Abstract
Here we identify Neuralized homologue 4 (Neurl4) as a protein that interacts with CP110, a centrosomal protein that regulates centrosome duplication. Neurl4 uses a Neuralized homology repeat to preferentially localize to procentrioles and daughter centrioles. Neurl4 depletion results in ectopic microtubular organizing centres (MTOCs), leading to accumulation of CP110 and recruitment of a cohort of centrosomal proteins. We show that these ectopic MTOCs persist through mitosis and assemble aberrant mitotic spindles. Interestingly, Neurl4 promotes ubiquitylation of CP110, thereby destabilizing this protein. Our results indicate that Neurl4 counteracts accumulation of CP110, thereby maintaining normal centriolar homeostasis and preventing formation of ectopic MTOCs.
Keywords: Neurl4, CP110, daughter centriole, centrosome amplification, ubiquitylation
Introduction
Centrosomes consist of two centrioles, an older ‘mother’ centriole and a younger ‘daughter’ centriole, surrounded by an electron-dense protein matrix known as pericentriolar material (PCM). Centrosomes function as primary microtubule organizing centres (MTOCs), organizing the assembly of a bipolar spindle essential for equal chromosome segregation. Cancer cells often show centrosome amplification, and recent experiments show that centrosome amplification can promote genomic instability [1].
CP110 was identified as a centrosomal protein whose suppression prevents centriole reduplication during prolonged S phase [2]. CP110 localizes to the distal tips of both parental centrioles and procentrioles. Depletion of CP110 in non-ciliated cells results in elongated centrioles and enhanced ciliogenesis in proliferating cells capable of ciliation [3–7]. Here we identify Neuralized homologue 4 (Neurl4), a daughter centriole-specific, CP110-interacting protein that destabilizes CP110 by promoting ubiquitylation. Further, we show that depletion of Neurl4 results in increased CP110 levels, promoting the formation of ectopic MTOCs that result in mitotic abnormalities.
Results and Discussion
Neurl4 interacts with CP110
We used immuno-affinity purification and mass spectrometric sequencing to identify a new CP110-interacting protein, Neuralized homologue 4 (Neurl4; see Methods). Neurl4, a member of the Neuralized family, is distinguished by the presence of six Neuralized Homology Repeats (NHR domain). The NHR domain is thought to mediate protein–protein interactions and is highly conserved from flies to mammals [8]. In humans, four Neuralized family members have been identified. Neurl1, 2 and 3 encode either a RING or SOCS domain and serve as E3 ubiquitin ligases involved in diverse processes [9] (supplementary Fig S1A online). Neurl4 and its fly homologue, Bluestreak, are members of the NHR-only subfamily and lack other recognizable domains.
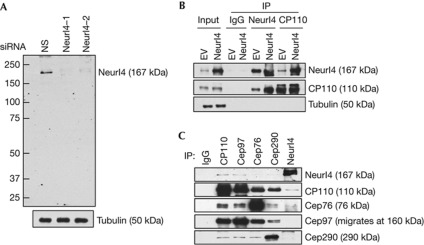
We generated polyclonal antibodies and showed that the affinity-purified antibody specifically recognized a band of ∼170 kDa on western blots of lysates from a variety of human cell lines (Fig 1A,B and supplementary Fig S6 online). Depletion of Neurl4 using two different small interfering RNAs (siRNAs) resulted in the disappearance of this band, confirming that the antibody specifically recognizes endogenous Neurl4 (Fig 1A). Next, we overexpressed Neurl4, performed anti-Neurl4 immunoprecipitations, and confirmed that Neurl4 and CP110 interact in vivo (Fig 1B). Further, immunoprecipitations with anti-Neurl4 antibody showed that endogenous Neurl4 associates with CP110 and, reciprocally, that CP110 antibodies immunoprecipitated endogenous Neurl4. We also found that antibodies recognizing two other CP110-interacting proteins, Cep97 and Cep76, immunoprecipitated Neurl4, albeit more weakly, perhaps due to indirect interactions through CP110 (Fig 1C). Moreover, we expressed Neurl4 and a series of deletion mutants and found that all fragments containing NHR repeat 3 (NHR3) interact with CP110 (supplementary Fig S1B online). The interaction between Neurl4 and CP110 is most likely direct, as an N-terminal fragment of CP110 produced in bacteria robustly and specifically bound to an in vitro translated fragment spanning NHR3–5 domains, but not a C-terminal Neurl4 fragment (supplementary Fig S1C online).
Figure 1.
Neurl4 is a novel NHR domain-containing protein that interacts with CP110. (A) Western blot detection of endogenous Neurl4. U2OS cells were transfected with a nonspecific control (NS) or two different Neurl4-specific siRNA as indicated. (B) 293T cells transfected with empty vector (EV) or Flag–Neurl4 were immunoprecipitated with rabbit IgG (control), anti-Neurl4 or anti-CP110 antibody and blotted with indicated antibodies. (C) Western blotting of endogenous CP110, Cep97, Cep76, Cep290 and Neurl4 after immunoprecipitation (IP) with indicated antibodies from 293T cell extracts. IgG, immunoglobulin G; siRNA, small interfering RNA.
Neurl4 associates with daughter centrioles
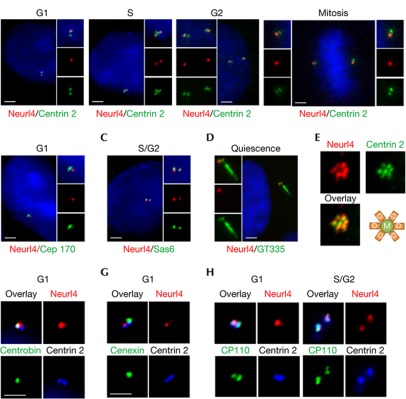
We visualized Neurl4 localization in U2OS and non-transformed human retinal pigment epithelial (RPE1) cells using antibodies against a panel of centriolar (centrin 2, CP110, c-NAP1 and polyglutamylated tubulin), procentriolar (Sas6), daughter centriole-specific (centrobin), mother centriole-specific (Cep170 and cenexin), PCM (γ-tubulin) and primary cilia (polyglutamylated tubulin) markers (Fig 2 and supplementary Fig S1D,E online). Neurl4 colocalized with procentriolar and daughter centriolar markers (Sas6 and centrobin) but did not colocalize with two mother centriole markers (Cep170 and cenexin), and Neurl4 was excluded from the mother centriole/basal body (Fig 2). Neurl4 also colocalized with daughter centrioles but not mother centrioles in a rosette pattern after Plk4 overexpression [10] (Fig 2E). These studies suggest that Neurl4 preferentially associates with daughter centrioles in growing and quiescent cells. Although we occasionally observed three Neurl4 dots specifically in S phase (data not shown), two Neurl4 dots were observed in subsequent phases of the cell cycle (Fig 2A). The subset of cells that express three Neurl4 dots might represent the two newly formed centrioles and the original daughter from which Neurl4 has not yet been displaced. As Neurl4 is present in two dots during G2 and mitosis, Neurl4 might disappear from the original daughter as it matures into a mother centriole, perhaps by means of degradation.
Figure 2.
Neuralized homologue 4 (Neurl4) preferentially localizes to the daughter centriole. (A) U2OS cells in diverse stages of the cell cycle were visualized by immunofluorescence with Neurl4 (red) and centrin (green) antibodies. (B–D) U2OS cells were visualized with Cep170 (green) and Neurl4 (red) antibodies (B) or Sas6 (green) and Neurl4 (red) antibodies (C). Quiescent retinal pigment epithelial 1 cells were stained with Neurl4 (red) and polyglutamylated tubulin (GT335, green) antibodies (D). Cell cycle stages for each cell are indicated. (E) Plk4 expression in U2OS cells produces a ‘rosette’ pattern with daughter centrioles, stained with Neurl4 antibodies, surrounding an unstained mother centriole in the centre. (F–H) Triple staining of Neurl4 (red)/centrobin (green)/centrin 2 (blue; F), Neurl4 (red)/cenexin (green)/centrin 2 (blue; G) and Neurl4 (red)/CP110 (green)/centrin 2 (blue; H). Scale bar, 2.5 μm.
Neurl4 is among a select group of proteins as, to our knowledge, only three other daughter-specific centriolar proteins, PARP3 [11], centrobin [12] and Cep120 [13], have been characterized. Neurl4 localization was observed with diverse fixation conditions, and it persisted after treatment with nocodazole, demonstrating that its localization is microtubule-independent (data not shown). We also examined the recruitment of Neurl4 after knocking down CP110, centrin-2 and Cep76, and in each case, centrosomal localization of Neurl4 was unaffected, indicating that Neurl4 localization is not dependent on these proteins (supplementary Fig S2A online).
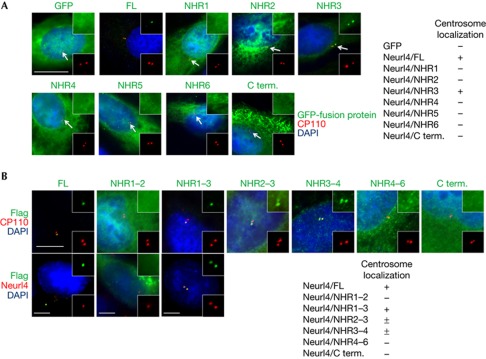
Several proteins have been shown to possess a centrosome localization signal, but these signals primarily target proteins to the PCM. Given its distinct localization, we attempted to determine which region of Neurl4 is responsible for targeting the protein to the daughter centriole. Expression of green fluorescent protein (GFP) fusion proteins containing individual NHR domains (or the C-terminus lacking NHR domains) showed that GFP–NHR3 colocalized with CP110 dots, while all other fragments were broadly distributed throughout the cell (Fig 3A). This demonstrates that the centriolar targeting region of Neurl4 lies within NHR3, although NHR3 alone did not show specific localization to the daughter centriole. Thus, we expressed a series of Flag–Neurl4 truncation mutants and examined colocalization with CP110 or a daughter-specific centriole marker (Neurl4). Interestingly, the NHR1–3 fragment localized to daughter centrioles, whereas NHR2–3 and NHR3–4 inefficiently localized to the centrosome, and NHR1–2, NHR4–6 and C terminus did not show specific localization (Fig 3B). As NHR3 alone localized to both mother and daughter centrioles and the deletion mutants that contain both NHR1 and 3 efficiently localize to the daughter centriole, it is tempting to speculate that NHR1 might have an ancillary role in NHR3-mediated daughter centriolar targeting of Neurl4.
Figure 3.
Neuralized homologue 4 (Neurl4) contains a novel daughter centriole targeting signal. (A) U2OS cells were transfected with plasmids expressing GFP–Neurl4 fragments and were stained with anti-GFP (green) and anti-CP110 (red) antibodies. Scale bar, 10 μm. (B) U2OS cells were transfected with plasmids expressing Flag–Neurl4 fragments and were stained with anti-Flag (green) and anti-CP110 (red) antibodies (upper panel) or anti-Flag (green) and anti-Neurl4 (red) antibodies (lower panel). Antibodies against Neurl4 do not recognize the truncation mutants. ± Denotes that a small percentage (1–5%) of cells expressing Flag–NHR2–3 and Flag–NHR3–4 show centrosome localization. Scale bar, 5 μm. C term., C-terminal; DAPI, 4,6-diamidino-2-phenylindole; FL, full-length Neurl4; GFP, green fluorescent protein; NHR, neuralized homology repeat.
Phenotype of Neurl4 depletion
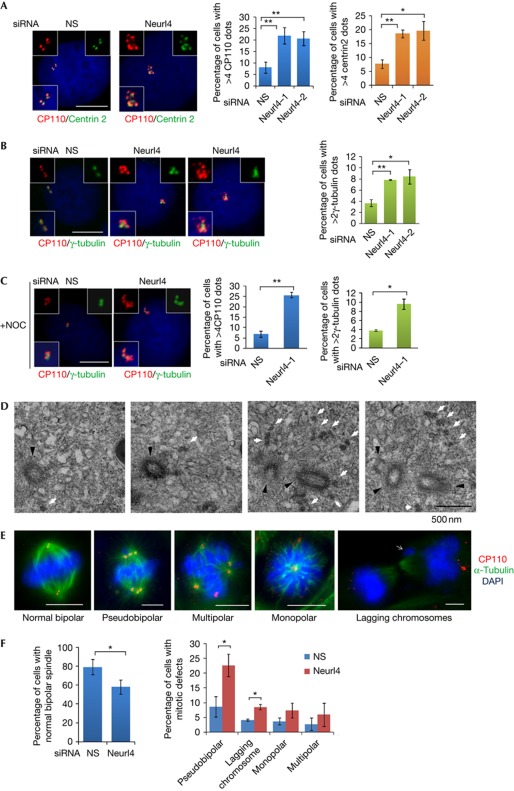
Next, we depleted Neurl4 using two distinct siRNAs in asynchronously growing U2OS cells. Importantly, the fluorescence intensity of Neurl4 on centrioles was reduced by >70% after treatment with Neurl4 siRNAs (supplementary Fig S1D online). Fluorescence-activated cell sorting analysis showed that Neurl4 depletion did not have an effect on cell cycle progression (supplementary Fig S3 online). Remarkably, Neurl4 depletion led to a two- to threefold increase in the number of cells with >4 CP110 or centrin dots or >2 γ-tubulin dots (Fig 4A,B). The majority of extra CP110 dots and all of the additional γ-tubulin dots were resistant to nocodazole treatment, indicating that they could be stable components of centrosomes (Fig 4C). We also used antibodies against a proximal centriolar marker (C-Nap1) and stabilized (polyglutamylated) tubulin to show that a subset of these extra CP110/centrin dots accumulated other centriolar markers and are stable structures (supplementary Fig S4A,B online). In contrast, the extra dots generated through Neurl4 ablation did not incorporate Sas6 (supplementary Fig S4C–E online). Interestingly, the same effect was observed with cyclin F depletion. The fact that several other centrosomal proteins, including γ-tubulin, were recruited to these extra CP110/centrin foci suggested that these structures could represent ectopic MTOCs. These results were not restricted to a single cell line, as we observed amplification of CP110 foci in HeLa and diploid RPE1 cells (supplementary Fig S6A,B online and data not shown).
Figure 4.
Depletion of Neurl4 results in the formation of aberrant CP110-positive assemblies and mitotic defects. U2OS cells were transfected with a nonspecific (NS) or two different siRNAs targeting Neurl4. The percentage of cells with >4 CP110 foci or centrin foci (A) or γ-tubulin foci (B) is analysed. In C, U2OS cells were transfected by indicated siRNA and treated with 10 mM nocodazole to induce microtubule depolymerization. (D) A set of representative electron microscopic images of U2OS cells treated with Neurl4 siRNA. A series of adjacent sections are shown. White arrows point to the extra electron-dense material observed after Neurl4 depletion. Black arrowheads point to normal centrioles. (E,F) U2OS were transfected with control or Neurl4 siRNA for 72 h. Mitotic cells were scored and representative pictures were shown. At least 100 cells for each siRNA transfection were scored for each experiment. Individual channels in E are also shown in supplementary Fig S7A online. Error bars in graphs represent standard error of the mean (s.e.m.). *P<0.05; **P<0.01. Scale bar, 10 μm. DAPI, 4,6-diamidino-2-phenylindole; Neurl4, Neuralized homologue 4; NS, nonspecific; siRNA, small interfering RNA.
We analysed extra CP110/centrin dots at the ultrastructural level by examining U2OS cells treated with a Neurl4 siRNA with serial section electron microscopy. In 35% of cells (10 of 29), we detected extra electron-dense material adjacent to structurally intact centriole pairs (Fig 4D and supplementary Fig S5 online), but we did not detect extra centrioles. In contrast, these structures were only observed in ∼9% of controls. Thus, the percentage of cells showing this phenotype closely mirrored our immunofluorescence studies (Fig 4). This analysis also allowed us to exclude the possibility that centriolar fragmentation occurred in Neurl4-depleted cells (supplementary Fig S5 online). Together, our immunofluorescence and electron microscopy analyses indicate that Neurl4 depletion promotes assembly of electron-dense foci containing multiple centrosomal proteins. We speculate that Neurl4 depletion generates additional CP110/centrin-positive assemblies that do not originate from Sas6-positive structures, reminiscent of structures observed during prolonged S-phase arrest in another study [14].
We also examined the impact of these extra CP110-containing structures on mitosis. First, we found that Neurl4 ablation led to a considerable increase in the percentage of cells showing abnormal mitoses in both U2OS and HeLa cells (Fig 4E,F and supplementary Fig S7 online). In particular, we observed that most of the extra CP110-positive structures formed pseudobipolar spindles in metaphase and anaphase, and there was a considerable increase in the proportion of cells with lagging chromosomes (Fig 4E,F and supplementary Fig S7 online). A modest increase in the frequency of cells with mono- or multi-polar spindles was also observed. This is consistent with recent data indicating that centrosome clustering is an effective strategy for cells to respond to centrosome amplification, allowing completion of mitosis. However, cells with extra MTOCs pass through transient multi- or mono-polar intermediates, which lead to lagging chromosomes as a result of merotelic and syntelic attachments [1]. Our results contrast with knockdown of Cep76, another CP110-associated protein, which resulted in transient, supernumerary centrin dots that disappeared during mitosis [15], as the loss of Neurl4 resulted in extra centrin, γ-tubulin and GT335 foci that persisted throughout mitosis (Fig 4 and supplementary Fig S7 online; data not shown). Importantly, our data indicate that a subset of the CP110-positive assemblies resulting from Neurl4 depletion forms functional MTOCs. Therefore depletion of Neurl4 has the potential to markedly enhance mitotic defects, and, potentially, aneuploidy, through unequal chromosome segregation after assembly of extra CP110-positive MTOCs.
Neurl4 regulates the abundance of CP110
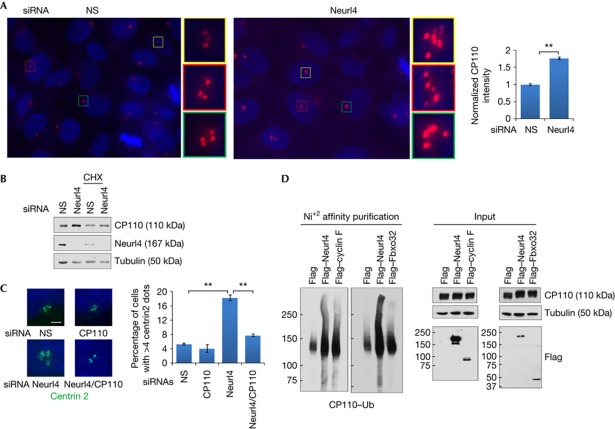
Next, we investigated the mechanisms through which Neurl4 regulates centrosome homeostasis. Interestingly, we found that Neurl4 depletion induced a substantial increase in CP110 levels by quantifying the intensity of CP110 immunofluorescence in the vicinity of centrosomes (Fig 5A). Similarly, an increase in CP110 protein levels was observed in lysates of U2OS and HeLa cells (Fig 5B and data not shown). We also measured CP110 protein levels in U2OS cells following treatment with Neurl4 or control siRNAs in HeLa cells synchronized by double thymidine block and release, which further attested to the fact that Neurl4 specifically stabilized CP110 protein levels (supplementary Fig S6C online). Moreover, while Neurl4 depletion provoked an increase in supernumerary CP110 foci, co-depletion of CP110 and Neurl4 completely reversed this effect, reinforcing the conclusion that this phenotype is dependent on elevated CP110 protein levels (Fig 5C and supplementary Fig S6A online).
Figure 5.
Phenotype associated with Neurl4 depletion is dependent on CP110 upregulation. (A) Quantifications of CP110 level on the centrosome in U2OS cells transfected with control or Neurl4 siRNA. CP110 fluorescence intensity at centrosomes was measured using Metamorph. CP110 was stained with anti-CP110 antibody (red). DNA was stained with DAPI (blue). (B) Western blot analysis of CP110 levels after Neurl4 depletion. Samples in the right-most two lanes were treated with 10 μM cycloheximide (CHX) for 5 h before collection. (C) U2OS cells were transfected with control siRNA or siRNAs targeting Neurl4, CP110 or both Neurl4 and CP110. Representative images of centrin staining and quantification of cells with >4 centrin foci are shown. Scale bar, 2 μm; **P<0.01. (D) 293T cells were transfected with haemagglutinin–CP110, His–ubiquitin and Flag-only vector control or Flag–Neurl4. Flag–cyclin F and Flag–Fbxo32 served as positive and negative controls, respectively. Cells were incubated with 10 μM MG132 for 4 h before collection. Ubiquitylated (Ub) CP110 protein was enriched with nickel agarose and analysed by western blots. DAPI, 4,6-diamidino-2-phenylindole; Neurl4, Neuralized homologue 4; siRNA, small interfering RNA.
Other members of the Neuralized family (Neurl1–3) have been shown to be E3 ubiquitin ligases. Thus, we speculated that Neurl4 could regulate CP110 levels, directly or indirectly, by promoting ubiquitylation of CP110 and targeting it for degradation. To this end, we asked whether CP110 could be ubiquitylated in a Neurl4-dependent manner. Remarkably, we observed that overexpression of Neurl4 promoted ubiquitylation of CP110 (Fig 5D). Cyclin F, a component of an SCFCyclin F E3 ubiquitin ligase that mediates ubiquitylation and degradation of CP110 [16], served as a positive control and Fbxo32, an unrelated F box protein, served as a negative control (Fig 5D). In addition, similar to depletion of Neurl4, overexpression of CP110 can induce several γ-tubulin dots (data not shown and D’Angiolella et al [16]), suggesting that one critical function of Neurl4 is to regulate CP110 levels and thereby regulate the centrosome duplication cycle. Further, we found that stable expression of Flag–Neurl4 led to a modest but statistically significant decrease in CP110 levels at centrioles (data not shown).
Next, although expression of either Neurl4 or cyclin F resulted in CP110 ubiquitylation, several lines of evidence indicated that Neurl4 might regulate CP110 stability through a distinct pathway. First, Neurl4 localizes to daughter centrioles throughout the cell cycle, whereas cyclin F appears at both centrioles and destabilizes CP110 exclusively in G2 phase. Thus, Neurl4 is likely to regulate CP110 stability in distinct but overlapping locations and cell cycle phases. Second, we found that even though both Neurl4 and cyclin F can coimmunoprecipitate with CP110, they did not interact with one another, indicating that Neurl4 and cyclin F form discrete and separable complexes with CP110 (supplementary Fig S8 online). Third, depletion of Neurl4 had no impact on the interaction between CP110 and cyclin F (data not shown), suggesting that Neurl4 depletion is not likely to directly affect the ubiquitylation of CP110 by cyclin F. Therefore, we propose that both Neurl4 and cyclin F regulate centrosome homeostasis through stabilization of CP110, but they are likely to operate in a distinct manner to modulate ubiquitylation of CP110. Future experiments will determine the exact mechanisms by which Neurl4 expression promotes ubiquitylation of CP110.
In summary, we have identified a novel CP110-interacting partner, Neurl4, as a centrosomal protein that specifically localizes to daughter centrioles through a new targeting motif, the NHR domain. Depletion of Neurl4 leads to the production of extra CP110/centrin dots, a subset of which recruit γ-tubulin, glutamylated tubulin and extra centrosome proteins, promoting the occurrence of mitotic abnormalities (supplementary Fig S9 online). Interestingly, it was recently shown that overexpression of centrin 2 generates supernumerary centriole-like dots, which were competent to form mitotic spindle poles [17]. Taken together with D’Angiolella et al [16] and Yang et al [17], we provide evidence of a new form of centrosome aberration that seems to lack the requirement of canonical centrioles per se, but is still capable of nucleating microtubules through recruitment of γ-tubulin. We speculate that the loss or overproduction of a single protein—such as Neurl4 or other proteins that amplify the level of CP110—could trigger these aberrations, thereby tipping a cell towards aneuploidy. Further studies will be required to examine the correlation between Neurl4 and CP110 levels and the ability to maintain centrosome homeostasis and genomic integrity in human tumours.
Methods
Identification of Neurl4. Neurl4 was initially identified as a CP110-interacting protein using immuno-affinity purification and tandem mass spectrometry as described previously [6]. Four non-overlapping Neurl4 peptides were identified using this method.
Cell culture, plasmids and antibodies. U2OS, HeLa, RPE1, and HEK293T were cultured in DMEM supplemented with 10% fetal bovine serum at 37°C in a humidified 5% CO2 atmosphere.
A Neurl4 (KIAA1787) complementary DNA was obtained from Kazusa Research Institute. The Neurl4 cDNA was PCR amplified and subcloned into pcDEF3 and pEGFP-N1. GFP–Neurl4 NHR constructs were prepared by PCR amplification of each NHR domain and subcloning into pEGFP-C1. Flag–Neurl4 deletion constructs were subcloned into pcDNA3.1 or pCMV5–Flag. Two different rabbit anti-Neurl4 antibodies were raised against bacterially produced glutathione-S-transferase fusion proteins containing N-terminal residues 1–120 and C-terminal residues 1,477–1,553 of Neurl4 (Cocalico Biologicals) and purified by affinity chromatography. Neurl4 antibody recognizing C-terminal residues was used for immunofluorescence and western blotting, whereas an N-terminally directed antibody was used for immunoprecipitation experiments.
RNA interference. Synthetic siRNA oligonucleotides were purchased from Dharmacon. Transfection of siRNAs was performed using siIMPORTER (Millipore) using manufacturer's instructions. Each knockdown experiment has been performed a minimum of three times. Western blots were performed to verify the efficacy of knockdown for each experiment.
A description of extra reagents and detailed protocols are available in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Sangeetha Vijayakumar for generating Neurl4 polyclonal antibodies and for initial effort on this project. We thank W. Lane (Harvard Microchemical Facility) and A. Spektor for assistance with mass spectrometric identification of CP110-interacting proteins and for identification of Neurl4 peptides, respectively, and M. Jelcic, I. Sanchez, W. Tsang, B. Bista, J. Jastrab, B. Weckselblatt, and S. Choi and members of the Dynlacht Laboratory for assistance with binding assays, immunoprecipitations and immunostaining. We thank V. D’Angiolella and M. Pagano for initial findings related to cyclin F, J. Salisbury, T. Tang, E. Nigg, C. Janke, Q. Gao for providing antibodies and reagents and C. Petzold, D. Kristen of NYULMC OCS Microscopy core for the electron microscopy services. B.D.D. was supported by a March of Dimes research grant. J.L. was supported by a DOD Prostate Cancer pre-doctoral fellowship. S.D. and N.K. were supported by National Institutes of Health R01 CA112598 and American Cancer Society grant RSG--07-075-01-MBC.
Author contributions: J.L., S.K., T.K., F.-X.L., N.K. and S.D. performed experiments. All authors analysed experiments. J.L. and B.D.D. wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ganem NJ, Godinho SA, Pellman D (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD (2002) CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell 3: 339–350 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD (2011) Regulating the transition from centriole to basal body. J Cell Biol 193: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD (2011) Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 145: 914–925 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P (2009) Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD (2007) Cep97 and CP110 suppress a cilia assembly program. Cell 130: 678–690 [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA (2009) Control of centriole length by CPAP and CP110. Curr Biol 19: 1005–1011 [DOI] [PubMed] [Google Scholar]
- He F et al. (2009) Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway. J Mol Biol 393: 478–495 [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev Cell 21: 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA (2007) Plk4-induced centriole biogenesis in human cells. Dev Cell 13: 190–202 [DOI] [PubMed] [Google Scholar]
- Augustin A et al. (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci 116: 1551–1562 [DOI] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q (2005) Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 171: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T (2010) Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol 191: 331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Straatman KR, Fry AM (2009) Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol 29: 1760–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Vijayakumar S, Bista BR, Li J, Sanchez I, Duensing S, Dynlacht BD (2009) Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev Cell 16: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M (2010) SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466: 138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Kasbek C, Majumder S, Yusof AM, Fisk HA (2010) Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell 21: 4361–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.