Abstract
Aneuploidy is frequently associated with disease and developmental abnormalities. It is also a key characteristic of cancer. Several model systems have been developed to study the role of chromosomal instability and aneuploidy in tumorigenesis. The results are surprisingly complex, with the conditions sometimes promoting and sometimes inhibiting tumour formation. Here, we review the effects of aneuploidy and chromosomal instability in cells and model systems of cancer, propose a model that could explain these complex findings and discuss how the aneuploid condition could be exploited in cancer therapy.
Keywords: aneuploidy, cancer, tumorigenesis, Down syndrome, genomic instability
See the Glossary for abbreviations used in this article.
Glossary.
- 17-AAG
17-N-Allylamino-17-demethoxygeldanamycin
- AICAR
5-Aminoimidazole-4-carboxamide ribotide
- APC/C
anaphase promoting complex/cyclosome
- ApcMin
adenomatous polyposis coli multiple intestinal neoplasia mouse model
- ATM
ataxia telangiectasia mutated
- BCR–ABL
Breakpoint Cluster Region–c-abl oncogene 1; the Philadelphia chromosome fusion
- Bub1
budding uninhibited by benzimidazoles 1 homologue (yeast)
- Bub1B/BubR1
budding uninhibited by benzimidazoles 1 homologue beta (yeast)
- Bub3
budding uninhibited by benzimidazoles 3 homologue (yeast)
- Cdc20
cell division cycle 20 homologue (S. cerevisiae)
- Cep57
centrosomal protein 57kDa
- CENP-E
centromere protein E
- Chk1/2
checkpoint kinase 1/2
- Dscr1
Down syndrome critical region 1
- ERBB2
v-erb-b2 erythroblastic leukaemia viral oncogene homologue 2, neuro/glioblastoma derived oncogene homologue (avian)
- EGFR
Epidermal growth factor receptor
- ETS2
v-ets erythroblastosis virus E26 oncogene homologue 2 (avian)
- H2A.X
H2A histone family, member X
- Hec1
NDC80 kinetochore complex component homologue (S. cerevisiae)
- Hsp90
Heat shock protein 90
- Kif11
kinesin family member 11
- KrasG12D
Kirsten rat sarcoma oncogene 2, expressed targeted mutation
- MAD2
mitotic arrest deficient-like 1 (yeast)
- p53
TP53; tumour protein 53
- Rae1
RNA export 1 homologue (S. pombe)
- RB1
retinoblastoma 1
- ROS
reactive oxygen species
- SA1/STAG1
stromal antigen 1
- SA2/STAG2
stromal antigen 2
- shRNA
short hairpin RNA
- Ts65Dn
Ts(1716)65Dn mouse model of Down syndrome
- UbcH10
ubiquitin-conjugating enzyme E2C
- UBP6
ubiquitin-specific protease 6
- VEGF
vascular endothelial growth factor
Introduction
Cancer cells contain a multitude of genetic lesions that endow them with increased proliferative potential and the means to evade elimination by apoptosis. Most transformed cells harbour mutations in multiple growth- and proliferation-promoting pathways, and the spectrum of genetic lesions varies significantly between individual cancers [1,2]. The genetic heterogeneity of cancer cells—both within tumours and between those derived from various individuals—makes targeting individual components of these pathways challenging [3]. Nevertheless, cancer cells also share features. Notably, the vast majority of solid tumours are aneuploid; they have an inappropriate number of chromosomes [4]. The definitive effects of aneuploidy on cell physiology are only beginning to be understood, and therefore its role in tumorigenesis and cancer remains unclear.
Here, we discuss recent data and propose hypotheses we deem particularly important in informing future studies of the role of aneuploidy in cellular physiology, tumorigenesis and cancer. First, we summarize recent advances in our understanding of the effects of aneuploidy on cellular and organismal physiology. We then discuss studies of mouse models of aneuploidy and chromosomal instability (CIN), which have revealed that the effects of aneuploidy and CIN on tumorigenesis are complex—sometimes promoting it and other times antagonizing disease initiation and progression. Finally, we propose a model that could explain these complex effects and discuss ways in which aneuploidy could be exploited for the development of new cancer therapeutics.
Aneuploidy, precisely defined
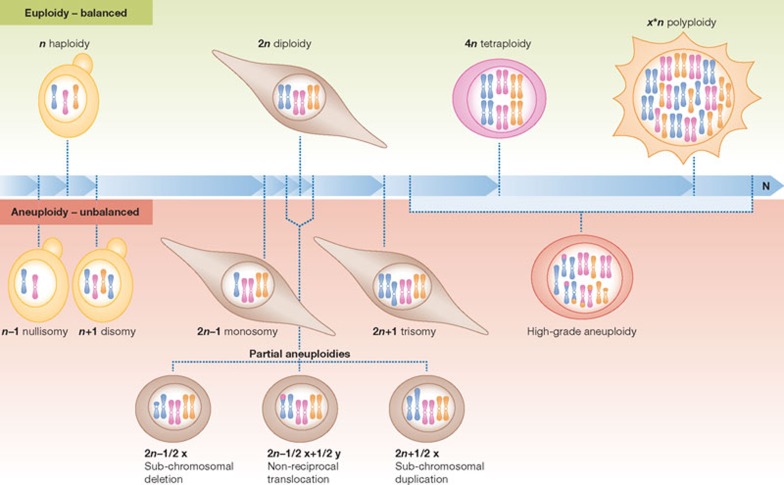
In the context of a discussion on aneuploidy and its role in tumorigenesis, it is especially important to distinguish between ‘aneuploidy’ and ‘polyploidy’. These terms describe two different cellular states that have distinct effects on cells and organisms. Aneuploidy—derived from the Greek an meaning ‘not’, eu meaning ‘good’, and ploos meaning ‘fold’—is a state in which a cell does not contain an exact multiple of the haploid chromosomal complement—literally ‘not the right fold’ (Fig 1). Therefore, aneuploidy refers to an unbalanced genomic state. By contrast, polyploidy refers to a state in which a cell contains a whole number multiple of the entire genome—literally ‘many fold’. Thus the genomic state is balanced in polyploid cells. Just as ‘polyploidy’ can describe cells with a range of ploidies, from diploid to tetraploid to octoploid and beyond, ‘aneuploidy’ is a general term that can describe a wide range of unbalanced karyotypes (Fig 1). We describe this spectrum of aneuploid karyotypes with two general terms, ‘high grade’ and ‘low grade’. High-grade aneuploidy describes the deviation of many chromosomes from the euploid chromosome number, whereas low-grade aneuploidy refers to the deviation of a few chromosomes from the euploid complement.
Figure 1.
Aneuploidy defined. Euploidy defines a species-specific karyotype. Depending on the species, euploidy can describe a haploid, diploid or polyploid karyotype. Euploidy refers to a balanced genomic state. By contrast, aneuploidy is an unbalanced genomic state that describes a range of karyotypes. Whole chromosomes can be lost (nullisomy or monosomy) or gained (disomy or trisomy). Additionally, sub-chromosomal regions can be amplified, deleted or translocated (partial aneuploidies). ‘High-grade aneuploidy’ occurs when complex aneuploidies are present, often a combination of chromosome losses and gains, as well as sub-chromosomal rearrangements.
In the strictest sense of the word, only changes in chromosome number that are not multiples of the haploid complement should be defined as aneuploidies. However, the term is also commonly used to describe genomic alterations that result in unbalanced copy numbers of sub-chromosomal regions. These copy number variations, non-reciprocal translocations and duplications or deletions of portions of chromosomes are termed ‘microaneuploidies’, ‘partial aneuploidies’ or ‘segmental aneuploidies’, to distinguish them from whole-chromosomal aneuploidies.
An accurate use of terms is particularly important when describing cancer cells. We suggest using the terms as they are defined and refraining from calling cancer cells ‘tetraploid’ or ‘polyploid’. Tetraploidization can precede the acquisition of aneuploidy in the development of some types of cancer [5], and many cancer cell lines have increased base-ploidy, meaning they contain a number of chromosomes that approaches a multiple of the haploid complement. However, cancer cells rarely—if ever—contain exact multiples of the haploid genome. Rather, cancer cells seem to invariably harbour complex aneuploid karyotypes—such as four copies of one chromosome, two of another, one of a third chromosome and so on [6,7,8]. Because the terms ‘tetraploid’ or ‘polyploid’ imply a sense of balance, we suggest describing cancer cells as ‘complex aneuploid cells’ or as ‘harbouring high-grade aneuploidy’ to both capture the complexity of their genomes and emphasize their unbalanced genomic complements, without stating their precise chromosomal composition (Fig 1).
Effects of aneuploidy on cell physiology
Two types of model are being used to analyse the effects of aneuploidy on cell physiology. Some studies analyse cells that contain defined chromosomal aneuploidies created through single-chromosome transfers or spontaneous meiotic non-disjunction. We refer to these systems as ‘chronic defined aneuploidies’ because the identity of the aneuploid chromosome is known and it is present from the genesis of the cell or organism. Other studies use cells that have CIN, that is, a high rate of chromosome missegregation due to mutations in genes required to ensure accurate chromosome segregation ([9]; see also reviews by Holland & Cleveland, and Swanton & colleagues in this issue of EMBO reports). We refer to aneuploid cells derived from CIN as ‘acute random aneuploidies’ because they are generated spontaneously as the cell divides, and the identity of the missegregated chromosome(s) varies with each non-disjunction event. In cells with CIN, it can be difficult to separate the effects of aneuploidy from other CIN-associated phenotypes, such as structural chromosomal aberrations, or from potential functions of mutated genes that induce chromosome missegregation. In addition, populations of cells with CIN continuously spawn new heterogeneous karyotypes, allowing for increased adaptive potential to selective pressures [10]. Nevertheless, general phenotypes emerge in many CIN models despite this complexity, providing insight into the cellular response to the induction of aneuploidy.
Studies of the effects of aneuploidy on cells and organisms have analysed whether gene expression is correlated with gene copy number in aneuploid cells or whether mechanisms exist that compensate for the gain or loss of chromosomes. In yeast and mammals, gene expression seems to correlate with gene copy number, at least in the case of chromosome gains. An increase in genomic material is accompanied by a corresponding increase in the transcription of those genes in excess, as observed in yeast cells with an extra chromosome, in trisomic mouse cells, in human cells with trisomy 21 [11], in yeast cells with complex aneuploid karyotypes [12] and in aneuploid human cell lines created by microcell-mediated chromosome transfer [13]. Whether mechanisms that compensate for the loss of an entire autosome exist remains unknown. However, a study in budding yeast that examined the effects of heterozygous deletions on protein expression levels showed that gene expression compensatory mechanisms are rare in this organism [14]. Consistent with this conclusion, when monosomy of a chromosome is induced in diploid yeast strains, endoreplication to duplicate the remaining chromosome or non-disjunction occurs, rebalancing the genome from 2n–1 to 2n [15]. However, the correlation between gene copy number and gene expression levels does not seem to be universal. Mechanisms that compensate for changes in chromosome copy number have been described in Drosophila and plants [16], suggesting that species-specific differences exist in the ability to respond to gene copy number variations.
Chronic defined aneuploidies. Analyses of chronic defined aneuploidies have provided insight into the consequences of changing the gene expression pattern of entire chromosomes in organisms that do not have prevalent compensatory mechanisms. A systematic analysis of the effects of chronic defined aneuploidies on Saccharomyces cerevisiae strains showed that strains harbouring an extra chromosome, known as disomes, have—in addition to chromosome-specific effects—an ‘aneuploidy stress response’ defined by defects in cell growth, altered metabolic properties and proteotoxic stress. In particular, proteotoxicity manifests itself as temperature sensitivity, sensitivity to protein folding and degradation inhibitors, and protein aggregate formation (Fig 2; [17,18]; A. Oromendia, unpublished results). In addition, evolution experiments showed that a mutation in UBP6, a ubiquitin-specific protease that antagonizes proteasome function, confers tolerance to some disomies in yeast [18].
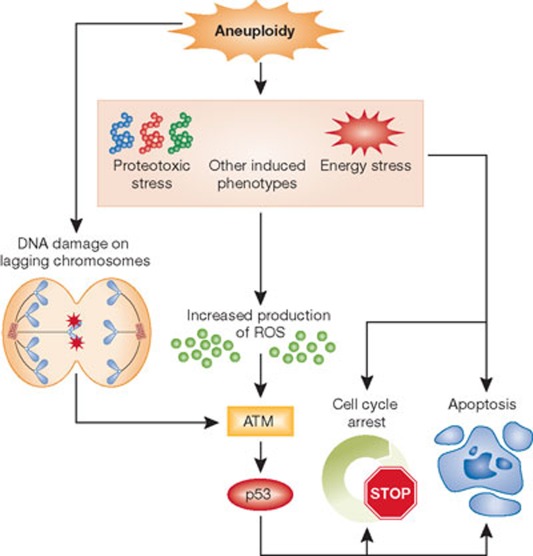
Figure 2.
The effects of aneuploidy on cell physiology. The generation of aneuploidy after chromosome missegregation has been proposed to trigger a checkpoint-like cellular response. Energy, proteotoxic and other aneuploidy-associated stresses have been proposed to increase the production of reactive oxygen species, which activate p53 through ATM [26]. DNA damage on lagging chromosomes during aberrant mitoses triggers a p53-response through ATM [30]. Depending on the level of aneuploidy, this p53 response can either trigger a cell-cycle arrest or promote apoptosis. Aneuploidy can also interfere with cell proliferation by p53-independent mechanisms, as trisomic MEFs do not mount a p53 response but have proliferation defects. ATM, ataxia telangiectasia mutated; MEF, mouse embryonic fibroblast; ROS, reactive oxgen species.
Chronic defined aneuploidies also have an adverse effect on mammalian cells. Trisomic mouse embryonic fibroblasts (MEFs) have growth defects, altered metabolism and differential kinetics of spontaneous immortalization in culture [19]. Skin fibroblasts isolated from Down syndrome individuals, which contain three copies of chromosome 21, proliferate more slowly [20]. Furthermore, comparative cytogenetic analysis of early- and late-stage human blastocysts derived from in vitro fertilizations that contain some aneuploid cells revealed that the percentage of aneuploid cells in the preimplantation embryo decreases with subsequent cell divisions. This suggests that in aneuploid and euploid mosaic embryos, euploid cells outcompete the aneuploid cells and could ultimately ‘normalize’ the embryo [21].
Similar to the disomic yeast strains, trisomic MEFs show signs of energy and proteotoxic stress (Fig 2; [22]). Specifically, they have increased sensitivity to the Hsp90 inhibitor 17-AAG and the autophagy inhibitor chloroquine, show basal activation of the autophagy pathway and express higher levels of the inducible chaperoneHsp72, which prevents protein aggregation [22]. However, increased sensitivity to proteasome inhibitors was not observed in these aneuploid MEFs [22], suggesting that proteasomal degradation is not rate-limiting in these cells. Rather, it seems that aneuploid mammalian cells are more reliant on autophagy to cope with the changes in protein homeostasis caused by aneuploidy.
Chronic defined aneuploidies are a major genetic perturbation, and collectively, these studies suggest that aneuploidy causes—among other detrimental outcomes—a set of shared phenotypes that are both independent of the specific set of genes amplified on the extra chromosome and are indicative of energy and proteotoxic stress. These general phenotypes are seen in addition to the chromosome-specific effects caused by amplification of individual genes and combinations of a small number of genes on the aneuploid chromosome.
Acute random aneuploidies. Cells that contain acute random aneuploidies due to CIN also have proliferation defects and show features of cellular stress. This was first noted in human cells using live cell-imaging and clonal-cell analyses, which showed that chemically induced chromosome missegregation compromises cell proliferation [23]. Subsequent studies in these cells showed that cells that become aneuploid as a result of chromosome missegregation activate p53, inducing cell-cycle arrest and apoptosis [24]. Consistent with these observations, MEFs that missegregate their chromosomes undergo apoptosis [25]
Other studies of the immediate cellular response to CIN-induced aneuploidy revealed similar phenotypes. Knockdown of the spindle assembly checkpoint (SAC) components BubR1 (encoded by the BUB1B locus), Mad2, or the centromere associated kinesin Cenp-E in near diploid HCT116 cells causes a p53 response [26]. MEFs with a mutation in the SAC component Bub1 or with a mutations that renders the checkpoint effector Cdc20 unresponsive to the checkpoint signal activate p53 during the genesis of aneuploidy. This activation depends on the DNA damage checkpoint kinase Atm but not on Chk1/Chk2 [26]. Interestingly, activation of Atm is not caused by DNA damage but by increased ROS levels, as the cell-cycle arrest observed in these aneuploid cells could be suppressed by treating the cells with ROS scavengers (Fig 2). Furthermore, the cellular response to CIN-induced aneuploidy differs depending on the degree of aneuploidy and leads to an enrichment of aneuploid cells, suggesting that massive chromosome missegregation induces p53-mediated apoptosis, whereas low levels of chromosome missegregation induce a p53-mediated cell-cycle arrest [26]. The graded response to the genesis of aneuploidy led to the proposal of an ‘aneuploidy checkpoint’ mediated by p53 and ROS (Fig 2). The increased energy demands generated by increased genomic content, the subsequently induced energy stress and proteotoxic stress, and other aspects of the aneuploid condition could trigger the production of ROS, which activate ATM [27] in a DNA-damage-independent manner. This in turn activates p53. Thus, this mechanism would limit the proliferation of aneuploid cells, and the degree of aneuploidy would toggle the strength of the cell-cycle arrest and induction of apoptosis so that the cell needs to reach a ROS threshold to activate p53 [28].
The prominent role of p53 in promoting cell death or cell-cycle arrest in response to aneuploidy was previously suggested. Embryos lacking both copies of the gene encoding the SAC component MAD2 die by embryonic day (E) 7.5, but deletion of p53 allows these embryos to survive until E10.5 [29]. However, other studies suggested that p53 activation as a consequence of chromosomal instability might not be due to the genesis of aneuploidy per se but could be a consequence of CIN. Chromosome missegregation leads to lagging chromosomes, which become damaged during cytokinesis and trigger a DNA damage response (Fig 2). Defects in chromosome attachment to the mitotic spindle after treatment with Monastrol—which inhibits the kinesin Kif11, arresting cells in mitosis—cause chromosomes to linger in the centre of the spindle [30]. The ensuing cytokinesis results in chromosome breaks on the lagging chromosomes, which elicit an Atm/Chk2-mediated p53 DNA damage response, characterized by increased levels of H2A.X foci and expression of a p53 reporter. Therefore, DNA damage could serve as a cellular sensor for detecting CIN and the ensuing aneuploidies. Errors in chromosome segregation could lead to anaphase bridges and chromosomes remaining in the spindle midzone, which break during cytokinesis and thus trigger a p53 response (Fig 2).
Several other studies in mice with an altered SAC also showed an increase in the frequency of lagging chromosomes during anaphase [25,31,32,33]. However, the DNA damage response in these cells has not yet been characterized. Additionally, cells heterozygous for a deletion in CENP-E do not have increased H2A.X expression after the induction of aneuploidy as a result of chromosome missegregation [34]. Therefore, further analysis is required to better understand the role of DNA damage in the induction of the response to CIN and the aneuploidy that accompanies it.
Several studies have implicated p53 in the response to the aneuploid state, but aneuploidy clearly interferes with cell proliferation through other mechanisms. Cells harbouring single constitutional chromosomal aneuploidies—trisomic MEFs—do not mount a p53 response yet have proliferation defects (Fig 2; [22]). In addition, p53 inactivation does not suppress the proliferation defect of trisomic MEFs when compared with wild-type controls. Although increased proliferation is observed in trisomic MEFs treated with a p53 shRNA, the fold change in proliferation is similar to that seen in wild-type cells treated with a p53 shRNA: there is no added effect in trisomic cells (S. Santaguida, personal communication). Furthermore, human HCT116 cells, in which p53 is disrupted, maintain a near diploid karyotype in continuous growth in culture for many passages [35]. Thus, mechanisms other than p53 must be preventing aneuploid cells from accumulating in these cell populations. Together, these data suggest that aneuploidy interferes with cell proliferation in both a p53-dependent and p53-independent manner. Identifying these unknown effectors of the aneuploidy-induced cellular response could provide crucial new targets in cancer therapy (see Sidebar A).
Sidebar A | In need of answers.
What is the functional role of aneuploidy in the brain and the liver?
What is the extent of proteotoxic stress in aneuploid mammalian cells?
Through which mechanisms does aneuploidy induce ROS and a p53 response?
What is the cause(s) of the p53-independent aneuploidy-associated proliferation defects in aneuploid cells?
Is p53 loss and the accumulation of other aneuploidy-tolerating genetic alterations a prerequisite for aneuploidy-induced tumorigenesis?
What other chemical compounds can be used to exaggerate aneuploidy-associated stress phenotypes in cancer cells?
Can pan-aneuploidy or chromosome-specific aneuploidy inhibitors be effectively used to treat cancer patients?
Can individual aneuploid chromosomes be targeted in the development of personalized cancer therapy?
Cells with CIN caused by chemically induced chromosome missegregation, by gain-of-function alleles of Cdc20 [26], or loss of function alleles of Bub1B [32], or by overexpressing the checkpoint factor Mad2 [36] proliferate poorly. However, not all cells with CIN-induced aneuploidy have been reported to have proliferation defects. Cells heterozygous for deletions in the SAC genes BUB3 or RAE1 [37], cells heterozygous for deletions in CENP-E [34] and cells that overexpress the ubiquitin-conjugating enzyme UbcH10 [33] become aneuploid in vitro but do not seem to slow cell proliferation. This apparent inconsequence of aneuploidy on cell proliferation could be due to several reasons. As observed in BUB1-deficient MEFs, perhaps the gene that is mutated is itself involved in promoting cell-cycle arrest and apoptosis when missegregation events occur [25]. Thus, even if cells acquire aneuploidies, they are not eliminated. It is also possible that in these mouse models of aneuploidy, only a subset of cells in the population acquire low-grade aneuploidies. Growth defects or death of a small fraction of the cell population could go unnoticed in population doubling measurements. Live cell analysis might be needed to detect proliferation defects of individual aneuploid cells.
When aneuploidy is normal
Chronic aneuploidy and chromosome missegregation adversely affect proliferation of cells in culture. It is thus of interest to note that some tissues are naturally aneuploid. This observation raises the question of whether aneuploidy is always detrimental. In mice and humans, approximately one-third of the dividing cerebral neuroblasts in the embryonic brain are aneuploid [38]. Many of these aneuploid cells are eliminated during the course of development, as there are fewer—around 10%—aneuploid cells in the adult brain [38,39,40]. Nevertheless, the aneuploid cells that survive into adulthood are functional, as judged by their ability to form synapses and contribute to the normal neuronal and glial population in the adult brain [41]. Single chromosomal abnormalities—monosomies and trisomies—are the predominant form of aneuploidy detected in the brain. The biological significance of this increased aneuploidy remains to be determined (see Sidebar A), but it has been suggested to enable specification of particular brain regions [42].
The mammalian liver also contains aneuploid cells, although the types of aneuploidy differ from those in the brain. Hepatocytes alter their ploidy both as part of normal development and in response to ageing, regenerative challenge, adaptation to toxic stresses and disease [43]. Instead of acquiring single chromosomal abnormalities, polyploidization is frequently observed, and is often the result of failed cytokinesis. Subsequent reductive cell divisions of these polyploid cells along multipolar mitotic spindles generate aneuploid cells [44,45]. Such divisions have been suggested to provide hepatocytes with a mosaic of genetically diverse cells so the liver can effectively adapt to stresses, such as nutritional and noxious challenges. However, it is also possible that the cell divisions leading to aneuploidy are a sign of ageing and deteriorating regenerative potential, and produce liver cells that are less fit than euploid cells. Only a careful comparison of the growth and metabolic properties of euploid and aneuploid liver cells will distinguish between these possibilities (see Sidebar A).
The apparent absence of the detrimental effects of aneuploidy in brain and liver cells could be due to the specific functions of these cell types. Neurons, once differentiated, will never divide again. In this post-mitotic state, the anti-proliferative effects of aneuploidy probably have a limited impact on cell function. Unlike neurons, liver cells retain the ability to proliferate even after they become aneuploid. However, it is important to note that in hepatocytes, aneuploidy often occurs in the context of polyploidy. Studies in yeast have shown that increased ploidy ‘buffers’ against the adverse effects of aneuploidy [11,46]. In this context, the genotypic variation generated by the aneuploid state has a net lower effect: in diploid cells, the presence of a trisomic chromosome induces a 1.5-fold change in the expression of that chromosome, whereas this is reduced to a 1.25-fold change in expression in tetraploid cells with an extra chromosome. Analogously, the lack of detrimental aneuploidy-associated phenotypes in hepatocytes could therefore be due to better tolerance of altered genomic content in the presence of higher ploidy. It is possible that liver cells might be naturally more tolerant to aneuploidy than other cell types. Consistent with this idea, hepatocytes are remarkably resistant to genomic insults such as telomere erosion and defects in the chromosome segregation machinery [47,48]. Furthermore, tetraploidy—which is not tolerated in most cell types [49]—is readily observed in hepatocytes, altering their genomic content from 2n to 8n without an apparent cell-cycle arrest [45].
Aneuploidy in the brain and the liver might allow cells to define and modify their functional capacities. If cancer cells use karyotype changes in a similar manner, aneuploidy could provide a convenient way to alter gene dosage. Such alterations could promote the development of cancer cell phenotypes—such as escape from growth control—and enable the acquisition of new functions, such as the ability to migrate or seed distant metastatic sites. Simultaneously, aneuploidy could mitigate the detrimental effects caused by changes in gene dosage by altering the balance of genes crucial for other cellular functions. Consistent with this idea, p53−/− mouse mammary epithelial cells chemically induced to become tetraploid, become chromosomally unstable and acquire new functions, such as the ability to form tumours in a xenograft assay [50], and primary human fibroblasts harbouring an extra copy of chromosome 8 have transformed cell phenotypes, such as loss of contact inhibition [51].
Aneuploidy and cancer
Aneuploidy can be used to generate genetic diversity in tissues to allow adaptation to challenges (see review by Holland & Cleveland in this issue of EMBO reports). This flexibility, however, comes at a price: the genetic imbalances induced by aneuploidy cause—among other deleterious outcomes—a disruption in protein and energy homeostasis and proliferation defects. These complex effects of aneuploidy are exemplified in its role in tumorigenesis.
Several approaches have been taken to examine the impact of aneuploidy and CIN on tumorigenesis. In asking how aneuploidy per se influences tumorigenesis, we are limited by the fact that only a small subset of aneuploidies is viable in mammals: two constitutional trisomies can survive infancy in humans whilst none survive embryonic development in the mouse. However, cancer karyotypes are complex and have high degrees of aneuploidy, calling into question the relevance of studies of single chromosomal aneuploidies in understanding the role of aneuploidy in tumorigenesis. Mouse models of CIN have been developed that recapitulate the more complex aneuploidies seen in cancer but, as discussed above, the other effects of the mutations causing CIN make it difficult to unambiguously determine how aneuploidy per se affects tumorigenesis. Therefore, the combined analysis of both models, chronic defined aneuploidies and CIN-induced random acute aneuploidies, is crucial in unravelling the role of aneuploidy in tumorigenesis. In what follows, we discuss studies of chronic defined aneuploidies and CIN in both humans and mice that shed light on the role of aneuploidy in cancer.
In humans, individuals trisomic for autosomes 13, 18 or 21 survive to birth in appreciable frequencies in the population. As in other organisms, constitutional trisomy in humans leads to developmental defects and increased risk of specific pathologies. Trisomy 18—also known as Edwards syndrome—has a prevalence between 1 in 3,000 and 1 in 8,000 live births, and about 90% of these individuals die in the first year of life, generally due to severe cardiovascular and brain defects [52]. Trisomy 13—or Patau syndrome—is the least frequently observed constitutional autosomal trisomy. Individuals with Patau syndrome have severe developmental abnormalities at birth, making their survival after the first year of life extremely rare, although a few individuals have been reported to live into their teens [53]. Trisomy 21—or Down syndrome (DS)—is observed with an incidence of about 1 in 700 live births, making it the most commonly observed constitutional autosomal trisomy [54]. The phenotypic manifestation of DS is complex and variable, but is commonly associated with mental retardation, heart defects, early onset of Alzheimer disease [55] and reduced life expectancy [56].
Evaluating the tumour spectra of individuals with DS and Edwards syndrome provides a means for observing the effect of chronic aneuploidy on tumorigenesis in humans. Because DS is the most prevalent chromosomal abnormality in the population and affords the longest life expectancy of all autosomal trisomies, there are substantially more data on the tumour profile of people with trisomy 21 than with trisomy 18. Individuals with trisomy 21 have an increased risk for acute lymphoblastic leukaemia and acute myeloid leukaemia—particularly acute megakaryocytic leukaemia (AMKL)—which is especially high in the first few years of life. In addition, lymphomas and germ-cell tumours are more frequently observed in DS individuals than in the general population. However, they have a decreased risk of developing solid tumours throughout life [57]. Although there are fewer individuals to evaluate, case studies reveal that Edwards syndrome predisposes affected individuals to Wilms' tumours and hepatoblastomas when compared with age-matched controls [58].
It seems that DS and Edwards syndrome increase the risk of developing childhood cancers [58], which might be explained by chromosome-specific effects. Consistent with this idea is the observation that the chromosome constitutionally trisomic in DS and Edwards syndrome is also found to be trisomic in sporadic cases of the same types of cancer. For example, acquired trisomy 21 is a prominent cytogenetic characteristic in many haematological neoplasms [59,60]—notably in many non-DS cases of AMKL [61]—suggesting that it could be an oncogenic event that promotes the development of acute leukaemia. Additionally, although trisomy 18 is not observed in non-Edwards syndrome hepatoblastomas [62], it is frequently observed in non-Edwards syndrome Wilms' tumours [63].
In contrast to these childhood cancers, the incidence of solid tumours is decreased in DS individuals. Due to short life expectancy, the tumour incidence of non-childhood solid tumours cannot be well examined in other viable trisomies. Because there are only data regarding the incidence of solid tumours in adulthood from one constitutional trisomy, it is difficult to distinguish chromosome-specific effects from general aneuploidy effects. Several tumour suppressors are encoded on human chromosome 21, which could account for the tumour-protective effect observed in DS. However, although some DS phenotypes can be explained by amplification of specific genes or sets of genes, others cannot [64]. Therefore, the decreased incidence of solid tumours in DS individuals might also reflect decreased cellular fitness associated with constitutional trisomy 21, and the aneuploid state could provide tumour protection throughout life. However, cancer is largely considered a disease of ageing and environment, and it has been suggested that the decreased incidence of solid tumours observed in the DS population is due to the decreased life expectancy of these individuals or environmental biases [57].
Several mouse models of DS have been used to analyse the effects of constitutional trisomy 21 on cancer incidence (summarized in Table 1). Most studies were conducted in the Ts65Dn and Tc1 mouse models. The Ts65Dn mouse harbours three copies of the subset of genes homologous to those encoded in human chromosome 21, which are encoded in the mouse in chromosome 16 [65]. This amplification includes all genes found in the Down syndrome critical region (DSCR)—including DSCR1 or RCAN1, which is a regulator of VEGF–calcineurin signalling in endothelial tissues that decreases tumour growth and angiogenesis when overexpressed [66,67]. The Tc1 mouse is a transchromosomal mouse model of DS that contains ∼90% of all genes on human chromosome 21 [68], but does not include the part of the DSCR that contains DSCR1.
Table 1. Mouse models of CIN or aneuploidy and cancer.
| Affected gene | Study | Karyotype effect | Effect on cell proliferation | Effect on tumorigenesis |
|---|---|---|---|---|
| BUB1 | [25] | CIN | None reported | Bub1+/− mice have decreased tumour incidence; Bub1+/H and Bub1H/H mice have increased tumour incidence and altered tumour spectrum |
| BUB1 overexpression | [31] | CIN | None reported | Increased tumour incidence and altered tumour spectrum |
| BUB1B (also known as BUBR1) | [32] | CIN | Bub1bH/H MEFs proliferate more slowly than WT by proliferation assays; Bublb−/H MEFs proliferate even more slowly | Bub1bH/H mice have early ageing and infertility phenotypes, no significant increase in tumour formation; Bub1b−/H mice die a few hours after birth |
| BUB3+/− | [37] | CIN | Growth rate not different from WT by proliferation assays | Increased incidence of tumours after carcinogen treatment |
| CDC20AAA | [87] | CIN | Cells proliferate more slowly than WT | Increased incidence of tumours and altered tumour spectrum |
| CENP-E | [34] | CIN | Growth rate not different from WT by proliferation assays | Altered spontaneous tumour spectrum and reduced tumour incidence after treatment with carcinogens |
| MAD2+/− | [88] | CIN | None reported | Increased tumour incidence and altered tumour spectrum |
| MAD2 overexpression | [36] | CIN | Cells proliferate more slowly than WT | Greatly increased incidence of spontaneous tumours, wide-ranging tumour spectrum |
| RAE1+/− | [37] | CIN | Growth rate not different from WT by proliferation assays | Increased incidence of tumours after carcinogen treatment |
| SA1+/− | [84] | CIN | Cells proliferate more slowly than WT | Increased incidence of tumours and altered tumour spectrum |
| UBCH10 overexpression | [33] | CIN | Growth rate not different from WT by proliferation assays | Increased incidence of spontaneous tumours and tumours after carcinogen treatment |
| CENP-E+/− p19Arf−/− | [34] | CIN | None reported | Decreased tumour incidence and size |
| MAD2 overexpression KrasG129D | [92] | CIN | None reported | Larger, more aggressive tumours that are prone to relapse observed |
| Bub1−/H p53−/− | [26] | CIN | None reported | Accelerated tumorigenesis compared to either single mutation |
| Cdc20+/AAA p53−/− | [26] | CIN | Cdc20AAA/AAA p53−/− MEFs and immortalize in culture | Accelerated tumorigenesis compared to either single mutation |
| Ts65Dn xenograft | [69] | DS | None reported | Decreased tumour burden and reduced tumour angiogenesis |
| Tc1 xenograft | [70] | DS | None reported | Decreased tumour burden and decreased tumour angiogenesis |
| Ts65Dn ApcMin | [72] | DS | None reported | Decreased tumour incidence and size |
| Ts65Dn NPcis | [73] | DS | None reported | Altered tumour spectrum and increased survival after tumour induction |
ApcMin, adenomatous polyposis coli multiple intestinal neoplasia mouse model; CIN, chromosomal instability; DS, Down syndrome; WT, wild-type
Two studies of these mice demonstrate that tumour angiogenesis is reduced in transplantable, subcutaneous lung carcinoma and melanoma tumour models [69,70]. Ts65Dn mice show decreased tumour burden—which is largely dependent on the expression of DSCR1 in a dose-dependent manner—and the tumours that arise have decreased microvessel density. Similar results were obtained with the Tc1 mouse [70]. However, DSCR1 is not contained in the amplified genomic region of the Tc1 mouse, suggesting that the decreased tumour angiogenesis observed is caused by altered dosage of other genes.
The effects of trisomy were also examined in mouse models of human cancer. The ApcMin model of small intestine and colon cancer [71] was examined in the Ts65Dn mouse [72]. The incidence of tumour formation and tumour size are reduced in Ts65Dn mice compared with their euploid littermates, an effect partly mediated by specific genes amplified in the DSCR, notably the transcription factor ETS2. Triplication of ETS2 largely, but not completely, accounts for the decreased tumour incidence. However, the Ts65Dn model of DS does not affect tumour growth in the aggressive Nf1+/− TP53+/− (NPcis) neurofibromatosis type 1 cancer model [73]. NPcis mice develop lymphomas, sarcomas or carcinomas with 100% penetrance due to loss of heterozygosity of the normal allele [74,75]. Trisomy does not decrease the incidence of tumour formation or reduce tumour size, but Ts65Dn mice have increased survival after tumour induction, an effect the authors propose is due to a shift in the observed tumour spectrum from mainly sarcomas to adrenal and brain tumours and lymphomas. However, the increased survival time is not attributable to ETS2 dosage. Furthermore, Ts65Dn NPcis mice do not have reduced tumour angiogenesis. It thus seems that the effects of trisomy of the genes, amplified in these two mouse models, have highly context-specific effects on tumorigenesis, but in both tumour models the trisomy inhibits rather than promotes tumorigenesis.
CIN and cancer
Studies in constitutionally aneuploid humans and mice describe how a specific chromosome affects tumorigenesis. Most human tumours, however, become aneuploid by a spontaneous aneuploidizing event, often harbour a complex and diverse assortment of chromosomes and experience continuous changes in karyotype due to CIN (see review by Swanton & colleagues in this issue of EMBO reports). In humans, there is only one known heritable syndrome—mosaic variegated aneuploidy (MVA)—with increased levels of random cellular aneuploidies. MVA results from biallelic loss-of-function mutations in the spindle checkpoint component BubR1—which leads to premature sister chromatid separation and frequent mitotic non-disjunction [76]—or from biallelic mutations in Cep57, a centrosomal protein involved in nucleating and stabilizing microtubules and therefore ensuring correct chromosomal division during mitosis [77]. Individuals with MVA have mental retardation and other developmental defects, as well as a predisposition to cancer [78,79]. Although this condition is extremely rare and few individuals with MVA have been reported to live past childhood, case studies reveal that rhabdomyosarcoma and Wilms' tumours are frequent early in life [80]. As with DS and Edwards syndrome, the increased incidence of these childhood cancers is probably the result of abnormal embryonic development due to changes in the gene dosage of specific gene products. Because the life expectancy of individuals with MVA is short, it is difficult to determine whether having constitutional premature sister chromatid separation would lead to cancer predisposition later in life. In the one reported case in which an individual survived beyond childhood [78], the generation of sporadic tumours later in life was not markedly increased, even though the aneuploidies generated in MVA are random and thus an appropriate gene combination could have been selected for that promotes tumour growth. This individual developed and died from acute myeloid leukaemia, suggesting there could be a bias towards haematological cancers as in DS. However, many more case studies are necessary to draw any definitive conclusions about the role of MVA-induced aneuploidy in tumorigenesis in adults.
Although BubR1 is mutated in MVA, mutations in SAC and other chromosome segregation factor encoding genes are rarely observed in sporadic cancers [81]. However, a recent study found that a diverse range of tumour types contains deletions or inactivating mutations in STAG2, a gene located on the X chromosome that encodes a subunit of one of the mammalian cohesin complexes [82]. Because cohesin complexes hold sister chromatids together, their proper function is critical for accurate chromosome segregation ([83]; see also review by Jessberger in this issue). Indeed, inactivation of STAG2 in diploid cell lines leads to significant aneuploidy, suggesting that aneuploidy promotes tumorigenesis. A mouse model of cohesion deficiency further supports the idea that cohesins are crucial for preventing tumour formation. Mice with a heterozygous deletion in the gene encoding the cohesin subunit SA1 lack cohesion at telomeres, leading to increased levels of aneuploidy and decreased cellular proliferation. Remarkably, however, these mice show an increased incidence of spontaneous tumours [84]. Nevertheless, it is important to bear in mind that cohesins do much more than ensure accurate chromosome segregation. During interphase, they have a crucial role in gene expression and repair of DNA damage [85]. Thus it is possible that the role of cohesins in these cellular processes, in addition to their roles in ensuring accurate chromosome segregation, contributes to their tumour suppressive function.
To understand the role of spontaneous whole-chromosomal aneuploidy in tumorigenesis, several mouse models with decreased chromosome segregation fidelity have been generated (summarized in Table 1). Such models of CIN use genetic alterations that interfere with either the chromosome segregation machinery itself or with SAC function. It is difficult to determine whether the genetic alterations used to induce chromosome missegregation, CIN itself or other potential genetic alterations resulting from CIN lead to a particular effect on tumorigenesis. However, these models provide invaluable insights into tumorigenesis and the effects that a potentially continuously changing genome has on disease progression.
Deficiency of BubR1—an essential component of the SAC [86]—has been analysed in mice at cellular and organismal levels [32] by using hypomorphic alleles. MEFs derived from BubR1-deficient mice have increased levels of chromosomal aneuploidy and an increased frequency of cellular senescence, and mice with decreased levels of BubR1 have early ageing and infertility phenotypes without a significant increase in tumour formation. Thus, reduced expression of the BubR1 protein does not seem to result in cancer predisposition in mice. Rather, BubR1-deficient mice show decreased cellular and organismal fitness.
Mice with a heterozygous deletion of CENP-E—which encodes an essential, centromere-associated, kinesin motor—are largely normal, despite the presence of aneuploid cells throughout the body [34]. They have an increased incidence of tumorigenesis in some tissues, such as the lungs and lymphoid cells, but a decreased incidence and reduced size of liver tumours. However, haploinsufficiency of CENP-E in cells confers a transformed phenotype in soft agar and increased tumorigenicity in xenograft assays. Thus, tissue-specific effects seem to modulate the consequences of reduced levels of CENP-E.
Cdc20 is the mitotic activator of APC/C, an E3 ubiquitin ligase that targets key mitotic substrates—notably cyclin B1 and securin—for degradation, allowing anaphase onset. The SAC inhibits the ability of Cdc20 to activate APC/C when chromosomes are not correctly attached to the mitotic spindle, but the Cdc20AAA allele cannot be inhibited by the SAC and therefore induces premature sister chromatid separation and subsequent aneuploidies [87]. This allele increases late-onset tumorigenesis in mice: heterozygous animals have an increased tumour incidence by 24 months of age, with 50% of the Cdc20+/AAA mice developing tumours compared with 10% of the wild-type control, and an altered tumour spectrum, as hepatomas are observed in the mutant but not the control population.
Mad2 is a key component of the SAC and has been both overexpressed to hyperactivate its activity and deleted to weaken it. Mad2 inhibits APC/C–Cdc20 and is thus the lynchpin of the SAC [66]. Deletion of one copy of Mad2 induces aneuploidy in vitro and in vivo and leads to a high frequency of mice with papillary lung adenocarcinomas, a tumour that is extremely rare in wild-type mice [88]. However, a more marked phenotype is observed when Mad2 is overexpressed. Overexpression of Mad2 delays rather than accelerates anaphase entry, and results in a greatly increased incidence of spontaneous tumours with a wide-ranging spectrum [36]. This oncogenic effect of a SAC component in mice is consistent with what is observed in human tumours. Loss-of-function mutations of SAC components are rarely observed in cancer cells, but overexpression is frequently seen. Loss of function of the tumour suppressor RB1 has been shown to lead to increased basal activation of the mitotic checkpoint [89]. Decreased Rb levels relieve inhibition of the transcription factor E2F, leading to overexpression of its direct target, Mad2. Mad2 inhibits APC/C–Cdc20, leading to prolonged metaphase arrest and consequent mitotic slippage, whereby cells exit from mitosis without undergoing chromosome segregation and become tetraploid or missegregate chromosomes [81]. Similar results are observed when the SAC is hyperactivated by overexpression of the outer kinetochore component Hec1 [90] or by overexpressing the SAC kinase Bub1 [31]. Both result in aneuploidies in vitro and cause an increase in tumour incidence and alteration of tumour spectra in vivo, although this increase is not as marked as that observed when Mad2 is overexpressed.
In summary, both weakening and hyperactivating the SAC is sufficient to generate aneuploidy and to induce tumorigenesis. However, although tumorigenesis is elevated, this increase is modest in many cases, particularly in mice with loss-of-function mutations in SAC genes. It has been suggested that—just as cells with defects in chromosome cohesion might have more marked phenotypes because cohesins have various functions—the range in severity of the phenotypes observed in cells with CIN differs depending on the number of processes that will be affected when such a mutation is incurred. If mutating a factor only affects one cellular process that promotes tumorigenesis, the effect will be less severe than if multiple tumour-promoting pathways are affected by a single alteration [91].
Interestingly, when CIN-inducing mutations are combined with p53 loss or other oncogenic mutations, the effects on tumorigenesis are significant. Crossing mice with mutated SAC components into mice homozygous for a p53 deletion has marked effects. p53−/−/Cdc20+/AAA and p53−/−/Bub1H/H mice have increased tumorigenesis and decreased survival compared with either mutation alone [26]. The combined overexpression of Mad2 and an inducible KrasG12D model also has dramatic effects, generating larger tumours that are more aggressive and have higher grade aneuploidy than tumours generated with oncogenic Kras expression alone [92]. Furthermore, these mice have ∼50% tumour recurrence after repression of Mad2 and KrasG12D expression, a phenotype which was never observed in mice with oncogenic Kras expression alone [92]. These results not only indicate that sustained overexpression of Mad2 is not required for tumour progression once tumours have developed, but also suggest that Mad2 overexpression leads to increased chromosomal instability, which overcomes addiction to the KrasG12D oncogene. By contrast, other oncogenic mutations do not lead to increased tumour formation in mice with increased chromosome missegregation frequencies. For example, deletion of the tumour suppressor p19Arf in CENP-E+/− mice leads to decreased tumour incidence and a reduction in tumour size [34]. Perhaps, loss of a tumour suppressor such as p53 is a prerequisite for the development of aneuploidy in human tumours, or an event required immediately after aneuploidy induction to promote tolerance to the aneuploid state. Combined models of inducible aneuploidy and inducible loss of tumour suppressors, such as the one described in [92], could be used to fully dissect this relationship.
Together, these results demonstrate that, similar to the range of tumorigenesis phenotypes observed in mouse models of CIN, introduction of CIN into mouse models of cancer has tumour-promoting and tumour-suppressive effects. However, when CIN-inducing mutations are combined with the loss of p53, more aggressive disease is consistently observed. Thus, in the absence of the gene that limits the proliferation of aneuploid cells, the tumorigenesis-promoting effects of CIN seem to reach their full potential. Exactly how CIN primes cells for tumorigenesis has not yet been elucidated. Below, we propose a model for how this might occur and discuss how CIN and aneuploidy could both promote and suppress tumorigenesis.
Aneuploidy and CIN in tumorigenesis—a model
The study of the effect of CIN and chromosomal trisomies on tumour formation in mouse models has revealed complex interactions between the aneuploid state and tumorigenesis. Aneuploidy seems to promote a dual cellular state: generally the presence of an unbalanced genome induces a cellular stress response and slow growth, whereas in rare selective circumstances—or in the presence of aneuploidy-tolerating mutations—aneuploidy can be beneficial and lead to increased cellular proliferation and cancer. We believe that the following general themes provide a working model for how CIN and aneuploidy impact tumorigenesis (Fig 3A).
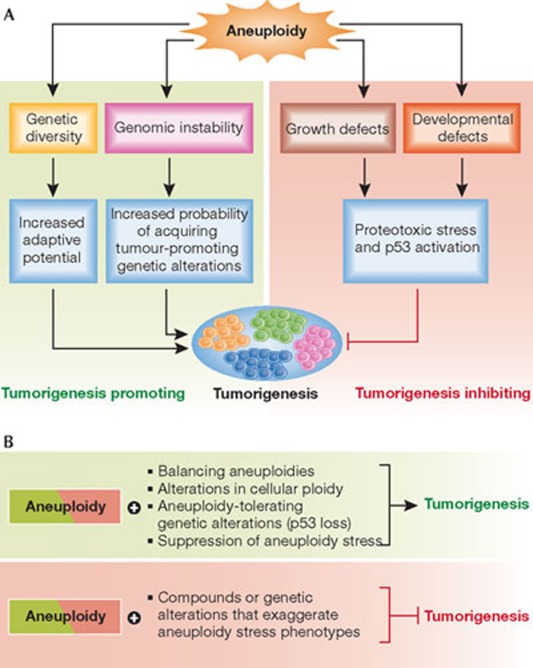
Figure 3.
A model for the role of aneuploidy in tumorigenesis. (A) Aneuploidy is most frequently associated with characteristic phenotypes, such as defects in cell proliferation and developmental defects in whole organisms. These phenotypes are often accompanied by particular cellular responses, such as increased proteotoxic stress and activation of p53. Generally, these adverse effects of aneuploidy serve to inhibit tumorigenesis. Aneuploidy can also generate genetic diversity, which can provide cells with increased adaptive potential when challenged and thus could be a means for promoting special aspects of tumorigenesis, such as metastasis. Finally, aneuploidy is also commonly associated with genomic instability, increasing the probability of acquiring tumour-promoting genetic alterations and thereby significantly contributing to tumorigenesis. (B) The adverse effects of aneuploidy can impair tumorigenesis, but in the presence of aneuploidy-tolerating mutations, increased ploidy or balancing aneuploidies, this anti-tumorigenic effect is lessened and the potential tumorigenesis-promoting effects of aneuploidy reach their full potential. Conversely, compounds or genetic alterations that enhance the adverse effects of aneuploidy could shift the equilibrium towards its anti-proliferative effects, thus preventing the growth of aneuploid cancer cells.
First, aneuploidy hinders cell proliferation in most cases. This anti-proliferative effect can be mitigated by genetic alterations that allow cells to tolerate the adverse effects of aneuploidy, and by mutating genes that restrict proliferation of aneuploid cells, such as p53. Second, chromosome missegregation is at least sometimes accompanied by DNA damage, because of events such as chromosome breakage during cytokinesis. In addition, the aneuploid state itself can induce genomic instability. Third, under specific selective pressures, aneuploidy can provide a survival advantage.
The results discussed above obtained in various mouse models indicate that not all CIN-inducing mutations have the same effect. Mutations that cause defects in sister chromatid cohesion before mitosis are tumorigenic, as evidenced by mice expressing a hypermorphic allele of Cdc20 [87], mice heterozygous for SA1 [84] and the STAG2 loss-of-function found in many human cancers [82]. SAC genes, the primary function of which is to ensure accurate chromosome segregation—and the inactivation of which has modest effects on sister chromatid cohesion before mitosis—are rarely found mutated in human cancers. Mouse models with loss-of-function mutations in SAC genes generally have a modest increase in tumour incidence.
The first conclusion we can draw from these observations is that CIN caused by the mutations that affect sister chromatid cohesion is not the only reason for the tumour-promoting effects of these mutations, and that the role of cohesin in controlling gene expression and DNA damage repair is probably crucial for its tumour-suppressive functions. The second conclusion we can draw is that CIN and the aneuploidies produced chromosome missegregation have only a moderately positive impact on tumorigenesis on their own. In the case of trisomy 21, aneuploidy even has a tumour-protective function—the tumours that are seen early in life are probably caused by developmental abnormalities due to imbalances in the dosage of specific genes. This is not surprising in light of the first conclusion, that aneuploidy generally interferes with cell proliferation (Fig 3A).
However, the picture changes when the anti-proliferative effects of CIN and aneuploidy are mitigated through the inactivation of p53. As mentioned above, mice with mutations that cause increased chromosome missegregation combined with loss-of-function p53 mutations show a marked increase in tumorigenesis. Mutations that improve the proliferation of aneuploid cells have been described in yeast [18]. Aneuploidy-tolerating mutations in mammalian cells could similarly increase the proliferative potential of aneuploid cells. Once the adverse effects of aneuploidy have been suppressed or ameliorated, the potential tumorigenesis-promoting effects of the condition could come into play (Fig 3B).
How could aneuploidy promote tumorigenesis? The genomic-instability-inducing effects of aneuploidy and chromosome missegregation recently described in yeast [46] and mammals [30,93], respectively, probably have a major impact on disease progression. Chromosome missegregation has been shown to cause DNA damage during the subsequent cytokinesis [30]. Increased double-strand breaks, as a result of inadequate replication in micronuclei, could be an important source of oncogenic mutations. A recent study in mammalian cell lines demonstrates that micronuclei that are generated after chromosome missegregation undergo substantial DNA damage during replication [93]. Cells that contain these micronuclei only persist when p53 function is absent, further emphasizing the importance of p53 in preventing genomic instability [93]. DNA from micronuclei can reincorporate into the nucleus, suggesting a mechanism by which a micronucleus could contribute to mutations and alterations in chromosomal composition. This mechanism of transient sequestration of chromosomes in micronuclei could explain the recently discovered process of chromothripsis, where hundreds of genomic rearrangements occur within one or few chromosomes [94], as is observed in 2–3% of all cancers (for example, medulloblastoma; [95]). In addition to chromosome missegregation leading to further aneuploidy, the aneuploid state itself has been shown to cause multiple forms of genomic instability in budding yeast, with an increased number of double-strand breaks observed in many aneuploid strains as well as aneuploid fission yeast cells [46]. DNA damage has a crucial role in tumour evolution [96], and we speculate that the DNA-damage-inducing features of whole-chromosome missegregation and the aneuploid state itself, could be a crucial aspect of the tumour-promoting effects of aneuploidy (Fig 3A).
Exploring aneuploidy: the significance of chromosomal imbalance.
This review series—published in this issue of EMBO reports—also includes:
A balancing act: focus on aneuploidy Nonia Pariente
Losing balance: the origin and impact of aneuploidy in cancer Andrew J Holland and Don W Cleveland
Cancer chromosomal instability: therapeutic and diagnostic challenges Nicholas McGranaham, Rebecca A Burrell, David Endesfelder, Marco R Novelli and Charles Swanton
Age-related aneuploidy through cohesion exhaustion Rolf Jessberger
Aneuploidy is a way to generate phenotypic diversity. This could have an important role in the stages of tumorigenesis in which a cancer cell must adapt to a new environment, such as during metastasis. Experimental evolution studies in microorganisms support this view. Yeast spontaneously acquire characteristic chromosome translocations in chemostat evolution experiments [97], segmental aneuploidies when acquiring resistance to anti-fungal agents [98] and whole-chromosome aneuploidies when challenged to evolve new traits [99]. Even MEF cell lines often become aneuploid when adapting to growth in culture [100].
In summary, we propose that CIN and aneuploidy have modest tumour-promoting abilities conferred through their associated genomic instability and their potential for generating new traits. However, these tumorigenic traits are offset by the antiproliferative effects associated with aneuploidy. When these anti-proliferative effects are suppressed—through aneuploidy-tolerating mutations, increased ploidy or balancing aneuploidies—the full tumorigenic potential of the condition is unleashed (Fig 3B). This hypothesis predicts that mutations that suppress the antiproliferative effects of CIN and aneuploidy are crucial factors in tumorigenesis (Fig 3B). Indeed, p53 is one of the most frequently mutated genes in human cancers, and the familial form of p53 loss—known as Li–Fraumeni syndrome—predisposes affected individuals to a wide spectrum of cancers [101]. p53 might be special in that it seems to protect cells not only from numerical chromosomal abnormalities such as aneuploidy [28] and tetraploidy [5,102,103], but also from structural aberrations through its central function in the DNA damage checkpoint pathway [104].
Cancer cells also seem to evolve karyotypes in which aneuploidies are mitigated by polyploidy and extra aneuploidies. Chromosome gains and losses often occur with other chromosome gains and losses, suggesting that these extra events are a compensatory mechanism that attempts to balance alterations in gene dosage caused by CIN [105]. Other aneuploidy-tolerating effects might also be important. Disomic yeast cells try to compensate for their altered gene dosage by degrading the excess of some proteins, especially of proteins found in large molecular complexes such as the ribosome [17,18]. Evolution experiments uncovered aneuploidy-tolerating mutations in proteins such as UBP6, a ubiquitin-specific protease that antagonizes the degradation of several proteasome substrates in yeast [18]. Identifying the genetic alterations that allow unbalanced, aneuploid mammalian cells to restore balance and tolerate aneuploidy could provide key insights into tumorigenesis and new targets for the development of cancer therapeutics (see Sidebar A).
Aneuploidy as a therapeutic target
Initial approaches to cancer treatment targeted a phenotype common to all cells: increased proliferation. Cancer is a disease of uncontrolled proliferation, and therefore, chemotherapeutics—which kill all rapidly dividing cells by interfering with DNA synthesis and chromosome segregation—are effective anti-cancer agents. However, cancer cells have a nearly relentless ability to adapt to their environments, mutate and survive in response to treatments [106]. Therefore, patients treated with chemotherapeutics often relapse due to the acquisition of chemotherapeutic resistance [107] or the presence of dormant tumour cells [108], and their cancers metastasize.
Subsequent approaches in cancer treatment targeted single-gene products, to which cancer cells are addicted. Imatinib (Gleevec) [109,110], which targets the hybrid kinase BCR–ABL and trastuzumab (Herceptin) [111], which targets the EGFR family member ERBB2, are among the most successful examples in this category of cancer therapeutics. However, even these targeted treatments eventually cease to be effective, as cells develop extra mutations to acquire resistance to these drugs [112]. Combination therapies that eliminate all tumour cells by targeting both specific genetic lesions and general cancer cell characteristics early during treatment seem to be the most promising method for exacting a cure. Thus, identifying as many differences between normal and tumour cells, and developing agents that selectively target as many of these differences as possible simultaneously, could prove to be a potent means of eliminating cancer cells.
Because adaptability is so important to cancer cell survival and higher degrees of aneuploidy are frequently associated with poor prognosis [4], aneuploidy should be considered as a therapeutic target. A proof-of-principle, small-scale screen for compounds that preferentially impair proliferation of trisomic MEFs compared with euploid MEFs, identified the energy-stress-inducing compound AICAR, the autophagy inhibitor chloroquine and the Hsp90 inhibitor 17-AAG as aneuploid-selective drugs [22]. Autophagy and Hsp90 are both required for eliminating protein aggregates and maintaining proteostasis, and their protein quality control functions seem to be rate-limiting in aneuploid cells. By contrast, proteasome inhibitors were not identified in this screen suggesting that, unlike in aneuploid yeast, proteasome activity is not limiting in trisomic MEFs. Interestingly, AICAR and 17-AAG inhibit the growth of highly aneuploid colon cancer cell lines that have CIN. AICAR has not yet been tested in clinical trials, but treatment of cancer patients with 17-AAG in phase II clinical trials either did not yield anti-cancer properties, despite activation of Hsp90 [113], or needed to be stopped due to adverse side effects [114]. Therefore, as with many cancer drug candidates, although these compounds seem to be effective in vitro, they might be ineffective in a therapeutic setting. Nevertheless, targeting aneuploidy for cancer therapy is worthy of further exploration and large-scale screens could identify new unanticipated sensitivities of aneuploid cells (Fig 3B; Sidebar A).
In addition to identifying pan-aneuploidy inhibitors, the isolation of compounds that selectively impair the proliferation of cells harbouring specific aneuploidies should also be explored (see Sidebar A). Many cancers frequently show gain or loss of a specific chromosome. For example, trisomy 8 is frequently observed in patients with acute myeloid leukaemia, and its presence is associated with poor survival when combined with other genetic aberrations [115]. Developing compounds that selectively eliminate cells with this particular aneuploidy could also provide another way to target specific cancers.
In summary, aneuploidy has a complicated but significant role in tumorigenesis. Finding treatments that exacerbate the phenotypes of aneuploid cells and selectively kill them could prove to be a fruitful endeavour in regards to cancer treatments.

Sarah J Pfau & Angelika Amon
Acknowledgments
We thank A. Oromendia and S. Santaguida for generously sharing unpublished results, and members of the Amon lab for critical reading of this manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Balmain A, Gray J, Ponder B (2003) The genetics and genomics of cancer. Nat Genet 33: 238–244 [DOI] [PubMed] [Google Scholar]
- Longo D (2012) Tumor heterogeneity and personalized medicine. N Engl J Med 366: 956–957 [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C (2004) Aneuploidy and cancer. Nature 432: 338–341 [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D (2007) Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 17: 157–162 [DOI] [PubMed] [Google Scholar]
- Davidson J, Gorringe K, Chin S, Orsetti B, Besret C, Courtay-Cahen C, Roberts I, Theillet C, Caldas C, Edwards P (2000) Molecular cytogenetic analysis of breast cancer cell lines. Br J Cancer 83: 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferti AD, Stamouli MJ, Panani AD, Raptis SA, Young BD (2004) Molecular cytogenetic analysis of breast cancer: a combined multicolor fluorescence in situ hybridization and G-banding study of uncultured tumor cells. Cancer Genet Cytogenet 149: 28–37 [DOI] [PubMed] [Google Scholar]
- Griffin CA, Morsberger L, Hawkins AL, Haddadin M, Patel A, Ried T, Schrock E, Perlman EJ, Jaffee E (2007) Molecular cytogenetic characterization of pancreas cancer cell lines reveals high complexity chromosomal alterations. Cytogenet Genome Res 118: 148–156 [DOI] [PubMed] [Google Scholar]
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR (2008) Defining ‘chromosomal instability’. Trends Genet 24: 64–69 [DOI] [PubMed] [Google Scholar]
- Foijer F, Draviam VM, Sorger PK (2008) Studying chromosome instability in the mouse. Biochim Biophys Acta 1786: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A (2008) Aneuploidy: cells losing their balance. Genetics 179: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T (2004) Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res 64: 6941–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Weissman JS, Kirschner MW (2010) A general lack of compensation for gene dosage in yeast. Mol Syst Biol 6: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJD, Sunjevaric I, Voth WP, Ciccone S, Du W, Olsen AE, Stillman DJ, Rothstein R (2008) Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes. Genetics 180: 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL (2001) Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234: 275–288 [DOI] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924 [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A (2010) Identification of aneuploidy-tolerating mutations. Cell 143: 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE (1974) Studies on Down's syndrome in tissue culture. I. Growth rates protein contents of fibroblast cultures. J Cell Physiol 83: 85–90 [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D (2008) Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod 23: 2596–2608 [DOI] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A (2011) Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA (2008) Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA (2010) Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM (2007) Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol 179: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P (2010) The ATM–p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA 107: 14188–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT (2010) ATM activation by oxidative stress. Science 330: 517–521 [DOI] [PubMed] [Google Scholar]
- Fang X, Zhang P (2011) Aneuploidy and tumorigenesis. Semin Cell Dev Biol 22: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burds AA, Lutum AS, Sorger PK (2005) Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci USA 102: 11296–11301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH (2011) Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333: 1895–1898 [DOI] [PubMed] [Google Scholar]
- Ricke RM, Jeganathan KB, van Deursen JM (2011) Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol 193: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ et al. (2004) BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749 [DOI] [PubMed] [Google Scholar]
- van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM (2010) Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol 188: 83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36 [DOI] [PubMed] [Google Scholar]
- Bunz F, Fauth C, Speicher M, Dutriaux A (2002) Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res 62: 1129–1133 [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R (2007) Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu J, Jeganathan K, Baker D, Wu X, Kang-Decker N, van Deursen J (2003) Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 160: 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J (2001) Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci USA 98: 13361–13366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK et al. (2005) Constitutional aneuploidy in the normal human brain. J Neurosci 25: 2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Peterson SE, Yung YC, Mutoh T, Barral S, Chun J (2008) Aneuploid mosaicism in the developing and adult cerebellar cortex. J Comp Neurol 507: 1944–1951 [DOI] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J (2005) Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci USA 102: 6143–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J (2010) Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol 518: 3981–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S (2000) Hepatic polyploidy and liver growth control. Semin Cancer Biol 10: 161–171 [DOI] [PubMed] [Google Scholar]
- Duncan AW, Hickey RD, Paulk NK, Culberson AJ, Olson SB, Finegold MJ, Grompe M (2009) Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet 5: e1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M (2010) The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM et al. (2011) Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, Celli G, de Lange T (2006) Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev 20: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth KG et al. (2006) Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol 172: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D (2007) Limiting the proliferation of polyploid cells. Cell 131: 437–440 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D (2005) Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Nawata H, Kashino G, Tano K, Daino K, Shimada Y, Kugoh H, Oshimura M, Watanabe M (2011) Dysregulation of gene expression in the artificial human trisomy cells of chromosome 8 associated with transformed cell phenotypes. PLoS ONE 6: e25319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-Y et al. (2006) Clinical characteristics and survival of trisomy 18 in a medical center in Taipei, 1988–2004. Am J Med Genet A 140: 945–951 [DOI] [PubMed] [Google Scholar]
- Hsu H-F, Hou J-W (2007) Variable expressivity in Patau syndrome is not all related to trisomy 13 mosaicism. Am J Med Genet A 143: 1739–1748 [DOI] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA (1984) Trisomy in man. Annu Rev Genet 18: 69–97 [DOI] [PubMed] [Google Scholar]
- Roizen NJ, Patterson D (2003) Down's syndrome. Lancet 361: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Glasson EJ, Sullivan SG, Hussain R, Petterson BA, Montgomery PD, Bittles AH (2002) The changing survival profile of people with Down's syndrome: implications for genetic counselling. Clin Genet 62: 390–393 [DOI] [PubMed] [Google Scholar]
- Satge D, Sommelet D, Geneix A, Nishi M, Malet P, Vekemans M (1998) A tumor profile in Down syndrome. Am J Med Genet 78: 207–216 [PubMed] [Google Scholar]
- Ganmore I, Smooha G, Izraeli S (2009) Constitutional aneuploidy and cancer predisposition. Hum Mol Genet 18: R84–R93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Heim S, Mandahl N (1990) Trisomy 21 in neoplastic cells. Am J Med Genet Suppl 7: 262–266 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang H, Wang H, Chen Z, Jin J (2009) Trisomy 21 in patients with acute leukemia. Am J Hematol 84: 193–194 [DOI] [PubMed] [Google Scholar]
- Hama A et al. (2008) Acute megakaryoblastic leukaemia (AMKL) in children: a comparison of AMKL with and without Down syndrome. Br J Haematol 140: 552–561 [DOI] [PubMed] [Google Scholar]
- Sainati L, Leszl A, Surace C, Perilongo G, Rocchi M, Basso G (2002) Fluorescence in situ hybridization improves cytogenetic results in the analysis of hepatoblastoma. Cancer Genet Cytogenet 134: 18–20 [DOI] [PubMed] [Google Scholar]
- Betts DR, Koesters R, Plüss HJ, Niggli FK (1997) Routine karyotyping in Wilms tumor. Cancer Genet Cytogenet 96: 151–156 [DOI] [PubMed] [Google Scholar]
- Korbel J et al. (2009) The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci USA 106: 12031–12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT (1995) A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet 11: 177–184 [DOI] [PubMed] [Google Scholar]
- Minami T et al. (2004) Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J Biol Chem 279: 50537–50554 [DOI] [PubMed] [Google Scholar]
- Ryeom S, Baek K-H, Rioth MJ, Lynch RC, Zaslavsky A, Birsner A, Yoon SS, McKeon F (2008) Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell 13: 420–431 [DOI] [PubMed] [Google Scholar]
- O'Doherty A et al. (2005) An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309: 2033–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K-H et al. (2009) Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459: 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LE et al. (2010) Tumour angiogenesis is reduced in the Tc1 mouse model of Down's syndrome. Nature 465: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256: 668–670 [DOI] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH (2008) Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature 451: 73–75 [DOI] [PubMed] [Google Scholar]
- Yang A, Reeves RH (2011) Increased survival following tumorigenesis in Ts65Dn mice that model Down syndrome. Cancer Res 71: 3573–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Shih T, Schmitt E, Santiago S, Reilly K, McLaughlin M, Bronson R, Jacks T (1999) Mouse models of tumor development in neurofibromatosis type 1. Science 286: 2172–2176 [DOI] [PubMed] [Google Scholar]
- Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF (1999) Mouse tumor model for neurofibromatosis type 1. Science 286: 2176–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S et al. (2004) Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36: 1159–1161 [DOI] [PubMed] [Google Scholar]
- Snape K et al. (2011) Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet 43: 527–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaja A, Vendrell T, Smeets D, Sarret E, Gili T, Català V, Mediano C, Scheres JM (2001) Variegated aneuploidy related to premature centromere division (PCD) is expressed in vivo and is a cancer-prone disease. Am J Med Genet 98: 216–223 [DOI] [PubMed] [Google Scholar]
- Callier P et al. (2005) Microcephaly is not mandatory for the diagnosis of mosaic variegated aneuploidy syndrome. Am J Med Genet A 137: 204–207 [DOI] [PubMed] [Google Scholar]
- Plaja A, Perez C, Miró R (2001) Chromosome aneuploidy and cancer: lessons from a chromosomal instability syndrome. Cancer Genet Cytogenet 131: 144–145 [DOI] [PubMed] [Google Scholar]
- Schvartzman J-M, Sotillo R, Benezra R (2010) Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 10: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA et al. (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A (2012) Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2090–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P (2009) Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol 185: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS et al. (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409: 355–359 [DOI] [PubMed] [Google Scholar]
- Hernando E et al. (2004) Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802 [DOI] [PubMed] [Google Scholar]
- Diaz-Rodriguez E, Sotillo R, Schvartzman J-M, Benezra R (2008) Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA 105: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, van Ree JH, van Deursen JM (2008) Whole chromosome instability and cancer: a complex relationship. Trends Genet 24: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman J-M, Socci ND, Benezra R (2010) Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K et al. (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T et al. (2012) Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G et al. (2008) Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleihues P, Schäuble B, Hausen zur A, Estève J, Ohgaki H (1997) Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol 150: 1–13 [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT (2004) Triggering p53 after cytokinesis failure. J Cell Biol 165: 607–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Pellman D (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5: 45–54 [DOI] [PubMed] [Google Scholar]
- Dalton WB, Yu B, Yang VW (2010) p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene 29: 1929–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozery-Flato M, Linhart C, Trakhtenbrot L, Izraeli S, Shamir R (2011) Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol 12: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Gottesman MM (2002) Mechanisms of cancer drug resistance. Annu Rev Med 53: 615–627 [DOI] [PubMed] [Google Scholar]
- Goss PE, Chambers AF (2010) Does tumour dormancy offer a therapeutic target? Nat Rev Cancer 10: 871–877 [DOI] [PubMed] [Google Scholar]
- Druker BJ et al. (2001) Efficacy and safety of a specific inhibitor of the BCR–ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344: 1031–1037 [DOI] [PubMed] [Google Scholar]
- Tuveson D, Willis N, Jacks T, Griffin J, Singer S, Fletcher C, Fletcher J, Demetri G (2001) STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene 20: 5054–5058 [DOI] [PubMed] [Google Scholar]
- Baselga J, Norton L, Albanell J, Kim Y, Mendelsohn J (1998) Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res 58: 2825–2831 [PubMed] [Google Scholar]
- Knight ZA, Lin H, Shokat KM (2010) Targeting the cancer kinome through polypharmacology. Nat Rev Cancer 10: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB et al. (2008) Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res 14: 8302–8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath EI et al. (2008) A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin Cancer Res 14: 7940–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman SR, Gundacker H, Appelbaum FR, Slovak ML, Southwest Oncology Group (2002) Impact of trisomy 8 (+8) on clinical presentation, treatment response, and survival in acute myeloid leukemia: a Southwest Oncology Group study. Blood 100: 29–35 [DOI] [PubMed] [Google Scholar]