Abstract
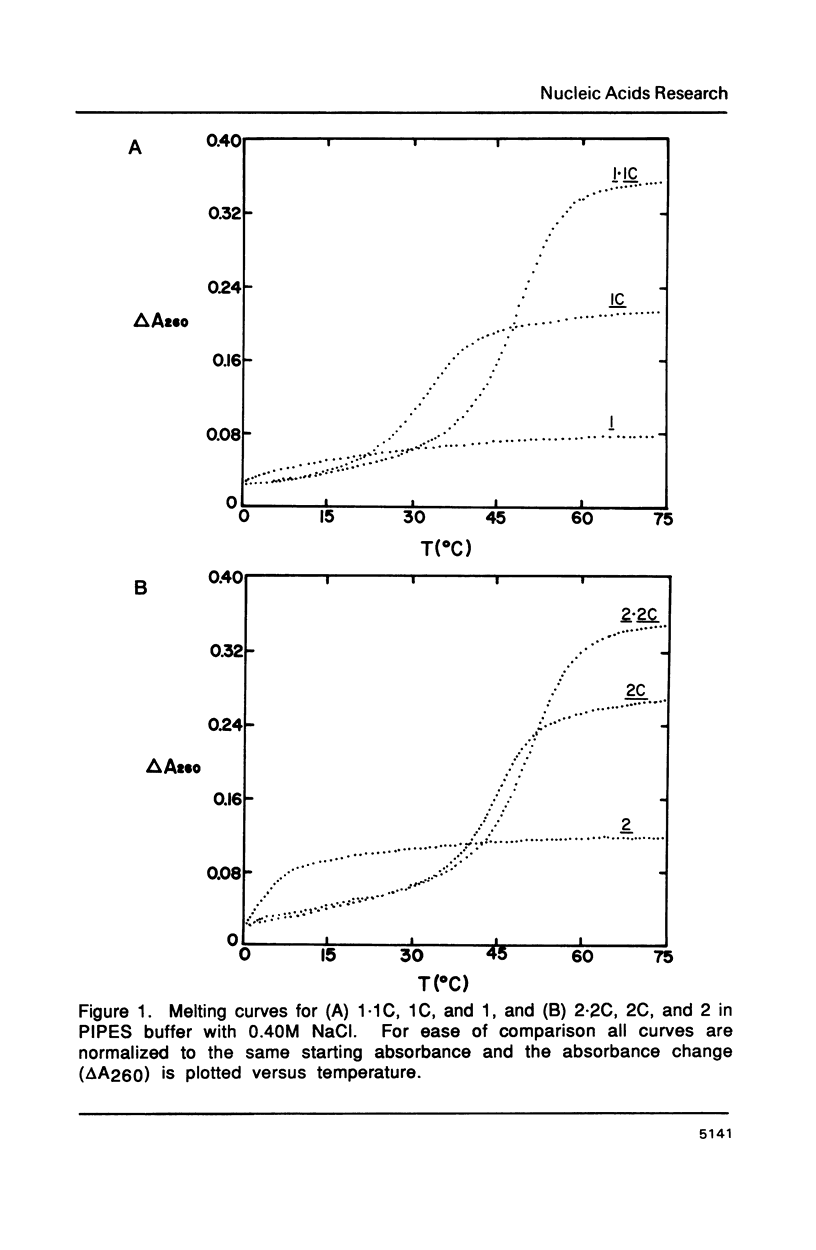
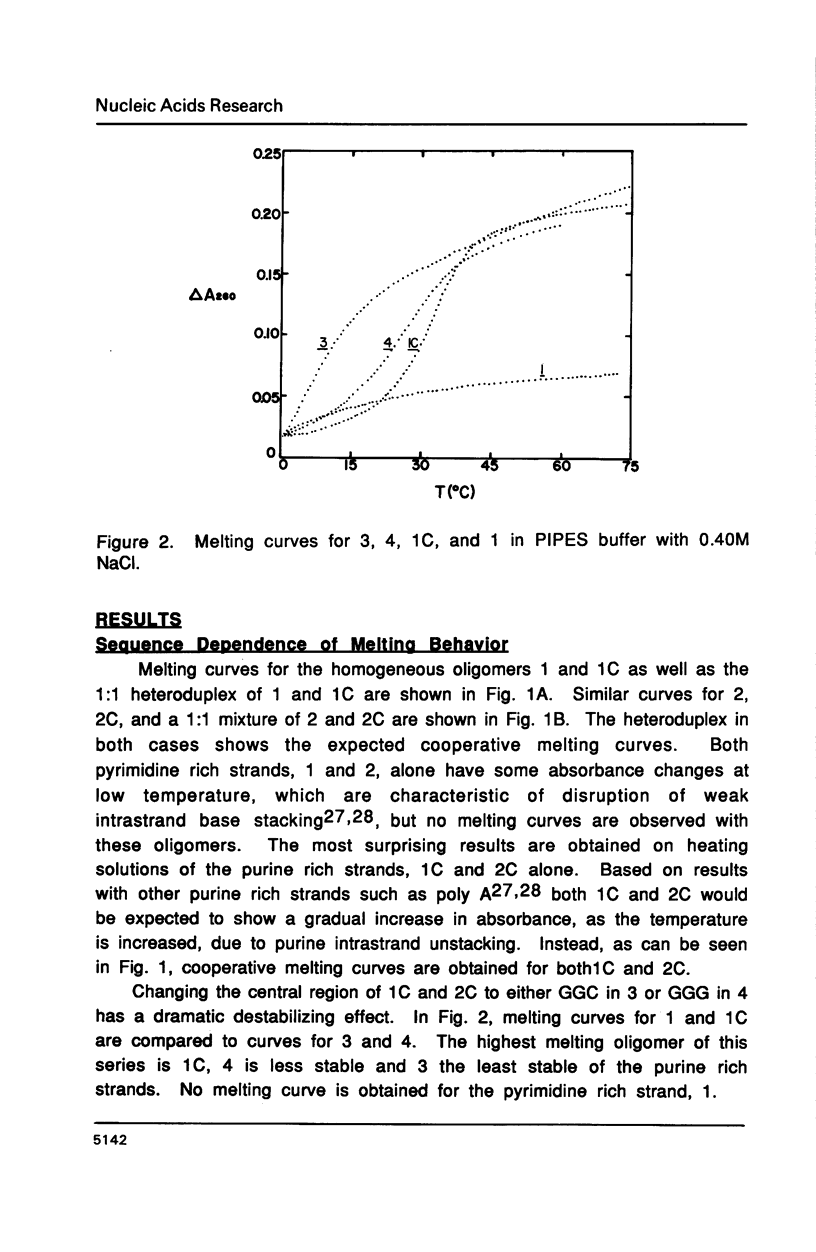
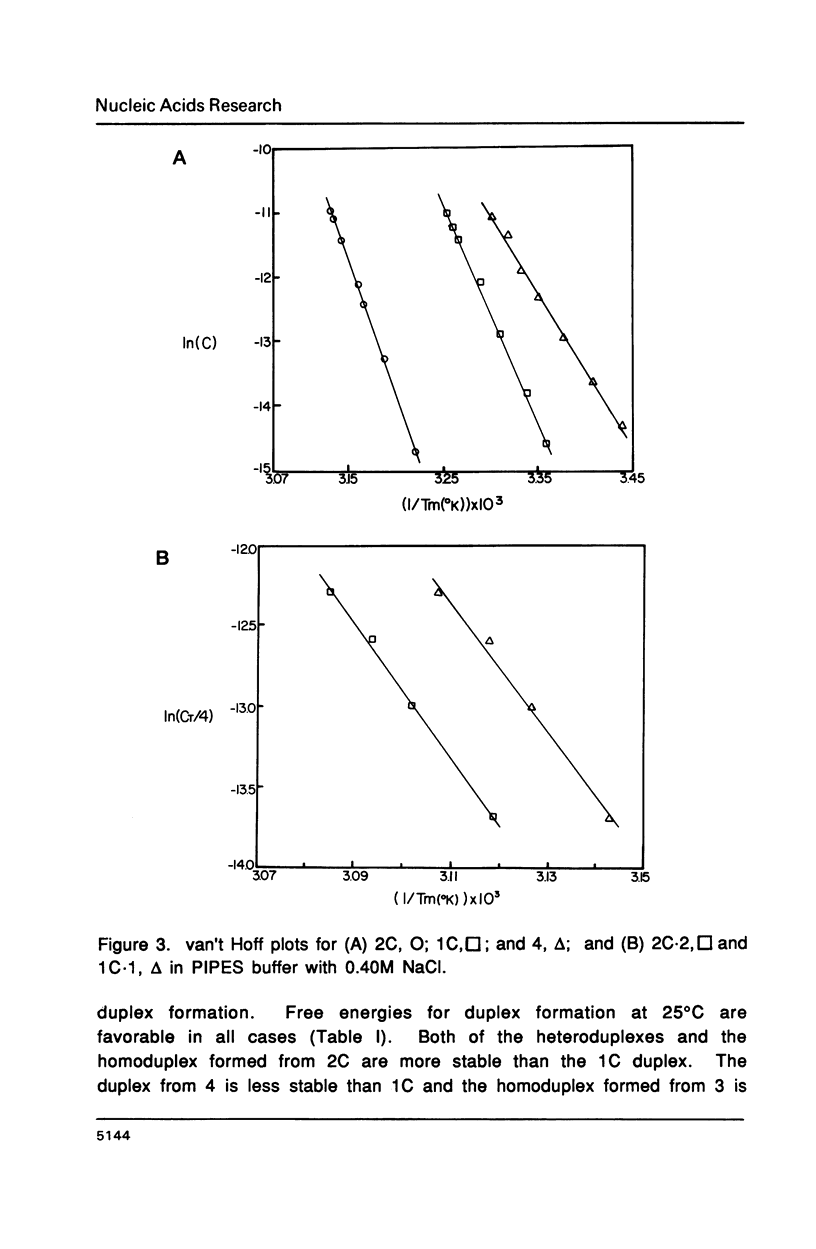
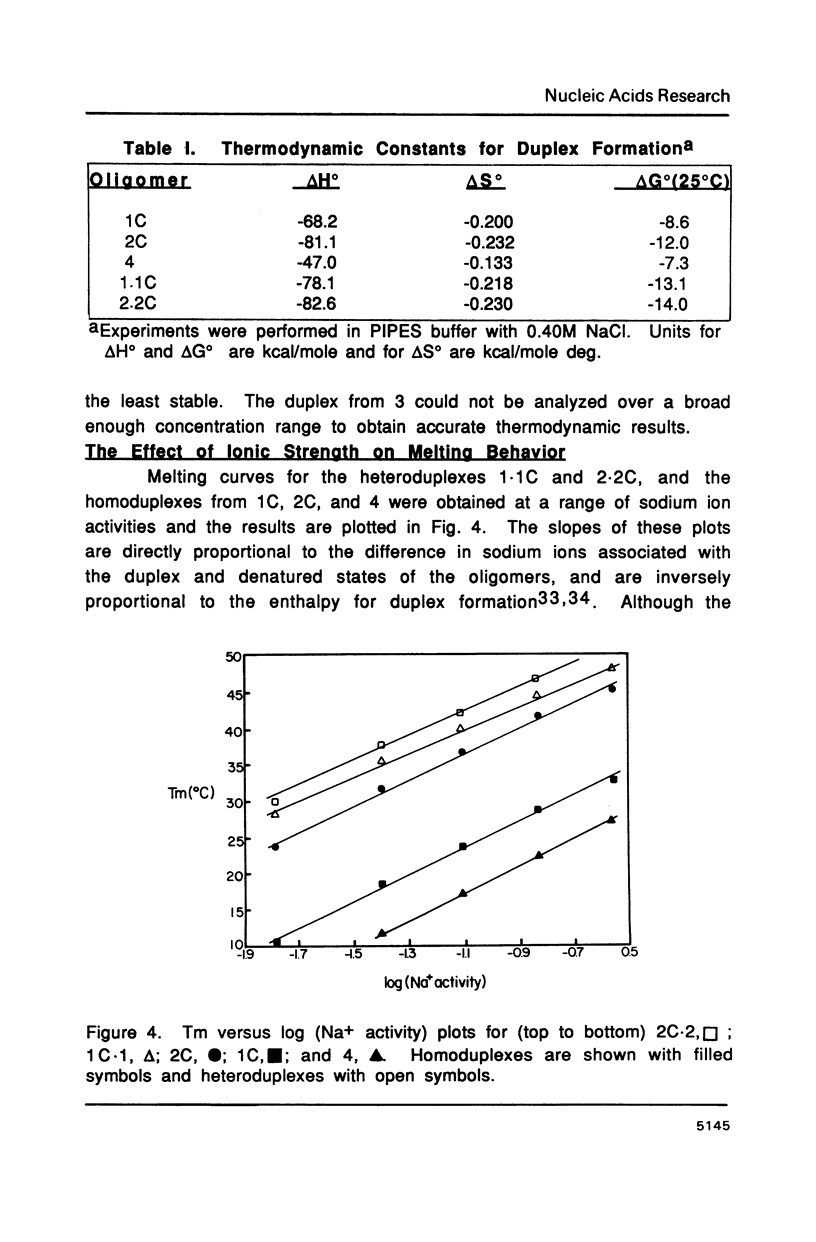
The purine rich oligodeoxyribonucleotides 1C, d(ATGACGGAATA) and 2C, d(ATGAGCGAATA) alone exhibit highly cooperative melting transitions. Analysis of the concentration dependence of melting, and electrophoretic studies indicate that these oligomers can form an unusual purine rich offset double helix. The unusual duplex is predicted to contain four A.T, two G.C, and four G.A mismatch base pairs as well as a single A base stacked on the 3' end of each chain of the helix. Other possible models for the duplex are unlikely because they are predicted to contain many base pairs of low stability. Changing the central sequence to CGG or GGG should destabilize the duplex and this is observed. The unusual duplex of 2C is more stable than the duplex of 1C indicating that the stability of G.A base pairs is quite sensitive to the surrounding sequence. Addition of 1C and 2C to their complementary pyrimidine strands results in normal duplexes of similar stability. We feel that the unusual duplexes are significantly stabilized by the intrinsic stacking tendency of purine bases.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- Bower M., Summers M. F., Kell B., Hoskins J., Zon G., Wilson W. D. Synthesis and characterization of oligodeoxyribonucleotides containing terminal phosphates. NMR, UV spectroscopic and thermodynamic analysis of duplex formation of [d(pGGAATTCC)]2, [d(GGAATTCCp)]2 and [d(pGGAATTCCp)]2. Nucleic Acids Res. 1987 Apr 24;15(8):3531–3547. doi: 10.1093/nar/15.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower M., Summers M. F., Powell C., Shinozuka K., Regan J. B., Zon G., Wilson W. D. Oligodeoxyribonucleoside methylphosphonates. NMR and UV spectroscopic studies of Rp-Rp and Sp-Sp methylphosphonate (Me) modified duplexes of (d[GGAATTCC])2. Nucleic Acids Res. 1987 Jun 25;15(12):4915–4930. doi: 10.1093/nar/15.12.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie J. S., Davidson D. S. Guanine modification during chemical DNA synthesis. Nucleic Acids Res. 1987 Oct 26;15(20):8333–8349. doi: 10.1093/nar/15.20.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Quignard E., Woisard A., Guschlbauer W., van der Marel G. A., van Boom J. H., Jones M., Radman M. Structures of mismatched base pairs in DNA and their recognition by the Escherichia coli mismatch repair system. EMBO J. 1986 Dec 20;5(13):3697–3703. doi: 10.1002/j.1460-2075.1986.tb04702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Alkema D., Sinclair A., Neilson T., Turner D. H. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985 Aug 13;24(17):4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- Henderson E., Hardin C. C., Walk S. K., Tinoco I., Jr, Blackburn E. H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987 Dec 24;51(6):899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kennard O. Structural features and hydration of d(C-G-C-G-A-A-T-T-A-G-C-G); a double helix containing two G.A mispairs. J Biomol Struct Dyn. 1986 Oct;4(2):173–191. doi: 10.1080/07391102.1986.10506338. [DOI] [PubMed] [Google Scholar]
- Ikuta S., Takagi K., Wallace R. B., Itakura K. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatched base pairs. Nucleic Acids Res. 1987 Jan 26;15(2):797–811. doi: 10.1093/nar/15.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer H., Sturtevant J. M. Heats of the helix-coil transitions of the poly A-poly U complexes. Biopolymers. 1968 Apr;6(4):491–512. doi: 10.1002/bip.1968.360060406. [DOI] [PubMed] [Google Scholar]
- LaPlanche L. A., James T. L., Powell C., Wilson W. D., Uznanski B., Stec W. J., Summers M. F., Zon G. Phosphorothioate-modified oligodeoxyribonucleotides. III. NMR and UV spectroscopic studies of the Rp-Rp, Sp-Sp, and Rp-Sp duplexes, [d(GGSAATTCC)]2, derived from diastereomeric O-ethyl phosphorothioates. Nucleic Acids Res. 1986 Nov 25;14(22):9081–9093. doi: 10.1093/nar/14.22.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. An NMR study of polymorphous behaviour of the mismatched DNA octamer d(m5C-G-m5C-G-A-G-m5C-G) in solution. The B-duplex and hairpin forms. Eur J Biochem. 1987 Dec 30;170(1-2):225–239. doi: 10.1111/j.1432-1033.1987.tb13690.x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984 Jul 3;23(14):3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Effects of DNA methylation on mismatch repair, mutagenesis, and recombination in Escherichia coli. Curr Top Microbiol Immunol. 1984;108:23–28. doi: 10.1007/978-3-642-69370-0_3. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Summers M. F., Powell C., Egan W., Byrd R. A., Wilson W. D., Zon G. Alkyl phosphotriester modified oligodeoxyribonucleotides. VI. NMR and UV spectroscopic studies of ethyl phosphotriester (Et) modified Rp-Rp and Sp-Sp duplexes, (d[GGAA(Et)TTCC])2. Nucleic Acids Res. 1986 Sep 25;14(18):7421–7436. doi: 10.1093/nar/14.18.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Zuo E. T., Jones R. L., Zon G. L., Baumstark B. R. Sequence dependent electrophoretic mobilities and melting temperatures for A-T containing oligodeoxyribonucleotides. Nucleic Acids Res. 1987 Jan 12;15(1):105–118. doi: 10.1093/nar/15.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]




