Abstract
The conserved MRE11–RAD50–NBS1 (MRN) complex is an important sensor of DNA double-strand breaks (DSBs) and facilitates DNA repair by homologous recombination (HR) and end joining. Here, we identify NBS1 as a target of cyclin-dependent kinase (CDK) phosphorylation. We show that NBS1 serine 432 phosphorylation occurs in the S, G2 and M phases of the cell cycle and requires CDK activity. This modification stimulates MRN-dependent conversion of DSBs into structures that are substrates for repair by HR. Impairment of NBS1 phosphorylation not only negatively affects DSB repair by HR, but also prevents resumption of DNA replication after replication-fork stalling. Thus, CDK-mediated NBS1 phosphorylation defines a molecular switch that controls the choice of repair mode for DSBs.
Keywords: NBS1, cyclin-dependent kinase, homologous recombination, end joining, replication restart
Introduction
DNA double-strand breaks (DSBs) are particularly toxic DNA lesions because they are difficult to repair and can lead to chromosome loss, translocations or truncations. Inherited defects in detecting, signalling or repairing DSBs cause pathologies, including cancer, neurodegenerative disease, immune deficiencies, infertility and premature ageing [1]. Consequently, eukaryotic cells have evolved genome surveillance pathways, collectively termed the DNA-damage response (DDR), to detect, signal and repair DSBs and other DNA lesions [2]. Of particular importance in the DDR is the MRE11–RAD50–NBS1 (MRN) complex, which promotes DSB repair and recruits and activates the checkpoint kinase ATM [3, 4], which then triggers DSB signalling by phosphorylating multiple protein targets [5]. Defects in NBS1 cause the human microcephaly, immune deficiency, radiosensitivity and enhanced cancer predisposition disease, Nijmegen breakage syndrome (NBS), whereas MRE11 and ATM defects cause neurodegeneration, radiosensitivity and elevated cancer predisposition in Ataxia telangiectasia (AT) and AT-like disease, respectively [6].
There are two principle mechanisms for DSB repair: non-homologous end joining (NHEJ), which is active throughout the cell cycle, and homologous recombination (HR), which is restricted to S and G2 cells [7]. For a given DSB, these repair modes are mutually exclusive, with the choice of repair pathway being in part governed by cell-cycle status. Thus, unlike NHEJ, HR generally requires a sister chromatid and depends on cell-cycle regulated 5′ to 3′ exonucleolytic processing (resection) of DNA ends that generates single-stranded DNA (ssDNA). DSB resection in human cells is promoted by ATM activity and other factors, including the MRN complex and CtBP interacting protein (CtIP), and is subject to cell-cycle control [8, 9, 10].
Once produced, ssDNA is bound by replication protein A (RPA), which is then replaced by the RAD51 protein that promotes DNA strand-invasion and ensuing HR [7]. RPA-coated ssDNA also mediates recruitment and activation of the DNA-damage checkpoint kinase ATR that phosphorylates various target proteins, including the downstream checkpoint kinase CHK1. In addition to responding to radiation- and chemical-induced DSBs, the above factors also function at sites of stalled DNA replication caused by agents such as hydroxyurea (HU) that reduces deoxyribonucleotide levels, thereby leading to replication-fork stalling [11]. While the precise functions for resection and HR proteins at stalled replication forks are not entirely clear, they are believed to stabilize replication forks and allow them to resume replication by recombination-dependent processes [12].
Results
NBS1 Ser 432 phosphorylation is CDK-dependent
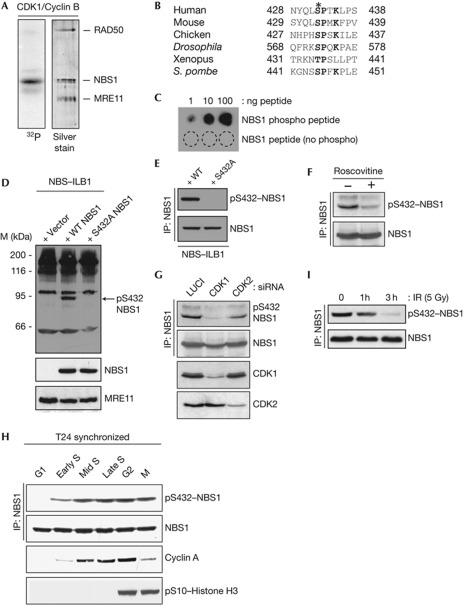
Cell-cycle control of resection and HR depends on cyclin-dependent kinase (CDK)-mediated phosphorylation of a conserved site on the MRN cofactor protein CtIP. Nevertheless, studies with a phospho-mimicking CtIP derivative (T847E–CtIP) have suggested that cell-cycle control of resection is also mediated by CDKs targeting other residues on CtIP and/or on additional proteins [10]. To investigate whether CDKs might target the MRN complex, we incubated a purified preparation of recombinant MRN with CDK1/CyclinB and 32P-ATP and tested for 32P incorporation into MRN components. As shown in Fig 1A, NBS1, but not MRE11 or RAD50, was phosphorylated by CDK1/CyclinB.
Figure 1.
NBS1 Ser 432 is a CDK substrate. (A) Biochemical kinase assay with recombinant CDK1/CyclinB and purified MRN as substrate. (B) Alignment of region surrounding putative phospho-site in NBS1 orthologues. (C) Characterization of the NBS1–Ser 432 phospho-specific antibody. (D) NBS1 phospho-Ser 432 antibody recognizes NBS1 in whole-cell extracts and (E) after immunoprecipitation. (F) U2OS cells were treated or mock-treated with 50 μM roscovitine for 4 h. (G) Depletion of CDK1 or CDK2 in U2OS cells reduces NBS1 Ser 432 phosphorylation. (H) NBS1 Ser 432 phosphorylation is cell-cycle regulated. (I) NBS1 Ser 432 phosphorylation is reduced after DNA-damage induction. CDK, cyclin-dependent kinase; IP, immunoprecipitation; IR, ionizing radiation; MRN, MRE11–RAD50–NBS1; NBS, Nijmegen breakage syndrome; siRNA, small-interfering RNA; WT, wild-type.
The NBS1 protein sequence contains a consensus CDK1/2 phosphorylation site (S/T-P-X-K/R, where X is any amino-acid residue [13]) that is conserved in most known NBS1 orthologues (Fig 1B). To assess whether this was a site of NBS1 phosphorylation, we generated a phospho-specific antibody (Fig 1C) and determined whether the antibody could recognize recombinant NBS1 expressed in patient-derived NBS1-deficient NBS–ILB1 cells. The antibody recognized a protein that migrated with the expected size for NBS1 in extracts of NBS–ILB1 cells expressing wild-type NBS1 (Fig 1D). Moreover, the antibody did not detect this protein in extracts of NBS–ILB1 cells transfected with a control vector or a vector expressing an NBS1 derivative, in which Ser 432 was mutated to a non-phosphorylatable Ala residue (S432A–NBS1; Fig 1D top; as shown in panels below, wild-type and mutated proteins were expressed to similar levels). In accord with these data, the phospho-specific antibody also recognized immunoprecipitated wild-type but not S432A–NBS1 (Fig 1E).
These findings suggested that NBS1 Ser 432 is a CDK target. Consistent with this, detection of NBS1 phospho-Ser 432 (pS432) was greatly diminished when cells were treated with the CDK inhibitor roscovitine (Fig 1F; [14]). Moreover, depletion of CDK1 or CDK2 by small-interfering RNAs also resulted in reduced NBS1 phospho-Ser 432 (Fig 1G). Furthermore, assessment of Ser 432 phosphorylation levels, as a function of cell-cycle progression in synchronized cultures of human T24 cells, revealed that NBS1 pS432 was not apparent in G1 cells but became detectable in early S phase, reached its maximum in mid S phase and then persisted throughout the rest of the cell cycle (Fig 1H). In addition, detection of NBS1 pS432 was decreased when U2OS cells were exposed to ionizing radiation (IR; Fig 1I), which leads to inhibition of CDK1/2 activity [15, 16].
Mutation of NBS1 Ser 432 impairs DNA-end resection
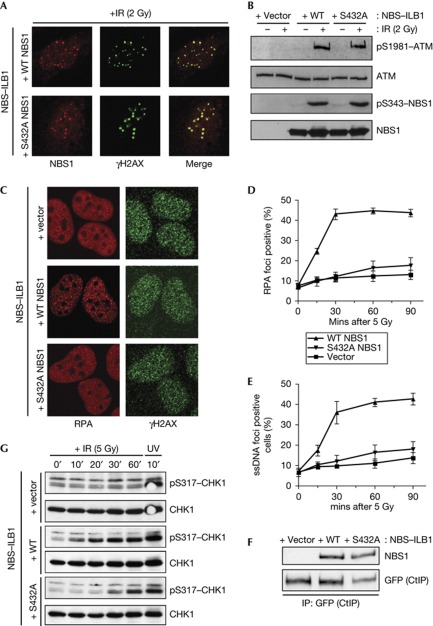
Given that MRN is recruited to DNA-damage sites, where it promotes ATM activation, we tested whether these properties were affected by NBS1 Ser 432 mutation. As shown in Fig 2A, like wild-type NBS1, S432A–NBS1 was recruited to sites of IR-induced DNA damage, where it colocalized with DNA damage–induced phosphorylated form of histone H2AX (γH2AX) [17]. Moreover, the mutated protein was competent at triggering ATM activation, as assessed by detecting ATM autophosphorylation on Ser 1981 [18] and by ATM-mediated phosphorylation of NBS1 Ser 343 (Fig 2B). Because CDK activity promotes DSB processing and ensuing HR, we speculated that NBS1 pS432 might affect MRN-dependent DSB resection. Indeed, IR-induced RPA focus formation, a marker of resected DSBs [19], was effective in NBS–ILB1 cells complemented with wild-type NBS1 but was impaired in NBS cells complemented with either empty vector or the S432A–NBS1 mutant (Fig 2C). Defective resection was not an indirect effect of reduced proliferation of NBS cells complemented with S432A–NBS1, as shown by assays measuring replication by incorporation of the nucleotide analogue EdU (supplementary Fig S1 online). Furthermore, time-course studies and quantification of RPA foci revealed that the RPA focus formation defect in cells expressing S432A–NBS1 was as severe as that of cells transfected with the vector control (Fig 2D). To substantiate these findings, we used a 5-bromodeoxyuridine-based detection procedure under non-denaturing conditions to directly measure ssDNA generation [9]. Unlike cells expressing wild-type NBS1, cells expressing the S432A–NBS1 mutant were defective in ssDNA generation (Fig 2E). Importantly, this defect was not due to an impaired interaction of S432A–NBS1 with CtIP (Fig 2F) [9]. Unfortunately, an S432E–NBS1 derivative did not work as a phospho-mimetic version of NBS1 and thus could not be used in these assays (data not shown).
Figure 2.
NBS1 S432A impairs DNA-end resection. (A) IR-induced focus formation in complemented NBS cells. Cells were collected 1 h after irradiation. (B) Immunoblots from complemented NBS cells after irradiation. Cells were collected 1 h after irradiation. (C) IR-induced RPA foci from complemented NBS cells. Cells were irradiated with 5 Gy IR and collected 1 h after irradiation. (D,E): Quantification of IF data. At least 100 cells were counted per condition. Error bars represent standard deviations. (F) NBS1 coimmunoprecipitates with CtIP regardless of S432A mutation. NBS cells were transfected with a plasmid containing GFP–CtIP. (G) Kinetics of IR-induced CHK1 phosphorylation in complemented NBS cells. Cells were irradiated with 5 J/m2 ultraviolet. GFP, green fluorescent protein; IF, immunofluorescence; IP, immunoprecipitation; IR, ionizing radiation; NBS, Nijmegen breakage syndrome; RPA, replication protein A; ssDNA, single-stranded DNA; UV, ultraviolet light; WT, wild-type.
In light of previous work showing that DSB resection triggers activation of ATR that then phosphorylates various proteins, including CHK1 on Ser 317 [8], we tested whether this response was affected by NBS1 Ser 432 mutation. Indeed, as shown in Fig 2G, IR-induced CHK1 Ser 317 phosphorylation was delayed in NBS–ILB1 cells expressing S432A–NBS1 (although not abolished as in cells containing an empty vector) in comparison to cells expressing wild-type NBS1. By contrast, cells expressing S432A–NBS1 or containing an empty vector were competent to trigger CHK1 Ser 317 phosphorylation after exposure to ultraviolet light (Fig 2G), which activates ATR without requiring MRN-mediated resection [20].
NBS1 Ser 432 regulates DSB repair pathway usage
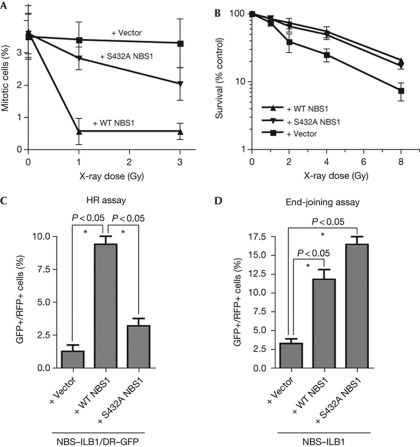
To explore the biological consequences of the NBS1 Ser 432 mutation, we assessed cell-cycle checkpoint responses and cell survival in response to IR. Consistent with the delayed CHK1 phosphorylation observed in S423A–NBS1 complemented NBS–ILB1 cells, while the mitotic index (as assessed by histone H3 phospho-Ser 10 staining) dropped rapidly after IR treatment of cells complemented with wild-type NBS1, this response was impaired in both vector- and S423A-complemented cells, albeit not to the same extent (Fig 3A). Nevertheless, while vector-complemented NBS–ILB1 cells were clearly hypersensitive to IR, cells expressing S432A–NBS1 were not or only marginally more IR sensitive than cells expressing wild-type NBS1 (Fig 3B). This absence of IR hypersensitivity in cells complemented with S432A–NBS1 suggested that these cells are able to mediate efficient DNA repair, either by HR or NHEJ.
Figure 3.
NBS1 S432A impairs HR without significantly affecting end joining. (A) G2/M assay in complemented NBS cells. Cells were collected 1 h after irradiation and measured for H3 pSer10 staining by flow cytometry. (B) IR survival. (C) HR assay in NBS/DR–GFP cells. (D) End-joining assay in complemented NBS cells. All quantitative data represent the average of three independent experiments (±s.d.). (*) Denotes statistically significant differences (P<0.05, unpaired t-test). HR, homologous recombination; GFP, green fluorescent protein; IR, ionizing radiation; NBS, Nijmegen breakage syndrome; RFP, red fluorescent protein; WT, wild-type.
Generation of RPA-coated ssDNA by DSB resection is promoted by MRN and represents a key early step in HR [21]. Owing to the resection defect observed in cells expressing S432A–NBS1, we speculated that CDK-mediated NBS1 Ser 432 phosphorylation would promote HR. To test this, we generated an NBS/ILB1-derived cell line, NBS–ILB1/DR–GFP, which contained a green fluorescent protein (GFP)-based HR reporter construct bearing a cleavage site for the I-SceI restriction endonuclease. I-SceI expression in such cells induces a DSB that can be repaired by HR-mediated gene conversion to produce GFP-positive cells [22]. NBS–ILB1/DR–GFP cells were transfected with I-SceI and red fluorescent protein in combination with an empty vector, wild-type NBS1 or S432A–NBS1. As shown in Fig 3C and Supplementary Fig S1C online, cells transfected with either empty vector or S432A–NBS1 displayed lower levels of HR-mediated repair than cells transfected with wild-type NBS1.
In addition to its roles in HR-mediated repair, NBS1 has also been linked to repair by end joining [6]. Thus, we transfected NBS–ILB1 cells with empty vector or with vectors expressing wild-type NBS1 or S432A–NBS1 together with a GFP expression plasmid that had been linearized with the restriction endonuclease EcoRI, uncoupling the transcriptional promoter from the GFP open reading frame. In this assay, perfect or near-perfect repair of the plasmid by end joining will restore GFP expression [23]. While NBS1-deficient cells transfected with empty vector displayed a low level of repair, expression of wild-type or S432A–NBS1 improved repair efficiency dramatically (Fig 3D). Collectively, these results demonstrated that, while NBS1 is required for efficient DSB repair by both HR and end joining, mutation of NBS1 Ser 432 impairs HR without significantly affecting end joining, thus leading to a model in which CDK-mediated NBS1 Ser 432 phosphorylation contributes to DSB repair pathway choice.
NBS1 Ser 432 promotes replication recovery
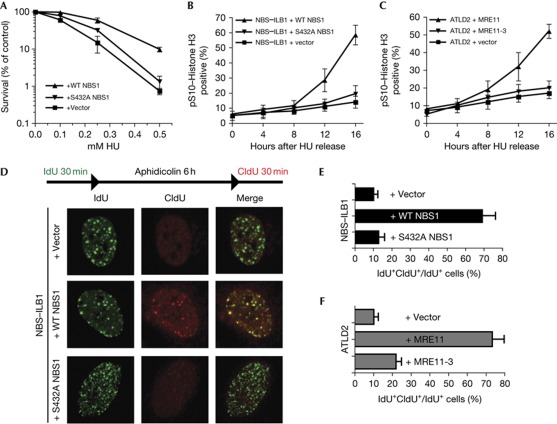
In addition to being hypersensitive towards DSB-inducing agents, NBS1-deficient cells also exhibit hypersensitivity to agents that result in replication-fork stalling [24]. To test for the potential importance of NBS1 Ser 432 phosphorylation in this scenario, we treated cells with HU. As shown in Fig 4A, NBS–ILB1 cells expressing S432A–NBS1 were substantially more sensitive to HU than cells expressing wild-type NBS1, although this sensitivity was not as great as that of vector-complemented cells. To further explore this phenotype, we examined the ability of complemented NBS–ILB1 cells to recover from HU-induced cell-cycle arrest. Strikingly, while cells expressing wild-type NBS1 recovered efficiently from the HU arrest and entered mitosis 12–16 h after HU release, both vector- and S432A–NBS1-complemented cells failed to enter mitosis within 16 h after HU removal (Fig 4B). Phenotypes associated with mutations in one subunit of the MRN complex are usually shared by mutations in any of the other components. In addition, phenotypes associated with MRE11 loss are also exhibited by cells expressing nuclease-defective MRE11 derivatives [25]. In line with these observations, by using MRE11-deficient ATLD2 cells complemented with either empty vector, wild-type MRE11 or a nuclease-inactive version of MRE11 (MRE11-3) [26], we found that efficient recovery from HU-induced arrest required MRE11 nuclease function (Fig 4C). Collectively, these data suggested a model in which CDK-mediated phosphorylation of NBS1 is as important as MRE11 nuclease activity for cells to recover efficiently from HU-induced cell-cycle arrest.
Figure 4.
NBS1 S432A impairs replication restart. (A) Cells were treated for 24 h with the indicated HU dose. Data represent the average of three independent experiments (±s.d.). (B) Mitotic entry after HU release in complemented NBS and (C) ATLD2 cells. Cells were treated with 2 mM HU for 24 h. Data represent the average of three independent experiments (±s.d.). (D) Replication restart after aphidicolin treatment in complemented NBS cells. (E) Quantification of IF data in complemented NBS and (F) ATLD2 cells. At least 100 cells were counted per condition. Error bars represent standard deviations. ATLD2, Ataxia telangiectasia-like disease; CldU, 5-chloro-2-deoxyuridine; HR, homologous recombination; HU, hydroxyurea; IdU, 5-iodo-2-deoxyuridine; IF, immunofluorescence; IR, ionizing radiation; NBS, Nijmegen breakage syndrome; RPA, replication protein A; UV, ultraviolet light; WT, wild-type.
Cellular sensitivity to HU and delayed entry to mitosis in NBS1-deficient cells could be explained by delayed DNA-damage repair in the absence of NBS1, but it could also be linked to a defect in recovering from replicative stress [27], a phenotype that is also observed in MRE11-deficient cells [28, 29]. To test whether NBS1 Ser 432 mutation might also cause such a phenotype, we tested the ability of control and complemented NBS–ILB1 cells to recover from replication stalling by using a technique that visualizes DNA replication by sequential incorporation of halogenated deoxyuridine derivatives [30]. Thus, we marked replicative cells by pulse labelling them with 5-iodo-2-deoxyuridine (IdU), arrested them with HU or with an inhibitor of DNA polymerase-α (aphidicolin) for 6 h, and then allowed replication restart by removing the drug in the presence of 5-chloro-2-deoxyuridine (CldU, see Fig 4D, top). Strikingly, all NBS1–ILB1 cells we analyzed restarted replication with normal kinetics after HU treatment, even those complemented with the empty vector (data not shown). However, whereas cells expressing wild-type NBS1 exhibited extensive replication recovery after aphidicolin treatment, both vector- and S432A-complemented cells displayed severe replication restart defects (Fig 4D,E). Similarly, analogous experiments with complemented ATLD2 cells demonstrated that cells expressing MRE11-3 were as impaired as vector-complemented cells in recovering from replication arrest (Fig 4F). Collectively, these data suggested that in cells treated with the replication inhibitor aphidicolin, both NBS1 Ser 432 phosphorylation and MRE11 nuclease activity are required for maintaining the integrity of stalled replication forks and/or for fork restart.
Discussion
CDK activity enhances DSB resection and HR by mediating CtIP Thr 847 phosphorylation during S and G2 phases of the cell cycle [10]. This and other CDK-dependent CtIP phosphorylations have been shown to be Mre11 dependent [31]. However, because a phospho-mimicking mutant of Thr 847 is not able to fully promote DNA-end resection in G1 cells, CtIP Thr 847 phosphorylation is not the only event that promotes HR by affecting resection [10]. Our data indicate that CDK-dependent NBS1 Ser 432 phosphorylation is also critical for promoting MRN-dependent resection and HR, and is also needed for MRN-dependent recovery from replication arrest. Indeed, cells expressing S432A–NBS1 exhibit a HR defect as pronounced as that seen in the absence of full-length NBS1, but display proficient end joining. Given the involvement of NBS1 in both HR and end joining [32], this suggests that CDK-dependent NBS1 Ser 432 phosphorylation acts as a molecular switch to funnel DSBs into either HR or end joining, depending on cell-cycle stage.
Although CDK activity is essential to initiate DNA replication [33], there is to our knowledge no direct evidence for its involvement in resumption of DNA replication after replication-fork stalling. The fact that NBS or ATLD2 cells fail to resume replication after aphidicolin treatment could indicate that stalled replication forks are collapsed—and are thus unable to restart replication—in the absence of MRN function. Replication-fork collapse under these conditions could result in activation of the intra-S phase checkpoint (mainly by ATR/CHK1), which would prevent rescue of collapsed forks by firing of otherwise dormant, neighbouring replication origins [34]. Given that the same is not observed when cells are treated with HU suggests that, although both drugs cause replication-fork stalling, there is a fundamental difference between them regarding the DDR they elicit. Whether this reflects the differing mechanisms-of-action of the drugs (direct DNA polymerase stalling caused by aphidicolin, and depletion of free deoxyribonucleotides by HU), or to other determinants such as different structures formed at stalled replication forks in response to either drug needs further investigation.
Our complementation assays also indicate that both MRE11 nuclease activity and CDK-dependent NBS1 Ser 432 phosphorylation are critical for this role of MRN in replication restart. Recent reports have suggested that MRE11 nuclease function is important for replication-fork stability, although tight control has to be exerted on this to prevent excessive nucleolytic activity at stalled replication forks that could otherwise collapse [35, 36, 37]. In light of this, it is tempting to speculate that CDK-mediated NBS1 Ser 432 phosphorylation, in conjunction with CDK-dependent phosphorylation of CtIP and perhaps additional proteins, could promote MRN nuclease activity. This would explain the lack of DNA-end resection and impaired replication restart in NBS cells complemented with S432A–NBS1. Whatever the precise mechanisms involved, our data highlight the crucial roles that CDK activity and cell-cycle stage have on DSB resection and repair, thus maximizing genome stability by allowing usage of the DSB repair pathway most appropriate to cell-cycle status.
Methods
Cell lines. NBS1-deficient NBS–ILB1 and MRE11-deficient ATLD2 human cells were cultured in DMEM supplemented with 15% fetal bovine serum (10% for T24 and U2OS) and penicillin–streptomycin–glutamine. T24 cells were synchronized as described previously [8]. NBS1 stable cell lines were generated by co-transfection of NBS–ILB1 cells with the expression plasmids pSG5, pSG5–Myc–NBS1 [4], or pSG5–Myc–NBS1–S432A together with pBABE-puro containing the puromycin resistance cassette. Human ATLD2 cells and their MRE11-reconstituted counterparts were kindly provided by Y Shiloh (Tel Aviv, Israel) [3].
Quantification of mitotic cells. Identification of mitotic cells was done by staining chromosomal DNA with propidium iodide combined with immunofluorescent detection of histone H3 pSer10 (rabbit; Millipore). When measuring mitotic entry after HU treatment, cells were incubated with 2 mM HU for 24 h. After extensive washing, cells were incubated in fresh medium containing nocodazole, and the proportion of mitotic cells at the indicated times was measured.
Cell survival assays. Cells were exposed to differing doses of IR or HU. For HU, cells were exposed to the drug for 24 h. After 10–14 days, colonies were stained with 0.5% crystal violet/20% ethanol, counted and normalized to plating efficiencies.
DNA repair assays. >HR and NHEJ assays were as described previously [22, 23].
Replication restart assays. Replicative cells were marked by pulse labelling for 30 min with CldU (50 μM) then arrested by treating with aphidicolin (10 μM) or HU (2 mM) for 6 h. After drug removal, cells were treated with IdU (50 μM) for 30 min (aphidicolin treatment) or 60 min (HU treatment) so that replication restart after stalling could be visualized. CldU and IdU detection was as described previously [27] with anti-5-bromodeoxyuridine antibodies, from Abcam and Becton-Dickinson, which crossreact with the indicated halogenated nucleotide. Replication recovery is represented by the overlap of CldU and IdU labelling.
Supplementary Material
Acknowledgments
J.F., M.M., J.L. and J.B. were supported by the Danish Cancer Society, the Danish National Research Foundation and the European Community (projects DDResponse and Biomedreg: CZ.1.05/2.1.00/01.0030). Research in the Jackson Laboratory is funded by Cancer Research UK Program Grant C6/A11224, the European Research Council and the European Community’s Seventh Framework Programme FP7/2007-2013 (GENICA and DDResponse, grant agreement No. HEALTH-F2-2012-259893). Funding for core infrastructure in the SPJ laboratory was provided by Cancer Research UK and the Wellcome Trust. S.P.J. receives his salary from the University of Cambridge, supplemented by Cancer Research UK.
Author contributions: J.F., J.V.F. and S.P.J. designed the experiments. J.F., J.V.F., M.M., J.L., J.B. and S.P.J. analysed the data. J.F., J.V.F. and J.C. performed the experiments. J.V.F. and S.P.J. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Uziel T et al. (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Matsuoka S et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JHJ (2011) The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Gómez-González B, Aguilera A (2009) DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A et al. (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Sartori A et al. (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson S (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Songyang Z et al. (1994) Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 4: 973–982 [DOI] [PubMed] [Google Scholar]
- De Azevedo WF et al. (1997) Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur J Biochem 243: 518–526 [DOI] [PubMed] [Google Scholar]
- Mailand N et al. (2000) Rapid destruction of human Cdc25A in response to DNA damage. Science 288: 1425–1429 [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842–847 [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP (2011) Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci USA 96: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei M et al. (2003) Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem 278: 14806–14811 [DOI] [PubMed] [Google Scholar]
- Huertas P (2010) DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol 17: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z et al. (2004) MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J Biol Chem 279: 46359–46362 [DOI] [PubMed] [Google Scholar]
- Brugmans L et al. (2009) NBS1 cooperates with homologous recombination to counteract chromosome breakage during replication. DNA Repair 8: 1363–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J et al. (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T et al. (2004) Nbs1 is required for ATR-dependent phosphorylation events. EMBO J 24: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H et al. (2009) PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28: 2601–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM (2000) Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol 2: 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Stoneham T, Spehalski E, Ferguson DO (2012) Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat Struct Mol Biol 19: 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G (2011) Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 711: 61–72 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX (2009) DNA replication as a target of the DNA damage checkpoint. DNA Repair 8: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA (2007) The ATR pathway: fine-tuning the fork. DNA Repair 6: 953–966 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri A, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K et al. (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V (2011) Brca2, Rad51 and Mre11: performing balancing acts on replication forks. DNA Repair 10: 1060–1065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.