Abstract
Chromosomal instability (CIN)—which is a high rate of loss or gain of whole or parts of chromosomes—is a characteristic of most human cancers and a cause of tumour aneuploidy and intra-tumour heterogeneity. CIN is associated with poor patient outcome and drug resistance, which could be mediated by evolutionary adaptation fostered by intra-tumour heterogeneity. In this review, we discuss the clinical consequences of CIN and the challenges inherent to its measurement in tumour specimens. The relationship between CIN and prognosis supports assessment of CIN status in the clinical setting and suggests that stratifying tumours according to levels of CIN could facilitate clinical risk assessment.
Keywords: cancer, chromosomal instability, genomic instability, aneuploidy, intra-tumour heterogeneity
See Glossary for abbreviations used in this article.
Glossary.
- 17-AAG
17-N-allylamino-17-demethoxygeldanamycin
- AURKA/B
aurora kinase A/B
- AMPK
5′ adenosine monophosphate-activated protein kinase
- AICAR
5-amino-1-β-D-ribofuranosyl-imidazole-4-carboxamide
- BUB1B
budding uninhibited by benzimidazoles 1 homologue beta (yeast)
- CCNB1/B2
cyclin B1/ B2
- CDC20
cell division cycle 20 homologue (yeast)
- CENP-E
centromere linked motor protein E
- H2AFX
H2A histone family, member X
- HER2
human epidermal growth factor receptor 2
- HSP90
heat shock protein 90
- MPS1
monopolar spindle 1-like
- NEK2
NIMA (never in mitosis gene a)-related kinase 2
- qPCR
quantitative PCR
- ZWINT
ZW10 interactor
Introduction
Genomic instability is a striking feature of human cancers [1,2] that results in genetic aberrations at many levels: from single nucleotide changes at the gene level, gross structural changes at the sub-chromosomal level to losses and gains of entire chromosomes. Chromosomal instability (CIN) is a form of genomic instability observed in a large proportion of solid tumours and many haematological malignancies. CIN describes a dynamic state in which cells continuously gain or lose whole chromosomes, or parts of chromosomes, at an elevated rate [3], and is therefore a principal mediator of aneuploidy and intra-tumour heterogeneity.
The clinical importance of CIN is underscored by its association with poor patient outcome in multiple cancer types, including lung, breast and colon cancer (Table 1; [4,5,6]). This association is probably driven by intra-tumour heterogeneity, facilitating the adaptation of tumours to environmental stress [7]. In addition to this, recent evidence suggests that this form of genomic instability might be linked to intrinsic multi-drug resistance [8]. However, despite the prevalence and the clinical relevance of CIN, a consistent basis for how it is generated at the molecular level is poorly understood (Sidebar A).
Table 1. Clinical relevance of CIN.
| Cancer type | Method of measuring CIN | Associated outcomes | Additional details | Reference |
|---|---|---|---|---|
| Lung cancer (NSCLC) | FISH (n = 63) | Poor prognosis (OS and DFS) | Korean patients | Choi et al [4] |
| FISH (n =47) | Poor prognosis (OS) | Korean patients | Yoo et al [51] | |
| FISH (n = 50) | Poor prognosis (OS) | Nakamura et al [52] | ||
| 12-gene signature (n = 647) | Poor prognosis (OS) | Multiple datasets | Mettu et al [66] | |
| CIN70 signature (n = 62) | Poor clinical outcome | Carter et al [5] | ||
| Breast cancer | SSI (n = 890) | Poor prognosis (OS) | CIN measured within diploid, tetraploid and aneuploid classified tumours | Kronenwett et al [55] |
| SNP (n = 313) | Poor prognosis (MFS) | Significant in ER-positive, luminal B and HER2-positive subtypes (not in ER-negative patients) | Smid et al [46] | |
| 12-gene signature (n = 469) | Poor prognosis (DFS and RFS) | Multiple datasets | Habermann et al [9] | |
| CIN70 signature (n = 1866) | Poor clinical outcome | Multiple datasets | Carter et al [5] | |
| FISH (n = 31) | Lymph node metastasis and ER negativity | Takami et al [50] | ||
| Myelodysplastic syndrome | FISH (n = 65) | Poor prognosis (DFS) | Heilig et al [102] | |
| Endocrine pancreatic tumours | CGH (n = 62) | Metastasis | Jonkers et al [79] | |
| Colon cancer | 12-gene signature (n = 92) | Recurrence of colon cancer | Multiple datasets | |
| Flow cytometry/image cytometry (n = 10126) | Poor prognosis | Meta-analysis | Walther et al [6] | |
| Ovarian cancer | 12-gene signature (n = 124) | Poor prognosis (RFS) | Mettu et al [66] | |
| Endometrial cancer | SNP (n = 31) | Poor prognosis (OS) | Murayama-Hosokawa et al [70] | |
| Synovial sarcoma | CGH (n = 22) | Poor prognosis (OS) | Nakagawa et al [71] | |
| Oral cancer (SCCs) | FISH (n = 77) | Poor prognosis (OS and DFS) | Sato et al [53] | |
| FISH (n =20) | (Loco) regional tumour outgrowth | Bergshoeff et al [103] | ||
| Diffuse large B-cell lymphoma | Anaphase segregation errors (n = 54) | Poor prognosis (RFS) | Bakhoum et al [48] |
CGH, comparative genome hybridization; DFS, disease-free survival; ER, oestrogen receptor; FISH, fluorescence in situ hybridization; MFS, metastasis-free survival; NSCLC, non-small-cell lung cancer; OS, overall survival; RFS, relapse-free survival; SCC, squamous cell carcinoma; SNP, single-nucleotide polymorphism; SSI, stem line scatter index.
Sidebar A | In need of answers.
What are the main molecular mechanisms that generate CIN in cancer?
Can structural and numerical CIN be considered in isolation, or are they governed by unifying mechanisms?
What is the best method to measure CIN in tumour specimens?
Can CIN status be used in the clinic for risk-stratification purposes?
Can we exploit CIN as a therapeutic target?
Characterization of the molecular mechanisms responsible for this pattern of genomic instability in tumours is hindered by the technical difficulties associated with measuring CIN in tumour specimens. Gene expression signatures provide a means to estimate levels of CIN from an aggregated population of cells [5,9], although such multiple-cell approaches capture neither the dynamic nature of CIN, nor the degree of intra-tumour heterogeneity. They therefore represent indirect, static measurements. By contrast, single-cell methods, such as fluorescence in situ hybridization (FISH), provide a more accurate measure of CIN; however, these are labour intensive, highlighting the need for alternative methods to assess CIN status in tumour specimens.
In this review, we outline the mechanisms that might contribute to CIN and discuss the clinical, pathological and molecular evidence that supports the need for assessment of CIN status in the clinical setting. We discuss the impact of CIN on patient outcome and highlight the challenges associated with determining CIN status in the clinical setting.
Classification of CIN
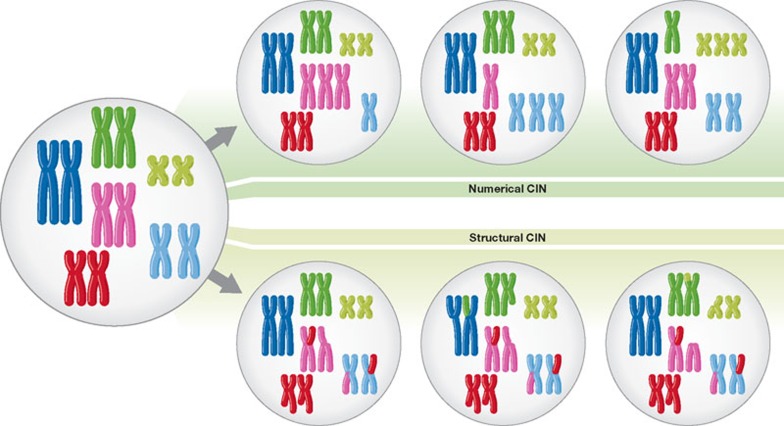
CIN tumour cells usually contain both numerical and structural chromosome changes (Fig 1; [10]). Numerical CIN (nCIN) refers to gain or loss of whole chromosomes at notably higher rates than normal cells. Structural CIN (sCIN), on the other hand, refers to an increased rate of formation of structurally abnormal chromosomes, resulting in gain or loss of chromosome fragments, translocations, deletions and amplifications of DNA [11]. Individual chromosomes in CIN cancer genomes are often aberrant at both the whole and sub-chromosomal level ([12], see also Reviews by Holland & Cleveland and Pfau & Amon, in this issue).
Figure 1.
Numerical and structural chromosomal instability. Scheme showing whole chromosome gains and losses (numerical CIN) and sub-chromosomal gains, losses, inversions and translocations (structural CIN).
CIN is thought to be a principal cause of aneuploidy, a state of abnormal chromosomal number [13]. Aneuploidy, however, is not synonymous with CIN; whilst CIN always results in aneuploidy, aneuploidy is not always associated with CIN. In human cancers, this is exemplified by hyperdiploid acute lymphoblastic leukaemia and near-triploid neuroblastoma, which have a stable aneuploid karyotype and a favourable prognosis [14,15]. The distinction between CIN and aneuploidy is important particularly when developing methods to measure CIN that must capture its dynamic nature (see CIN—diagnostic methods below), and also when exploring its impact on prognosis (see Clinical impact of CIN below). The distinction is also important when considering mechanisms that lead to CIN, which must consider both the structural and numerical chromosome aberrations observed in CIN cells.
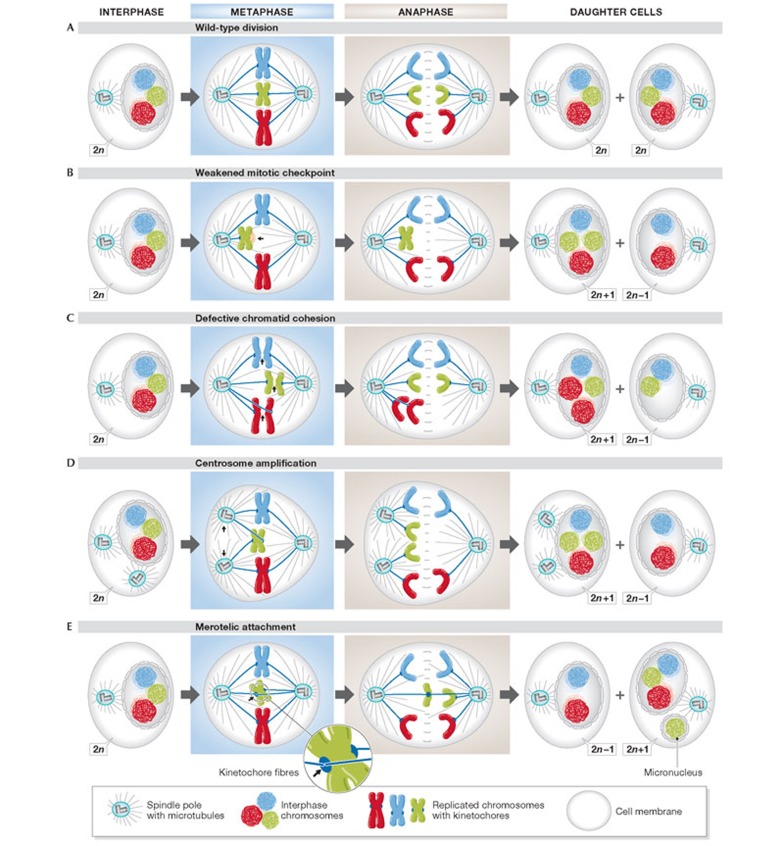
Efforts to understand nCIN have focused on elucidating the mechanisms that lead to mitotic errors and the uneven distribution of chromosomes between daughter cells (Fig 2A,B). Chromosomally stable cells rarely undergo chromosome missegregation events (only in approximately 1% of cell divisions [16]), in contrast to CIN cancer cells, in which chromosomes missegregate in excess of every fifth division [16,17]. Putative mechanisms that might contribute to nCIN include weakening of the mitotic checkpoint (Fig 2B; [18,19,20,21]); aberrant sister chromatid cohesion (Fig 2C; [22,23,24,25]); centrosome amplification (Fig 2D; [26]) and improper attachment of chromosomes to the mitotic spindle (Fig 2E; [27,28]). Recent evidence has linked whole-chromosome missegregation in mitosis to DNA damage and structural aberrations in the subsequent interphase [29,30]. These mechanisms are discussed in detail in the Reviews by Holland & Cleveland and Pfau & Amon in this issue of EMBO reports.
Figure 2.
Mechanisms of chromosome missegregation in mitosis. (A) A wild-type division, producing identical daughter cells. (B–E) Mitotic errors that result in aneuploid daughter cells.
CIN genomes are characterized by many forms of structural genomic aberration, including reciprocal and non-reciprocal translocations, amplifications, insertions and deletions [31]. A key structural feature associated with sCIN is the formation of ‘reactive’ chromosomes after chromosome breaks. These ‘reactive’ chromosomes can result in breakage–fusion–bridge cycles (BFB), which propagate extensive genomic rearrangements (see [32] for a review). BFB cycles have been linked to sCIN and associated with intra-tumour heterogeneity [11,33], and can lead to whole-chromosome missegregation [34,35]. Accordingly, there is a need to understand the mechanisms by which BFB cycles can be initiated. Three main explanations have been offered to account for the formation of breaks in chromosomes: telomere dysfunction [34,36,37,38],fragile sites [39,40] and aberrant DNA repair pathways [31,41].
Taken together, the studies of both nCIN and sCIN highlight the diversity and complexity of the mechanisms that contribute to CIN, and hint at the multitude of genetic alterations that could be responsible for these phenotypes, even within one individual tumour [42,43]. Further work will be needed to analyse which mechanisms are the most frequent, which mechanisms occur in vivo and whether distinct mechanisms interact. A significant correlation has been found between nCIN and sCIN in human cancer cell lines derived from a range of tissues [44], and recent studies have revealed that these two forms of CIN might be generated by common mechanisms [29,30,35,45]. A reductionist view of CIN—whereby numerical and structural CIN are considered in isolation—is therefore probably incomplete (Sidebar A).
CIN—diagnostic methods
The CIN status of tumour specimens is not routinely assessed in the clinical setting. Although existing parameters, such as tumour grade, often correlate with CIN [46,47], these are not direct measures of genomic instability. Ultimately, a direct measurement of CIN requires the determination of cell-to-cell variability in chromosome number and structure within a tumour cell population, as well as an assessment of the rate at which these chromosomal changes occur. Broadly speaking, diagnostic methods can be divided according to whether they directly or indirectly measure levels of CIN. Indirect methods rely on proxies for CIN. These can be further sub-classified according to whether they measure intra-tumour heterogeneity and unstable aneuploidy, or karyotypic complexity, on the basis of the average numerical or structural chromosomal changes across the cell population of the tumour. As these methods have been evaluated in retrospective studies, there remains a need for prospective validation of methods to identify CIN in tumours.
Direct approaches for measuring CIN. Due to its dynamic nature, direct methods for determining CIN from fixed tumour tissue are limited to the assessment of the frequency of anaphase segregation errors. This method therefore captures the dynamic nature of CIN and has been used to assess CIN status in diffuse large B-cell lymphoma specimens [48]. However, this approach might be challenging in tumours with lower proliferation indexes and in tumours with morphology unsuited to scoring anaphases.
Intra-tumour heterogeneity measures of CIN. An alternative method for assessing CIN from tumour specimens involves measuring the level of cell-to-cell variability in chromosome number and structure. In this manner, one can quantify both aneuploidy and intra-tumour heterogeneity simultaneously and thereby infer CIN status. Such methods can distinguish between stable and unstable aneuploidy, the latter of which equates to CIN.
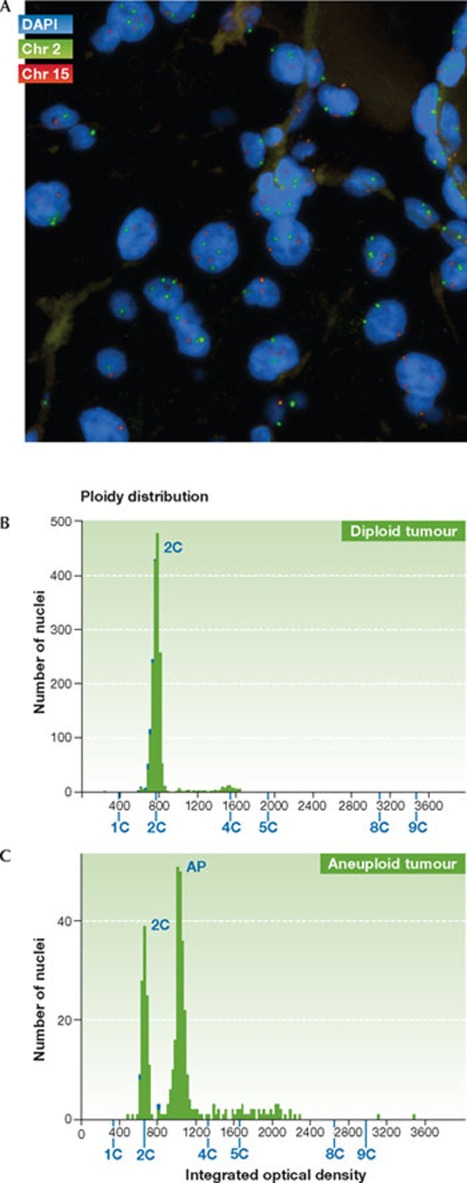
FISH has been adopted as one of the main methods for the assessment of CIN status in tumours (Fig 3A). Variations in chromosome copy number across the cell population can be quantified using fluorescently labelled DNA probes that bind to the centromeres of specific chromosomes [49]. FISH thus allows the assessment of the chromosomal state of hundreds of cells, and the rate of change can be inferred from the cell-to-cell variability in chromosome number [3]. The usefulness of FISH in assessing CIN status has been demonstrated in several retrospective studies, including of breast, lung and oral cancer (see Table 1 and The clinical impact of CIN; [4,50,51,52,53]). However, this technique has limitations. In particular, overlapping nuclei in tumour sections cannot be quantified using FISH, and thin (4 μm) tumour sections will artificially increase variation in chromosome copy number through bisection of tumour nuclei (although this limitation can be accounted for in the analysis and processing of the data). The main drawback of this technique is the fact that most tumours cannot be scored in an automated fashion, which renders FISH extremely labour intensive, limiting its potential use in the clinical setting.
Figure 3.
Direct methods to assess tumour CIN status. (A) Section from a renal cell carcinoma biopsy, hybridized to two fluorescently labelled centromere probes—chromosome 2 (green) and chromosome 15 (red). Variation in centromere copy number is evident between the nuclei. (B) DNA image cytometry profile for a diploid tumour (diploid DNA content determined relative to control diploid cells). (C) DNA image cytometry profile for an aneuploid tumour (determined relative to control diploid cells).
Perhaps the most widely used tools for assessing CIN status are flow and DNA image cytometry (for a review, see [54]). Both methods measure cellular DNA content through the use of dyes that bind stoichiometrically to DNA, allowing DNA cell cycle distribution and ploidy to be determined. CIN status can be inferred from aneuploid DNA content and a tumour cell population with high levels of clonal heterogeneity, as measured by the stemline scatter index (SSI) [9,55]. DNA image cytometry is amenable to relatively high-throughput analysis as compared with FISH, and is relatively inexpensive. The ability of cytometry techniques to identify CIN tumours is supported by the observation that anaphase bridges were only observed in tumours defined as CIN by cytometry in a small cohort of sarcomas, colorectal and pancreatic carcinomas [56]. Nevertheless, given that cytometry provides an overview of cellular DNA content, it has predominantly been used for identification of tumour aneuploidy and polyploidy rather than CIN (Fig 3B,C). Furthermore, its accuracy can be affected by a variety of factors, including section thickness and tissue preservation.
New technologies will probably arise in the future for the determination of CIN status. Single-cell, comparative genomic hybridization (CGH) [56] can yield information on both numerical and structural chromosomal aberrations at a single-cell level, and heterogeneity can then be quantified by comparing multiple cells. CGH, however, cannot readily detect structural rearrangements that do not involve genomic imbalances, such as balanced chromosome inversions or translocations. In addition, single-cell CGH is not amenable to high-throughput analysis and its cost is substantial. A more detailed picture of the genomic landscape can be obtained through next-generation sequencing systems, which adopt massively parallel sequencing techniques. Single-nucleus sequencing—which combines flow sorting with whole-genome amplification and next-generation sequencing—has been recently used to quantify genomic copy number profiles of individual tumour cells at a high resolution (50 kb) [57]. A comparison of the profiles of multiple cells allows inference of the levels of intra-tumour heterogeneity. It seems probable that such single-cell, next-generation sequencing approaches to define tumour CIN status will become more prevalent as costs decrease and technology improves.
Multi-cell, population-based measures of CIN. Karyotypic complexity measures of CIN are generally performed on an aggregated population of cells. Therefore, these techniques provide a static picture of chromosome complexity or of a CIN-associated gene-expression signature, yielding limited direct evidence of cell-to-cell heterogeneity or rates of structural or numerical chromosomal change.
Conventional array CGH makes use of DNA from multiple cells, and can be used to define the structural chromosomal complexity and copy number changes of a tumour sample [58]. More recently, high-density, single-nucleotide polymorphism (SNP) microarrays have been used to define genome-wide copy number changes. This technique provides superior resolution to conventional array-based methods, and has the advantage of being able to detect loss of heterozygosity (LOH) events, including copy-neutral LOH [59]. Quantifying the proportion of the genome that shows copy number aberrations has allowed the definition of genome integrity indexes [60] and CIN scores [46, 61] that have prognostic value. An advantage of these indexes is their ability to capture both numerical and structural chromosome complexity, which is difficult to achieve using the more direct CIN-detection methods described above.
SNP microarray analysis can also be complemented by massively parallel paired-end sequencing of rearrangements, which provides a more complete, higher resolution picture of cancer-specific genome aberrations [62]. By using such an approach, a striking new mechanism for genomic instability—termed chromothripsis—has been recently uncovered [59], whereby a chromosome undergoes tens to hundreds of rearrangements in a single cellular event. Furthermore, the degree of heterogeneity can be estimated by using next-generation sequencing data through the detection of low frequency mutations, which are indicative of sub-clonal architecture [63,64]. However, whether these mutations arise through CIN remains unclear. Given the burgeoning data from next-generation sequencing technologies, it will become increasingly important to establish new methods that use next-generation sequencing datasets to assess the levels of intra-tumour heterogeneity and CIN. A recent study by our group [64] used multi-region, next-generation sequencing to sequence multiple spatially separated regions from individual clear-cell renal tumours. This uncovered branched evolutionary tumour growth and striking mutational intra-tumour heterogeneity, coupled with tumour aneuploidy and karyotypic complexity that was heterogeneous between distinct biopsies.
An appreciation of the inherent heterogeneity in cancer has led to the development of gene-expression-based approaches that might reflect CIN status. A summary of the total level of chromosomal aberrations in a given tumour at the transcriptional level, total functional aneuploidy (tFA), has been used to measure chromosomal imbalance [5]. By correlating this proxy of aneuploidy with expression measures, a set of 70 CIN genes that consistently correlated with tFA across several cancer types was identified. The value of the CIN70 signature as a proxy for CIN has since been further validated, as it correlates with both structural complexity (defined from SNP microarrays) and numerical CIN (defined by DNA image cytometry) [61]. Interestingly, among the 70 genes in the list were several key regulators that facilitate the maintenance of faithful replication and segregation of chromosomes, including AURKB, AURKA, NEK2, H2AFX, CDC20, ZWINT, CCNB1 and CCNB2. The prognostic ability of the CIN70 signature is probably not only based on its detection of proliferation rate; when genes defined as cell-cycle-regulated were not included in the signature, it still had prognostic ability.
A 12-gene expression signature associated with genomic instability in breast cancer has been recently defined; the signature was derived by identifying a set of 12 genes whose expression can discriminate between stable and unstable aneuploidy (defined by DNA image cytometry) in a cohort of 48 breast cancer specimens [9,55]. The biological and prognostic value of this gene expression signature has been demonstrated across a range of cancer types [9,66]. Interestingly, MammaPrint® and Oncotype DX®—two clinically used breast cancer prognostic gene expression signatures—reflect genomic instability levels as measured by the 12-gene signature [9]. There is no overlap between the genes in the CIN70 signature and the 12-gene signature. However, the expression of the signatures are highly correlated [47], and AURKA—a gene identified in the CIN70 signature and previously linked to aneuploidy [67]—has significantly increased expression in the genomically unstable tumours as defined by the 12-gene genomic instability signature [9].
Conceivably, high-throughput qPCR-based approaches could be derived from such CIN expression signatures. Such approaches might be applicable to the analysis of paraffin-embedded material, to rapidly estimate CIN status of clinical samples. Although the dynamic nature of CIN cannot be fully captured by multi-cell, population-based approaches, these techniques can facilitate studies exploring the genetic and molecular changes underlying CIN. Furthermore, given the correlation of gene expression signatures and copy-number-based scores with direct measures of CIN [61], these could be used to facilitate the assessment of the clinical consequences of CIN in a higher throughput and less labour-intensive manner than FISH-based approaches.
Consequences of CIN
CIN has profound effects on the genome of a cell. Structural rearrangements can result in distinct molecular genetic alterations, such as the formation of fusion gene products, the amplification of some genes and the deletion of others [31]. The repercussions of numerical chromosome changes include alterations to gene dosage and LOH events [68]. Tumour chromosomal instability would be therefore expected to be directly associated with patient prognosis.
The clinical impact of CIN. The relationship between CIN and cancer prognosis has been explored across a range of cancer types, using various methods. In several retrospective studies, summarized in Table 1, CIN has been consistently associated with poor prognosis, providing additional prognostic information beyond conventional clinical parameters such as tumour grade.
In non-small-cell lung cancer (NSCLC), CIN is related to unfavourable prognosis [4,5,51,52]. In three separate studies, all using FISH to measure CIN, the association between CIN status and overall survival was shown to be significant, independent of conventional risk factors such as tumour stage, age and sex in multivariate analyses [4,51,52]. The association between CIN and poor prognosis in lung cancer was also found to hold when using gene expression signatures to assess CIN status [5,66].
In a meta-analysis of 10,126 colon cancer patients, tumour aneuploidy was used as a surrogate measure for CIN and found to be associated with a worse prognosis, either in terms of overall survival or, if not available, progression-free survival [6]. Aneuploid status—quantified by either flow cytometry (9,526 patients) or image cytometry (600 patients)—could stratify colon cancer patients independently of standard pathological staging, and irrespective of ethnic background, anatomical location and treatment with 5-fluorouracil (5-FU)-based adjuvant chemotherapy. By using a 12-gene genomic instability signature, CIN predicted colon cancer recurrence independently of tumour stage [66].
In breast cancer, CIN has been quantified by using FISH and DNA image cytometry techniques, as well as summary gene expression and SNP-array methods. By using the SSI as a means to assess CIN (see Intra-tumour heterogeneity measures of CIN), CIN was linked to a worse prognosis, in terms of cancer-specific survival [55]. CIN status provided additional information to the ploidy status of a tumour, emphasizing the need to distinguish between aneuploidy or polyploidy and CIN when considering CIN assessment in clinical samples. FISH-based measures have found CIN to be associated with lymph node metastasis and prevalence of oestrogen receptor (ER) negativity [50,69]. SNP-based measures of chromosomal aberrations are associated with more aggressive subgroups of breast cancer—such as ER-negative and triple-negative [46]. Interestingly, CIN scores are prognostic in certain subgroups (ER-positive, luminal B and HER2), but not others (ER-negative), suggesting a complex relationship between CIN and outcome in breast cancer.
Furthermore, studies in ovarian, endometrial, synovial and oral cancers, as well as diffuse B-cell lymphoma, have suggested that CIN is associated with poor prognosis [48,53,66,70,71].
Overall, these studies associate CIN and poor prognosis across a range of cancer types, and highlight the relationship between CIN and multiple clinical parameters, including the probability of disease recurrence. However, it is important to emphasize that the studies have used different techniques for the measurement of CIN; in particular, some studies have used surrogates for CIN, such as aneuploidy status, whereas others have measured CIN more directly. Indirect measures of CIN increase the probability of confounding variables and could mask more subtle relationships between CIN and prognosis. Indeed, given that CIN probably has negative effects on cellular fitness, its impact on prognosis is probably not straightforward, as will be discussed further below (see Exploiting CIN in the clinical setting).
A darwinian perspective on the clinical impact of CIN. The mechanisms by which CIN contributes to cancer outcome can be considered from a darwinian point of view. Selection depends on cell-to-cell phenotypic variation. CIN is characterized by gain and loss of chromosomes or parts of chromosomes, which gives rise to genetic variation, and thus generates distinct cellular phenotypes. This intra-tumour heterogeneity could facilitate darwinian selection, thereby providing a means for the tumour to adapt to environmental and stromal pressures [7,72]. Consistent with this hypothesis, findings in mouse models suggest that CIN might promote early tumour relapse [73] and studies in yeast have found aneuploidy can generate important phenotypic variation that might lead to fitness gains under stressful conditions [74]. Furthermore, evolutionary models posit that CIN might be selected for by increasing the rate at which tumour suppressor genes are lost from the genome [75].
Given the heterogeneity that arises as a consequence of CIN, and the propensity for selection and adaptation, it seems probable that the relationship of elevated levels of CIN and poor prognosis could ultimately be explained by the emergence of drug resistance [8,76,77,78] and an increased capacity to metastasize to distant sites [50,79]. Consistent with this explanation, CIN cancer cell lines acquire multi-drug resistance at an elevated rate compared with chromosomally stable, diploid cells [77].
Furthermore, CIN has also been shown to be associated with intrinsic taxane resistance in ovarian cancer patients and cancer cell lines [76]. More recently, we have found that CIN is associated with intrinsic multi-drug resistance in colorectal cancer: CIN cells are intrinsically multi-drug resistant compared with diploid cell lines, independent of somatic mutation status—according to the mutation status of 20 recurrently mutated genes assessed in colorectal cancer—and proliferation rate [8]. A meta-analysis of tumour drug response in colorectal cancer also supported a relationship between tumour CIN status and drug resistance [8].
From a darwinian perspective, a drug treatment might be viewed as a selective pressure. Increased heterogeneity resulting from CIN could increase the probability of resistant sub-clones arising in the tumour before drug therapy. The emergence of drug-resistant disease in this case might be a direct consequence of tumour heterogeneity resulting from CIN. An alternative, but not mutually exclusive explanation based on the ability of CIN tumours to survive the impact of repeated extensive genome remodelling, is the existence of a CIN survival phenotype [43]. This survival phenotype is postulated to reflect an adaptation to CIN, which could also facilitate intrinsic drug resistance. According to this hypothesis, intrinsic drug resistance could be a feature of cells with elevated levels of CIN, as opposed to being purely a consequence of the intra-tumour heterogeneity promoted by CIN.
Taken together, these studies suggest CIN could have a central role in determining patient outcome: increased levels of CIN might facilitate darwinian adaptation and selection, thus having a negative impact on patient outcome. This association highlights the clinical relevance of CIN in cancers and suggests CIN status could be exploited in the clinical setting.
Exploiting CIN in the clinical setting
Given the prevalence and prognostic impact of CIN in human cancers, there is a clinical need to exploit tumour CIN status. CIN status could be used to assist prognostic predictions and could be a therapeutic target.
CIN as a prognostic tool. The development of robust clinical tools to assess CIN status could assist in risk stratification. However, as has been discussed, there are multiple possible tests to measure CIN, each with its own advantages and disadvantages. Thus, a central goal will be to determine which test most accurately determines CIN status of tumour specimens in prospective studies, whilst balancing practical and economic considerations. Different tests might perform better in different cancer types. Furthermore, given the potential variability in the degree of CIN, a clinically relevant test must include defined thresholds that allow the oncologist to distinguish between patients who have good or poor prognoses. Defining thresholds to identify CIN tumours will be challenging, particularly as the relationship between CIN and prognosis is probably complex.
Crucially, a threshold for CIN must balance the clinically important concept that it might have negative effects on tumour fitness with its potential to enhance tumour growth. Experimental studies in yeast and murine systems have demonstrated that an abnormal chromosomal number can be deleterious to normal cells, negatively affecting proliferation [80,81]. It remains unclear, however, whether the growth of aneuploid tumour cells is hindered in a similar manner, or whether this has been overcome during tumorigenesis. A recent study suggested that chromosome gains and losses might serve to redress imbalances in the stoichiometry of protein complexes, which can result from aneuploidy [82], raising the possibility that CIN might enable cells to overcome the negative effects of aneuploidy. However, in addition to the potential negative effects of having an abnormal chromosome complement, CIN cells probably accumulate deleterious genomic events fostered by continuing gross numerical or structural chromosomal changes, occurring from one cell division to the next [83]. It has thus been suggested that there is an optimal level of CIN in tumours required for tumour adaptation and progression, beyond which CIN becomes unfavourable [7,84]. Such a scenario, whereby extreme CIN is deleterious for a tumour, might be comparable to ‘mutational meltdown’ in bacteria [85], or error catastrophe in viruses [86].
Evidence substantiating the idea that excessive levels of CIN might have adverse consequences for a tumour derives from mouse models and studies on human cancer cell lines. Mice with reduced levels of CENP-E have CIN and develop tumours [13]. However, intriguingly, CENP-E-induced CIN was found to have tumour-suppressive abilities in some contexts—in tissues that had a pre-existing level of aneuploidy, which was increased through depletion of CENP-E. This suggests that excessive CIN might be disadvantageous for a tumour. Consistent with this observation, excessive CIN, introduced by inactivation of spindle assembly checkpoint components, leads to excessive aneuploidy and cell death in human cancer cells [87], and multipolar cell divisions generate highly aneuploid, non-viable cells [26].
Exploring aneuploidy: the significance of chromosomal imbalance.
This review series—published in this issue of EMBO reports—also includes:
A balancing act: focus on aneuploidy Nonia Pariente
Losing balance: the origin and impact of aneuploidy in cancer Andrew J Holland and Don W Cleveland
Chromosomal instability and aneuploidy in cancer: from yeast to man Sarah J Pfau and Angelika Amon
Age-related aneuploidy through cohesion exhaustion Rolf Jessberger
Further supporting this hypothesis, a recent retrospective study has suggested a nonlinear relationship between CIN and cancer outcome [61]. Specifically, in a cohort of 265 patients with ER-negative breast cancers, extreme CIN—defined as tumours in the upper quartile of CIN70 [5] expression—was associated with a significantly better prognosis, in terms of recurrence-free or distant metastasis-free survival, compared with patients having tumours in the third CIN70 expression quartile. The association between extreme CIN and improved prognosis was also observed in ovarian, gastric and non-small-cell lung cancers, but not in ER-positive breast cancers. These findings have been further validated in a study of ER-negative breast cancers, in which CIN was assessed by dual centromeric FISH analysis [88]. This paradoxical observation indicates that the relationship between CIN and prognosis is probably complex. Hence, defining thresholds of CIN that are associated with poor or, conversely, improved prognosis, might be challenging. Furthermore, it suggests that the relationship of CIN with prognosis might vary between tumour types and subgroups. It is interesting to note that measuring aneuploidy alone would probably not have detected this relationship, emphasizing that CIN status can provide further prognostic information. Nevertheless, despite this intriguing association, these data are derived from small retrospective analyses, and validation in larger cohorts using pre-defined CIN thresholds is required before such observations are suitable for clinical implementation.
CIN as a therapeutic target. Drug resistance and poor prognosis associated with CIN tumours renders their treatment a considerable challenge. However, the frequency and clinical significance of CIN and its restriction to neoplastic tissue suggests the CIN phenotype could be an attractive therapeutic target [89]. In this regard, the direct targeting of CIN by anti-cancer therapeutics is a subject of active research [90,91,92].
Given the plethora of mechanisms that have been attributed to CIN, several possible strategies have been suggested. For example, supernumerary centrosomes observed in CIN cells could be targetable by a drug that impedes centrosome clustering and enforces lethal multipolar cell divisions [93]. Observations that extreme CIN might be detrimental for cancer growth suggest potential therapeutic approaches. To this end, tumour cells can be selectively killed by the anti-mitotic drug paclitaxel if levels of the essential mitotic regulators MPS1 and BUB1B are first reduced, inducing excessive chromosome missegregation [94].
In addition, studies in cancer cell lines and yeast have indicated that there could be targetable cellular adaptations to the aneuploid and CIN state [16,80,95,96,97]. The NCI-60 drug discovery panel of human cancer cell lines was analysed to uncover potential anticancer agents that specifically target CIN cancer cells [91,98]. Seven groups of compounds that could preferentially target unstable cell lines through growth inhibition were uncovered by using this approach, and further studies are warranted to examine the activity of these compounds. A recent study identified three energy and proteotoxic stress-inducing compounds—AICAR, 17-AAG and chloroquine—that showed selectivity against aneuploid primary mouse embryonic fibroblasts carrying Robertsonian fusion chromosomes [99]. Of these, AICAR (AMPK activator) and 17-AAG (HSP90 inhibitor) could also target human aneuploid cancer cells. Interestingly, it has been shown recently that inhibition of HSP90 in yeast induces CIN and aneuploidy [100]. It is tempting to speculate, on the basis of these observations, that HSP90 inhibition might induce excessive CIN, resulting in cell lethality in cells with a pre-existing level of aneuploidy or CIN, similar to previous findings [94]. These results emphasize that further work is warranted to assess the efficacy of these drugs against aneuploid cells, and to understand the precise relationship between these compounds and protein overload [101]. However, it is unclear whether targeting aneuploidy will be sufficient to target CIN cells, as these cells must tolerate ongoing structural and numerical chromosone aberrations. Efforts to identify mechanisms of CIN tolerance need to consider both ongoing structural and numerical chromosome aberrations observed in CIN genomes.
Conclusion
A significant obstacle to the exploitation of CIN in the clinical setting results from our limited understanding of the mechanisms by which this form of genomic instability is driven. The molecular mechanisms that contribute to structural and numerical aberrations in CIN tumours have proved elusive. This problem has been compounded by a lack of understanding as to whether structural and numerical changes are initiated by unifying mechanisms. Further research is needed to elucidate crucial drivers and suppressors of CIN, and to explore whether subtypes of CIN with distinct patterns of chromosomal aberrations exist across cancer types.
The relationship between CIN and prognosis is evidently complex; CIN has been associated with adverse patient survival and linked with both intrinsic and acquired drug resistance. By contrast, emerging evidence suggests that extreme CIN might be associated with improved patient outcome. Such a paradoxical relationship between CIN status and clinical outcome creates a challenge both for the targeting of this pattern of genome instability, and in defining CIN thresholds to predict prognosis. A key impediment to the implementation of CIN assessment in the clinical setting remains the technical difficulty associated with its measurement. Improvements in clinicopathological methods to identify CIN tumours are essential, alongside efforts to target this pattern of genomic instability. CIN-specific drug discovery approaches developed in parallel with CIN-stratification criteria will be crucial to improve patient survival outcome in this high-risk disease cohort.

Nicholas McGranahan

Rebecca A Burrell

David Endesfelder

Marco R Novelli

Charles Swanton
Acknowledgments
C.S. is funded by Cancer Research UK and the Medical Research Council and is an MRC Senior Research Fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Negrini S, Gorgoulis VG, Halazonetis TD (2010) Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR (2008) Defining ‘chromosomal instability’. Trends Genet 24: 64–69 [DOI] [PubMed] [Google Scholar]
- Choi CM, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, Lee DS, Lee SD (2009) Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer 64: 66–70 [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Walther A, Houlston R, Tomlinson I (2008) Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut 57: 941–950 [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Swanton C (2010) How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer 103: 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ et al. (2011) Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 71: 1858–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann JK et al. (2009) The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer 124: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D (2012) Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13: 189–203 [DOI] [PubMed] [Google Scholar]
- Gollin SM (2005) Mechanisms leading to chromosomal instability. Semin Cancer Biol 15: 33–42 [DOI] [PubMed] [Google Scholar]
- Beroukhim R et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36 [DOI] [PubMed] [Google Scholar]
- Paulsson K, Johansson B (2009) High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 48: 637–660 [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Knudson AG (2000) Mechanism and relevance of ploidy in neuroblastoma. Genes Chromosomes Cancer 29: 89–95 [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA (2008) Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1997) Genetic instability in colorectal cancers. Nature 386: 623–627 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S et al. (2004) Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36: 1159–1161 [DOI] [PubMed] [Google Scholar]
- Grabsch H, Takeno S, Parsons WJ, Pomjanski N, Boecking A, Gabbert HE, Mueller W (2003) Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer—association with tumour cell proliferation. J Pathol 200: 16–22 [DOI] [PubMed] [Google Scholar]
- Wang XH, Jin DY, Ng RWM, Feng HC, Wong YC, Cheung ALM, Tsao SW (2002) Significance of MAD2 expression to mitotic checkpoint control in ovarian cancer cells. Cancer Res 62: 1662–1668 [PubMed] [Google Scholar]
- Barber TD et al. (2008) Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA 105: 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang NG et al. (2008) Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA 105: 13033–13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R, Fofanov V, Panigrahi A, Merchant F, Zhang NG, Pati D (2009) Overexpression and mislocalization of the chromosomal segregation protein separase in multiple human cancers. Clin Cancer Res 15: 2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaizumi M et al. (2009) Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut 58: 249–260 [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang LJ, Tolic-Norrelykke IM, Cimini D (2011) Merotelic kinetochore attachment: causes and effects. Trends Cell Biol 21: 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC (2009) Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 21: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH (2011) Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333: 1895–1898 [DOI] [PubMed] [Google Scholar]
- Crasta K et al. (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayani J, Selvarajah S, Maire G, Vukovic B, Al-Romaih K, Zielenska M, Squire JA (2007) Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin Cancer Biol 17: 5–18 [DOI] [PubMed] [Google Scholar]
- Jones RN (2005) McClintock's controlling elements: the full story. Cytogenet Genome Res 109: 90–103 [DOI] [PubMed] [Google Scholar]
- Gisselsson D et al. (2000) Chromosomal breakage–fusion–bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA 97: 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewenius Y, Gorunova L, Jonson T, Larsson N, Hoglund M, Mandahl N, Mertens F, Mitelman F, Gisselsson D (2005) Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci USA 102: 5541–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J, Soler D, Genescà A, Tusell L (2010) Whole chromosome loss is promoted by telomere dysfunction in primary cells. Genes Chromosomes Cancer 49: 368–378 [DOI] [PubMed] [Google Scholar]
- Feldser DM, Hackett JA, Greider CW (2003) Telomere dysfunction and the initiation of genome instability. Nat Rev Cancer 3: 623–627 [DOI] [PubMed] [Google Scholar]
- Perera SA et al. (2008) Telomere dysfunction promotes genome instability and metastatic potential in a K-ras p53 mouse model of lung cancer. Carcinogenesis 29: 747–753 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Murnane JP (2006) Telomeres, chromosome instability and cancer. Nucleic Acids Res 34: 2408–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B (2011) Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell 43: 122–131 [DOI] [PubMed] [Google Scholar]
- Dereli-Öz A, Versini G, Halazonetis TD (2011) Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol 5: 308–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Alt FW (2003) The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev 194: 77–95 [DOI] [PubMed] [Google Scholar]
- Boland CR, Komarova NL, Goel A (2009) Chromosomal instability and cancer: not just one CINgle mechanism. Gut 58: 163–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland SE, Burrell RA, Swanton C (2009) Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle 8: 3262–3266 [DOI] [PubMed] [Google Scholar]
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR (2003) Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res 63: 8634–8647 [PubMed] [Google Scholar]
- Martinez C, van Wely KHM (2010) Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle 9: 2275–2280 [DOI] [PubMed] [Google Scholar]
- Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y, Foekens JA, Martens JWM (2011) Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat 128: 23–30 [DOI] [PubMed] [Google Scholar]
- Endesfelder D, McGranahan N, Birkbak NJ, Szallasi Z, Kschischo M, Graham TA, Swanton C (2011) A breast cancer meta-analysis of two expression measures of chromosomal instability reveals a relationship with younger age at diagnosis and high risk histopathological variables. Oncotarget 2: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA (2011) Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res 17: 7704–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher MR, Carter NP (2005) The new cytogenetics: Blurring the boundaries with molecular biology. Nat Rev Genet 6: 782–792 [DOI] [PubMed] [Google Scholar]
- Takami S, Kawasome C, Kinoshita M, Koyama H, Noguchi S (2001) Chromosomal instability detected by fluorescence in situ hybridization in Japanese breast cancer patients. Clin Chim Acta 308: 127–131 [DOI] [PubMed] [Google Scholar]
- Yoo JW, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, Lee DS, Lee SD, Choi CM (2010) The relationship between the presence of chromosomal instability and prognosis of squamous cell carcinoma of the lung: fluorescence in situ hybridization analysis of paraffin-embedded tissue from 47 Korean patients. J Korean Med Sci 25: 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Saji H, Idiris A, Kawasaki N, Hosaka M, Ogata A, Saijo T, Kato H (2003) Chromosomal instability detected by fluorescence in situ hybridization in surgical specimens of non-small cell lung cancer is associated with poor survival. Clin Cancer Res 9: 2294–2299 [PubMed] [Google Scholar]
- Sato H, Uzawa N, Takahashi KI, Myo K, Ohyama Y, Amagasa T (2010) Prognostic utility of chromosomal instability detected by fluorescence in situ hybridization in fine-needle aspirates from oral squamous cell carcinomas. BMC Cancer 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Halicka HD, Zhao H (2010) Analysis of cellular DNA content by flow and laser scanning cytometry. Adv Exp Med Biol 676: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenwett U et al. (2004) Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res 64: 904–909 [DOI] [PubMed] [Google Scholar]
- Fiegler H, Geigl JB, Langer S, Rigler D, Porter K, Unger K, Carter NP, Speicher MR (2007) High resolution array-CGH analysis of single cells. Nucleic Acids Res 35: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Albertson DG (2005) Array comparative genomic hybridization and its applications in cancer. Nat Genet 37: S11–S17 [DOI] [PubMed] [Google Scholar]
- Stephens PJ et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin SF et al. (2007) High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol 8: R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ et al. (2011) Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 71: 3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ et al. (2009) Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC et al. (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38: 468–473 [DOI] [PubMed] [Google Scholar]
- Gerlinger M et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML et al. (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettu RK, Wan YW, Habermann JK, Ried T, Guo NL (2010) A 12-gene genomic instability signature predicts clinical outcomes in multiple cancer types. Int J Biol Markers 25: 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R (2010) Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 10: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM (2009) Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R et al. (2011) Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev 20: 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama-Hosokawa S et al. (2010) Genome-wide single-nucleotide polymorphism arrays in endometrial carcinomas associate extensive chromosomal instability with poor prognosis and unveil frequent chromosomal imbalances involved in the PI3-kinase pathway. Oncogene 29: 1897–1908 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Numoto K, Yoshida A, Kunisada T, Ohata H, Takeda K, Wai D, Poremba C, Ozaki T (2006) Chromosomal and genetic imbalances in synovial sarcoma detected by conventional and microarray comparative genomic hybridization. J Cancer Res Clin Oncol 132: 444–450 [DOI] [PubMed] [Google Scholar]
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C (1999) Genetic instability and darwinian selection in tumours. Trends Biochem Sci 2 4: M57–M60 [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R (2010) Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N (2004) Does cancer solve an optimization problem? Cell Cycle 3: 840–844 [PubMed] [Google Scholar]
- Swanton C et al. (2009) Chromosomal instability determines taxane response. Proc Natl Acad Sci USA 106: 8671–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P, Stindl R, Hehlmann R (2000) Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy. Proc Natl Acad Sci USA 97: 14295–14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Hehlman R, Sachs R, Duesberg P (2005) Chromosomal alterations cause the high rates and wide ranges of drug resistance in cancer cells. Cancer Genet Cytogenet 163: 44–56 [DOI] [PubMed] [Google Scholar]
- Jonkers YMH et al. (2005) Chromosomal instability predicts metastatic disease in patients with insulinomas. Endocr Relat Cancer 12: 435–447 [DOI] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924 [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozery-Flato M, Linhart C, Trakhtenbrot L, Izraeli S, Shamir R (2011) Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol 12: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA (2001) A mutator phenotype in cancer. Cancer Res 61: 3230–3239 [PubMed] [Google Scholar]
- Komarova NL, Wodarz D (2004) The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc Natl Acad Sci USA 101: 7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Bürger R, Butcher D, Gabriel W (1993) The mutational meltdown in asexual populations. J Hered 84: 339–344 [DOI] [PubMed] [Google Scholar]
- Eigen M (2002) Error catastrophe and antiviral strategy. Proc Natl Acad Sci USA 99: 13374–13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G, Foltz DR, Cleveland DW (2004) Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA 101: 8699–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R et al. (2011) Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev 20: 2183–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, Burrell RA, Futreal PA (2011) Breast cancer genome heterogeneity: a challenge to personalised medicine? Breast Cancer Res 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa LT, Lengauer H (2002) Exploring and exploiting instability. Cancer Biol Ther 1: 212–225 [DOI] [PubMed] [Google Scholar]
- Roschke AV, Kirsch IR (2005) Targeting cancer cells by exploiting karyotypic complexity and chromosomal instability. Cell Cycle 4: 679–682 [DOI] [PubMed] [Google Scholar]
- Burrell RA, Juul N, Johnston SR, Reis JS, Szallasi Z, Swanton C (2010) Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem 111: 782–790 [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D (2008) Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22: 2189–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Kops G, Medema RH (2009) Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA 106: 19108–19113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A (2010) Identification of aneuploidy-tolerating mutations. Cell 143: 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D (2006) Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547 [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA (2010) Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Kirsch IR (2010) Targeting karyotypic complexity and chromosomal instability of cancer cells. Current Drug Targets 11: 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A (2011) Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R (2012) Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Malumbres M (2011) Targeting aneuploidy for cancer therapy. Cell 144: 465–466 [DOI] [PubMed] [Google Scholar]
- Heilig CE, Loffler H, Mahlknecht U, Janssen JWG, Ho AD, Jauch A, Kramer A (2010) Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med 14: 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergshoeff VE, Hopman AHN, Zwijnenberg IR, Ramaekers FCS, Bot FJ, Kremer B, Manni JJ, Speel EJM (2008) Chromosome instability in resection margins predicts recurrence of oral squamous cell carcinoma. J Pathol 215: 347–348 [DOI] [PubMed] [Google Scholar]