Receptor biologists have traditionally focused on receptors that are physiologically coupled to inductive responses such as gene expression, cytokinesis, secretion, or entry into the cell cycle. It has generally been assumed that such inductive responses were terminated by ligand decay or receptor desensitization, which lead to cessation of signaling. While such mechanisms are clearly operative, a number of recent experiments have demonstrated that inhibitory receptors exist that function to attenuate inductive signals and further indicate that these receptors play a very important regulatory role in biology. Relatively few inhibitory receptors have been defined in part because the effects of these receptors can only be detected against the backdrop of an inductive signal. However, this situation is rapidly changing as recent pioneering studies have provided new approaches to isolation of such receptors. Three reports published in recent months (1–3), one by Kubagawa et al. in the Proceedings (1), have identified two new extended families of cell surface proteins that may function as inhibitory receptors. These findings are discussed below in the context of known inhibitory receptors and their mode of action (Table 1).
Table 1.
The growing inhibitory receptor family
| Receptor/structure | Ligand | Cellular expression | Function | Likely effectors |
|---|---|---|---|---|
| FcγRIIB | IgG | B(T?)cells/mast cells, etc. | Inhibition of antigen/Fc receptor signaling | SHP-1, SHP-2, SHIP |
| CD22 | α2,6 sialogly- coproteins | B cells | Inhibition of antigen receptor signaling | SHP-1 |
| Killer inhibitory receptors (KIRs) (hp58/p70, mLy49, etc. >11 members) | MHC class I | T cells/NK cells | Inhibition of antigen/Fc receptor signaling | SHP-1, SHP-2 |
| CTLA4 | CD80, CD86 | T cells | Inhibition of antigen receptor signaling | SHP-2 |
| gp49B1 | ? | Mast cells/Nk cells | Inhibition of antigen/Fc receptor signaling | ? |
| PIR-B | ? | B cells | ?? | ? |
| Signal-regulatory Proteins (SIRPs) (SHPS-1, etc., >15 members) | ? | “Ubiquitous” | Inhibition of PDGF, EGF and insulin receptor signaling | SHP-1, SHP-2 |
One of the first important insights regarding mechanisms of inhibitory signaling came from observations that signal transduction by tyrosine kinase-coupled receptors can be terminated by receptor association with phosphotyrosine phosphatases. A notable example is the termination of erythropoietin receptor signaling as a consequence of receptor phosphotyrosine binding to the hematopoietic lineage restricted phosphatase SHP-1 (previously known as HCP, SHPTP1, PTP1C, and SHP) (4). It was subsequently shown that FcγRIIB, a receptor for immunoglobulin G constant (Fc) regions known to mediate inhibition of antigen receptor activation of B cells, could recruit SHP-1 as well as the closely related and ubiquitiously expressed phosphotyrosine phosphatase SHP-2 (previously known as SHPTP2, PTP1D, and Syp) to the receptor complex upon coligation with the antigen receptor (5, 6). SHP-1 expression was found to be necessary for FcγRIIB inhibition of antigen receptor activation of B cell proliferation. Based in part on these findings, the role of these phosphatases in inhibitory signaling by CD22 (7), the newly described killer inhibitory receptors (KIRs) (8, 9), and CTLA4 (10) was explored. Activated receptors and/or phosphopeptides containing the cytoplasmic sequences of these molecules were found to bind SHP-1, SHP-2 or both phosphatases. More recently, Fujioka et al. (2) and subsequently Kharitonenkov et al. (3) isolated and cloned potential new inhibitory receptors based on their ability to coimmunoprecipitate with SHP-2. Fujioka et al. (2) isolated a protein they named SHP substrate 1 (SHPS-1) from v-src-transformed rat fibroblasts and subsequently cloned human and mouse SHPS-1 homologues. Using the same strategy Kharitonenkov et al. (3) isolated a family of proteins they named SIRPs (signal-regulatory proteins) of which SHPS-1 appears to be a member. SIRPs appear to be a broadly expressed multigene family with more than 15 members. SIRPα1 was shown to be tyrosine phosphorylated following cell stimulation with epidermal growth factor, insulin, or platelet-derived growth factor. Similarly, SHPS-1 was shown to be phosphorylated upon stimulation with insulin, serum, or lysophosphatidic acid. In their phosphorylated state, SIRPα1 and SHPS-1 bind SHP-2 and SHP-1 and act as SHP substrates. Overexpression of SIRPα1 led to decreased responsiveness to epidermal growth factor, insulin, and platelet-derived growth factor, suggesting that SIRPs have inhibitory function and indicating that multiple receptor-tyrosine kinase coupled pathways are SIRP targets. In the most recent chapter of this quest for novel inhibitory receptors, Kubagawa et al. (1) have cloned genes encoding two novel surface molecules, PIR-A and PIR-B, expressed on B lymphocytes and myeloid lineage cells, based on homology to the mouse Fcα receptor.
The supposition that PIR-B and SIRP are receptors is based on their content of extracellular domains and the fact that these extracellular domains exhibit sequence variability consistent with their being determinants of ligand specificity. Although the ligand specificity of SIRPs, PIR-A and PIR-B, are unknown, activation of SIRP phosphorylation by growth factors and lysophosphatidic acid (2, 3) is most consistent with the possibility that the SIRP ligands are the respective receptors themselves. In this regard the situation is similar to CD22, a known inhibitory receptor that is rapidly phosphorylated upon B cell antigen receptor aggregation and binds SHP-1 (7). Thus a component of the B cell antigen receptor (BCR) complex may be a CD22 ligand.
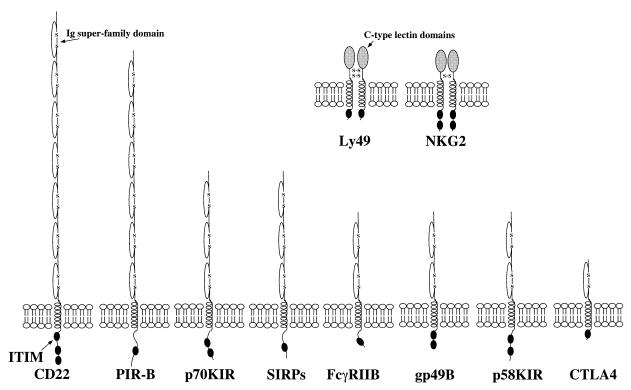
The inhibitory receptors defined thus far fall into two structural families (Fig. 1). Most are monomeric proteins that contain multiple immunoglobulin super-family (IgSF) domains in their extracellular regions. Surprisingly, some of the KIRs are homodimers containing c-type lectin domains in their extracellular regions. Thus, KIR can recognize its ligand, major histocompatiblity complex (MHC) class I molecules, using very different structures (11). All of the inhibitory receptors contain single transmembrane spanning regions and cytoplasmic tails ranging from 35 to 178 amino acids in length.
Figure 1.
Schematic diagram of inhibitory receptors.
The contextual sequence surrounding the sites of tyrosine phosphorylation of the inhibitory receptors has proven to be one of the best predictors of the ability of candidate receptors to associate with SHP-1 and SHP-2 and to function in an inhibitory capacity. SHP-1 binding activity has been localized to specific tyrosines in FcγRIIB1, p58.183 (tyrosine 1 and 2), p58.EB6, p70 (tyrosine 1 and 2), and CD22 (tyrosine 2, 5, and 6) shown in Fig. 2 (7–9). Analysis of these sequences suggests a consensus for SHP-1 SH2 binding that is I/VxYxxL. This motif has been referred to as the immunoreceptor tyrosine-based inhibitory motif or ITIM (5, 12). This consensus is also found in gp49B, SIRPs, Ly49, and NG2A. Importantly, Vely et al. (13) have recently shown that mutation of the I/V position in the motif disrupts its ability to bind SHP-1 and SHP-2. The requirement for a hydrophobic residue in this position is consistent with the occurrence of a hydrophobic cleft in the c-SH2 domain of SHP-2 that may be properly positioned to interact with Y-2 residue when the phosphotyrosine binding site is occupied (14). This feature may be responsible for the restricted SH2 domain binding activity of phosphorylated ITIM peptides (5).
Figure 2.
ITIM sequences in inhibitory receptors.
Among the inhibitory receptors, only CTLA4 does not contain a canonical ITIM motif. Unlike other members of the family, CTLA4 reportedly binds to SHP-2 but not SHP-1 (10). Further, tyrosine phosphorylation of CTLA4 has not been formally demonstrated. Thus, the mode of action of CTLA4 may be different from more conventional ITIM-containing receptors.
Relatively little is known regarding the mechanisms by which ITIM-containing inhibitory receptors mediate their inhibitory effects. The best studied model is FcγRIIB. Co-crosslinking of FcγRIIB with antigen receptors (BCR) on B cells or FcɛRI on mast cells inhibits signaling by BCR and FcɛRI (5, 12, 15). The FcγRIIB ITIM binds to SHP-1, SHP-2, and SHIP (5, 6, 16, 17). SHIP is an SH2 containing phosphatidylinositol (3,4,5)P35′ inositol phosphatase (17). In coimmunoprecipitation experiments, association of activated FcγRIIB with SHIP is more easily demonstrated than association with SHP-1 or SHP-2 leading to the suggestion that SHIP may be the primary mediator of inhibitory FcγRIIB function. Interestingly, B cells from SHP-1 knockout mice exhibit defective FcγRIIB signaling while mast cells from these mice do not. Further, other ITIM-containing receptors such as, p58 and p70 KIRs, do not bind to SHIP, and KIR signaling is blocked by dominant negative mutants of SHP-1 (13). Minimally one must conclude that multiple effectors—e.g., SHP-1, SHP-2, and perhaps SHIP—must be capable of mediating the inhibitory signals transduced via ITIMs. The pathway that is operative may depend on relative cellular expression of effectors and other as yet undefined parameters of cell phenotype.
The downstream targets of the effectors of this inhibitory receptor family are similarly poorly defined. Involvement of KIR in the context of natural killer cell activation leads to reduced phosphorylation of multiple proximal signaling molecules including receptor ζ chains, ZAP-70, and PLCγ, and inhibition of InsP3 production and calcium mobilization (18, 19). CTLA4 signaling reportedly leads to reduced T cell antigen receptor mediated phosphorylation of multiple early effectors but Shc is implicated as a specific SHP-2 substrate (9). In B cells, FcγRIIB co-crosslinking with antigen receptors leads to a selective reduction in phosphorylation of CD19, an accessory molecule involved in PI3-kinase activation by the antigen receptor (20). This failed phosphorylation or dephosphorylation of CD19 and resultant failed PI3-kinase activation appears causally linked to attenuated inositol trisphosphate generation and calcium mobilization responses (21). This is suggested by findings that the antigen receptor mediated response of B cells from CD19 knockout mice and normal B cells are pretreated with the PI3-kinase inhibitors wortmannin or Ly 294002 is inhibited equivalently to that seen in normal B cells following FcγRIIB co-crosslinking with BCR. Interestingly, FcγRIIB coligation with antigen receptors also leads to reduced p21ras activation and a modest reduction in PLCγ phosphorylation (22, 23). The relationship of these effects and their causality in FcγRIIB-mediated reduction of proliferation is unknown. Finally, it is unclear whether these FcR effects are mediated by SHP-1, SHP-2, or SHIP or some as yet undefined effector. Interesting in this regard is a recent report by Musci et al. (24) in which membrane localization of SHP-1 is shown to inhibit T cell antigen receptor-mediated inositol trisphosphate production and Ca2+ mobilization without affecting PLCγ phosphorylation. Thus, most evidence supports a role for SHP-1 in mediating inhibitory signals in cells of the hematopoietic lineage. Although SHP-2 has been implicated as an important positive player inductive signaling by some growth factor receptors, it may be an inhibitory effector of SIRPs in nonhematopoietic lineage cells that express little or no SHP-1.
Recent reports by Kubagawa et al. (1), Fujioka et al. (2), and Kharitonenkov et al. (3) suggest that this family of inhibitory receptors is extensive, coupling a large number of ligands to cytoplasmic phosphatases—e.g., SHP-1 and SHP-2 that mediate inhibitory signaling. Elucidation of this receptor family, their ligands and their mode of signal transduction promises to be an interesting and challenging endeavor. Clearly, these receptors and coupled pathways may contain excellent targets for therapeutic intervention in a variety of pathologies.
References
- 1.Kubagawa H, Burrows P D, Cooper D A. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, Kasuga M. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. Nature (London) 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 4.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Cell. 1995;80:729–736. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 5.D’Ambrosio D, Hippen K L, Minskoff S A, Mellman I, Pani G, Siminovitch K A, Cambier J C. Science. 1995;268:293–297. doi: 10.1126/science.7716523. [DOI] [PubMed] [Google Scholar]
- 6.D’Ambrosio D, Fong D C, Cambier J C. Immunol Lett. 1996;54:77–82. doi: 10.1016/s0165-2478(96)02653-3. [DOI] [PubMed] [Google Scholar]
- 7.Doody G M, Justement L B, Delibrias C C, Matthews R J, Lin J, Thomas M L, Fearon D T. Science. 1995;260:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 8.Burshtyn D N, Scharenberg A M, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet J-P, Long E O. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olcese L, Lang P, Vely F, Cambiaggi A, Marguet D, Blery M, Hippen K L, Biassoni R, Moretta A, Moretta L, Cambier J C, Vivier E. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 10.Marengere L E M, Waterhouse P, Duncan G S, Mittrücker H-W, Feng G-F, Mak T W. Science. 1986;272:1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 11.Vivier, E. & Daëron, M. (1997) Immunol. Today, in press. [DOI] [PubMed]
- 12.Daeron M, Latour S, Malbec O, Espinosa E, Pina P, Pasmans S, Fridman W H. Immunity. 1995;3:635–646. doi: 10.1016/1074-7613(95)90134-5. [DOI] [PubMed] [Google Scholar]
- 13.Vely, F., Olivero, S., Olcese, L., Moretta, A., Damen, J. E., Liu, L., Krystal, G., Cambier, J. C., Daëron, M. & Vivier, E. (1997) Eur. J. Immunol., in press. [DOI] [PubMed]
- 14.Eck M J, Pluskey S, Trub T, Harrison S C, Shoelson S E. Nature (London) 1996;379:277–280. doi: 10.1038/379277a0. [DOI] [PubMed] [Google Scholar]
- 15.Daeron M, Malbec O, Latour S, Arock M, Fridman W H. J Clin Invest. 1995;95:577–585. doi: 10.1172/JCI117701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono M, Bolland S, Tempst P, Ravetch J V. Nature (London) 1996;383:263–265. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 17.Damen J E, Liu L, Rosten P, Humphries R K, Jefferson A B, Majerus P W, Krystal G. Proc Natl Acad Sci USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binstadt B A, Brumbaugh K M, Dick C J, Scharenberg A M, Williams B L L, Colonna M, Lanier L L, Kinet J P, Abraham R T, Leibson P J. Immunity. 1996;5:629–638. doi: 10.1016/s1074-7613(00)80276-9. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman D A, Schoon R A, Robertson M J, Leibson P J. Proc Natl Acad Sci USA. 1995;92:6484–6488. doi: 10.1073/pnas.92.14.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiener P A, Lioubin M N, Rohrschneider L R, Ledbetter J A, Nadler S G, Diegel M L. J Biol Chem. 1997;272:3838–3843. doi: 10.1074/jbc.272.6.3838. [DOI] [PubMed] [Google Scholar]
- 21.Hippen, K. L., Buhl, A. M., D’Ambrosio, D., Nakamura, K., Persin, C. & Cambier, J. C. (1997) Immunity, in press. [DOI] [PubMed]
- 22.Tridandapani S, Chacko G W, Van Brockly J R, Coggeshall K M. J Immunol. 1997;158:1125–1132. [PubMed] [Google Scholar]
- 23.Sarkar S, Schlottmann K, Cooney D, Coggeshall K M. J Biol Chem. 1996;271:20182–20186. doi: 10.1074/jbc.271.33.20182. [DOI] [PubMed] [Google Scholar]
- 24.Musci M A, Beaves S L, Ross S E, Yi T, Koretzky G A. J Immunol. 1997;158:1565–1571. [PubMed] [Google Scholar]