneumoniae 232 | 0.35 | 1572.03 | 4491.901 | 1.70 E-22 |
| AADB02010034.1/410071-410401 | 7SK RNA | H. sapiens (human) | 6.51 | 1665.87 | 255.914 | 4.19 E-22 |
| EU714234.1/1-1496 | Bacterial small subunit ribosomal RNA | M. hyorhinis | 1.4 | 980.26 | 700.25 | 4.02 E-21 |
| AK292656.1/2-181 | U11 spliceosomal RNA | H. sapiens (human) | 0.35 | 1082.40 | 3092.831 | 3.16 E-20 |
This table is a partial representation of the output table produced by RandA. Base mean 1 and 2 represent the normalized read count mean for each condition, namely uninfected and infected, respectively.
Mycoplasma ncRNA transcript expression
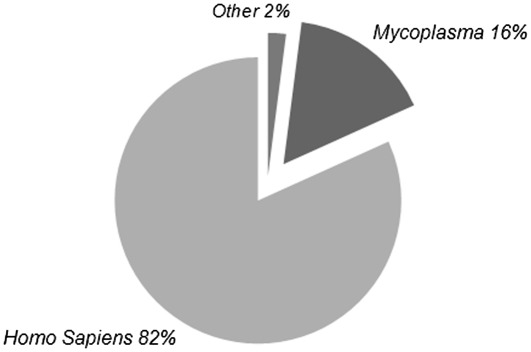
Using RandA, we demonstrated that expression of Mycoplasma derived transcripts was indeed significantly different between samples. These transcripts were highly expressed in the infected sample in opposition to the uninfected sample in which they were not detected. The analysis resulted in 2748 different RNA transcripts detected in at least one of the samples. Out of these, 273 transcripts exhibited significantly different expression (P < 0.01), of which 148 had a base mean count of over 100. Of these 148 transcripts, 121 were human transcripts, 24 Mycoplasma and only 3 from other bacterial sequences (Figure 1).
Figure 1.
Distribution of organisms when running the deep sequencing output against all human transcripts combined with all bacterial transcripts. Abundance of Mycoplasma derived sequences within the most differentially expressed transcripts (P < 0.01) with a base mean of over 100 demonstrates its' presence in the infected sample. Mycoplasma infection was previously validated (29).
Human ncRNA transcript profile during HIV infection
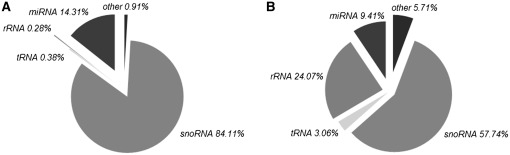
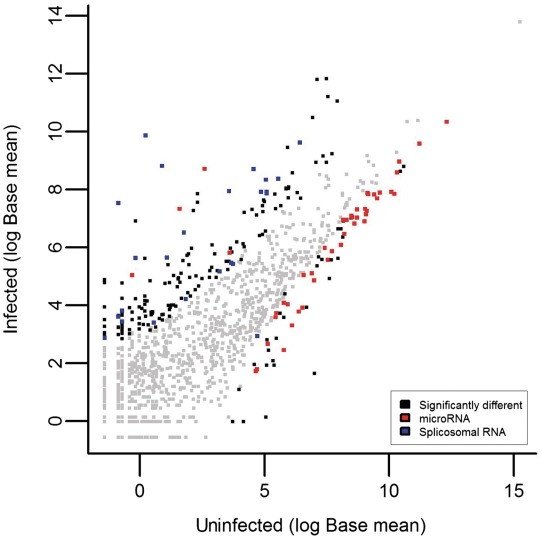
Running RandA with a defined database which includes viral transcript sequences did not result in any HIV-related ncRNA transcripts (data not shown), despite its presence in the samples. This might be due to the scarcity of HIV-related sequences in the sample tested or the shortage of these in the Rfam database. Yet, we asked whether the profile of the human ncRNA transcripts in the infected sample can demonstrate features that strongly support an HIV infection (Figure 2). When focusing only on the miRNA transcripts in both samples, we noticed a substantial decrease in expression in a large proportion of miRNAs (96%) demonstrating a significant down-regulation in miRNA levels compared to other ncRNAs (Fisher's exact test; P < 0.0001; Figure 3). This was confirmed by real time PCR on the six most significantly differentially expressed miRNAs (P < 0.0001) that are found in both Rfam and in the commercial real time PCR array used (Table 2). The decreased miRNA expression in HIV infected human cells coincides with previously reported studies (30–32) and can be attributed to the suspected Dicer-suppressive effect exerted by HIV-1 Tat protein and/or TAR RNA (33). We further examined the most significantly decreased miRNAs [using DIANA-mirPath (34)] and observed a noteworthy enrichment (P < 0.001) of miRNA-targeted genes in the mitogen activated protein kinase (MAPK) pathway. The MAPK pathway modulates and induces HIV infectivity (35–37). Thus, we speculate that this decrease in MAPK pathway genes-targeting miRNAs could serve as a viral mechanism to induce pathway activity and subsequent increased infectivity. This requires further experimental validation. MiRNA expression was not the only ncRNA group that changed after infection. We identified an enrichment of spliceosomal RNAs in the infected versus non-infected samples (Fisher's exact test; P < 0.01; Figure 3). Since HIV is known to disrupt the process of splicosome assembly in the nucleus (38,39), enrichment of such spliceosomal RNA fragments might support the presence of HIV in the infected samples. This could be a direct spliceosomal outcome or via changing the stability of splicing factors. Again, further experimentation is required for validation. Finally, 7SK RNA demonstrated a significant increase in expression in the infected sample (fold change >200; P < 4 ×e−22), suggesting a cellular antiviral defense mechanism given its reported disruption of HIV transcription (40).
Figure 2.
Distribution of human RNA transcripts in the uninfected (A) and infected (B) samples. The chart includes only transcripts with a base mean of more than 20. The various ncRNA types demonstrate variable relative expression which can be partly attributed to the HIV infection. This stresses the importance of a comprehensive ncRNA transcriptome overview to achieve an accurate sample assessment.
Figure 3.
ncRNA transcript expression in the uninfected versus the infected samples (Base mean 1 and Base mean 2, respectively). This figure demonstrates the reduction of miRNA expression (red) and the induction of splicosomal RNA expression (blue) when inspecting the significantly different transcripts (non-gray; P < 0.01).
Table 2.
Six most significantly down-regulated miRNAs detected by both RandA and real time PCR in our experiments
| miRNA | RandA fold change | P-value | Real time PCR RQ |
|---|---|---|---|
| mir-342 | 0.113 | 1.01 E-08 | 0.398 |
| mir-423 | 0.145 | 4.05 E-07 | 2.19 E-05 |
| mir-197 | 0.138 | 5.11 E-07 | 0.459 |
| mir-92 | 0.177 | 1.66 E-05 | 0.419 |
| let-7 | 0.245 | 1.12 E-04 | 1.23 E-03 |
| mir-101 | 0.244 | 6.43 E-04 | 1.93 E-04 |
The table describes the fold change between uninfected and infected samples as detected by RandA with its corresponding P-value, and the relative quantification (RQ; see Methods) between samples as detected by real time PCR.
ncRNA expression analysis following EBV infection
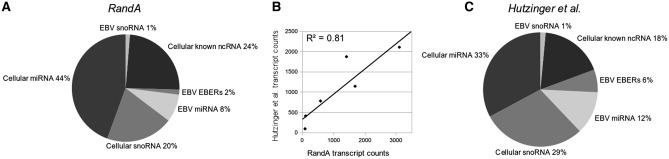
We further demonstrate the utility of RandA for ncRNA transcript analysis by application to publicly available data from a deep sequencing experiment in which Epstein Barr virus (EBV) infected human cells were sequenced [accession number SRA010803.5;(9)]. In this work, Hutzinger et al. sequenced a specialized cDNA library, comprising of EBV encoded ncRNAs and host EBV-induced ncRNAs. Sequence reads were computationally analyzed using an assortment of alignment tools and ncRNA databases. Using RandA we preformed the same essential analysis steps. Briefly, all samples were clipped and trimmed by quality threshold of 30. The reads were then aligned against an RandA generated database comprising of all the human and EBV-related ncRNA transcripts. Reads mapping to multiple transcripts were incorporated to the transcript count by the SEQ-EM algorithm. The sequences were mapped to 334 different known ncRNA transcripts (as opposed to 274 detected by Hutzinger et al.). Comparison of the ncRNA transcript type composition derived from each method demonstrated high similarity between the two (R2 = 0.81; Figure 4). In order to perform ncRNA differential expression analysis on EBV-infected and uninfected samples, Hutzinger et al. generated a custom-designed ncRNA-microchip. In this context of ncRNA transcript differential expression we have demonstrated that RandA is highly applicable and thus we suggest implementing a combination of deep sequencing and RandA analysis as an alternative to microchip-based methods.
Figure 4.
NcRNA family distribution in human with and without EBV derived transcripts. RandA ncRNA distribution (A) and Hutzinger et al. data (C) show high similarity (B); Pearson correlation coefficient 0.899; P < 0.01) demonstrating the utilization of RandA in a single sample, multi-species ncRNA expression analysis.
In summary, utilizing RandA, we were able to comprehensively examine the ncRNA transcriptome and provide a broad perspective on transcript expression. We show a multitude of ncRNA shifts in pathogen-infected samples exemplifying the value of this type of analysis during cellular processes.
FUNDING
Chief Scientist Office, Ministry of Health, Israel; Israel Cancer Association and the Wolfson family Charitable Fund (to The Shomron Laboratory); I-CORE Program of the Planning and Budgeting Committee; The Israel Science Foundation (grant number 41/11); Fellowship from the Edmond J. Safra Bioinformatics program at Tel-Aviv University (to O.I.). Funding for open access charge: Chief Scientist Office, Ministry of Health, Israel.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Tel Aviv University Genome High-Throughput Sequencing Laboratory staff, Drs Varda Oron-Karni, Orly Yaron and Nitzan Kol, for their dedicated and professional work. Authors also thank Dr Eran Halperin and Dr Bogdan Pasaniuc for technical assistance. This work was performed in partial fulfillment of the requirements for a Ph.D. degree of O.I. at the Sackler Faculty of Medicine, Tel Aviv University.
REFERENCES
- 1.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 4.Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- 5.Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Song M, Cui ZS, Li CY, Xue XX, Yu M, Lu Y, Zhang SY, Wang EH, Wen YY. Down-regulation of TSG101 by small interfering RNA inhibits the proliferation of breast cancer cells through the MAPK/ERK signal pathway. Histol. Histopathol. 2011;26:87–94. doi: 10.14670/HH-26.87. [DOI] [PubMed] [Google Scholar]
- 7.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 8.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 9.Hutzinger R, Mrázek J, Vorwerk S, Hüttenhofer A. NcRNA-microchip analysis: a novel approach to identify differential expression of noncoding RNAs. RNA Biol. 2010;7:586–595. doi: 10.4161/rna.7.5.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin H, Lebanony D, Rosenwald S, Cohen L, Gibori H, Barabash N, Ashkenazi K, Goren E, Meiri E, Morgenstern S, et al. A diagnostic assay based on microRNA expression accurately identifies malignant pleural mesothelioma. J. Mol. Diagn. 2010;12:771–779. doi: 10.2353/jmoldx.2010.090169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Rukov JL, Shomron N. MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol. Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 14.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 15.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.t’ Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen J. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics. 2010;11:77. doi: 10.1186/1471-2164-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronen R, Gan I, Modai S, Sukacheov A, Dror G, Halperin E, Shomron N. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics. 2010;26:2615–2616. doi: 10.1093/bioinformatics/btq493. [DOI] [PubMed] [Google Scholar]
- 20.Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;38:W385–W391. doi: 10.1093/nar/gkq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38:W392–W397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsingh G, Koboldt DC, Trissal M, Chiappinelli KB, Wylie T, Koul S, Chang LW, Nagarajan R, Fehniger TA, Goodfellow P, et al. Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood. 2010;116:5316–5326. doi: 10.1182/blood-2010-05-285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paşaniuc B, Zaitlen N, Halperin E. Accurate estimation of expression levels of homologous genes in RNA-seq experiments. J. Comput. Biol. 2011;18:459–468. doi: 10.1089/cmb.2010.0259. [DOI] [PubMed] [Google Scholar]
- 27.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isakov O, Modai S, Shomron N. Pathogen detection using short-RNA deep sequencing subtraction and assembly. Bioinformatics. 2011;27:2027–2030. doi: 10.1093/bioinformatics/btr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang K-T. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang D. RNA Viruses: Host Gene Responses to Infections. World Scientific Publishing, UK; 2009. [Google Scholar]
- 32.Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang K-T. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennasser Y, Le SY, Benkirane M, Jeang K-T. Evidence that HIV-1 Encodes an siRNA and a Suppressor of RNA Silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 35.Emerman M, Malim MH. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 36.Jacqué JM, Mann A, Enslen H, Sharova N, Brichacek B, Davis RJ, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Gabuzda D. Regulation of human immunodeficiency virus Type 1 infectivity by the ERK mitogen-activated protein kinase signaling pathway. J. Virol. 1999;73:3460–3466. doi: 10.1128/jvi.73.4.3460-3466.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Si ZH, Rauch D, Stoltzfus CM. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kjems J, Sharp PA. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6.U5 small nuclear ribonucleoprotein in spliceosome assembly. J. Virol. 1993;67:4769–4776. doi: 10.1128/jvi.67.8.4769-4776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]