Ixodes and Dermacentor ticks harbor Borrelia, Anaplasma/Ehrlichia, Bartonella, and Babesia species.
Keywords: Ixodes persulcatus, Dermacentor reticulatus, Borrelia burgdorferi sensu lato, Anaplasma phagocytophilum, Ehrlichia muris, Bartonella, Babesia canis canis, nested PCR, sequencing, research
Abstract
Ixodes persulcatus (n = 125) and Dermacentor reticulatus (n = 84) ticks from Western Siberia, Russia, were tested for infection with Borrelia, Anaplasma/Ehrlichia, Bartonella, and Babesia spp. by using nested polymerase chain reaction assays with subsequent sequencing. I. persulcatus ticks were infected with Borrelia burgdorferi sensu lato (37.6% ± 4.3% [standard deviation]), Anaplasma phagocytophilum (2.4% ± 1.4%), Ehrlichia muris (8.8% ± 2.5%), and Bartonella spp. (37.6% ± 4.3%). D. reticulatus ticks contained DNA of B. burgdorferi sensu lato (3.6% ± 2.0%), Bartonella spp. (21.4% ± 4.5%), and Babesia canis canis (3.6% ± 2.0%). Borrelia garinii, Borrelia afzelii, and their mixed infections were observed among I. persulcatus, whereas B. garinii NT29 DNA was seen in samples from D. reticulatus. Among the I. persulcatus ticks studied, no Babesia spp. were observed, whereas B. canis canis was the single subspecies found in D. reticulatus.
Ticks are second only to mosquitoes as vectors of bacterial, viral, and protozoan agents (1). Among tickborne bacteria, extracellular spirochetes of the genus Borrelia are widely spread and most studied. Some of these, those that belong to the Borrelia burgdorferi sensu lato complex, are causative agents of Lyme borreliosis (1,2). Other known pathogenic bacteria transmitted by ticks are intracellular alpha proteobacteria, which includes the families Anaplasmataceae, Bartonellaceae, and Rickettsiaceae (3). Members of the genera Anaplasma and Ehrlichia, from the family Anaplasmataceae, infect mainly monocytes and granulocytes and cause human and animal anaplasmoses and ehrlichioses (4). Bacteria of the genus Bartonella infect erythrocytes and endothelial cells. Different species of Bartonella are the etiologic agents of cat-scratch disease, trench fever, and Carrión disease (5,6). The tickborne protozoa of the genus Babesia reproduce in erythrocytes, thus causing babesiosis among humans as well as wild and domestic animals (7).
Prevalent tick species for forest-steppe zones of Western Siberia, Russia, are taiga ticks (Ixodes persulcatus Schulze) and meadow ticks (Dermacentor reticulatus) (Acarina: Ixodidae) (1,2,8). They parasitize many ground-foraging bird species and virtually all the terrestrial mammals (8); both species are able to feed on humans (8,9) and transmit different tickborne infections (2).
The I. persulcatus habitat is the southern part of the forest zone of Eurasia (1,2). Until 1987, only tickborne encephalitis virus was thought to be associated with taiga ticks, but extensive studies have shown their competence in the transmission of pathogenic spirochetes, Borrelia garinii and Borrelia afzelii (2). Recently, Anaplasma/Ehrlichia was found in I. persulcatus ticks (10). The main Ehrlichia species found in I. persulcatus ticks is a recently characterized species, Ehrlichia muris, isolated from a wild mouse in Japan (11–13). The etiologic agent of human granulocytic anaplasmosis, Anaplasma phagocytophilum, has also been found in I. persulcatus ticks (14–16). Infection of ixodid ticks with Bartonella spp. has recently been described in the United States (17), Europe (18,19), and Western Siberia (20). Infection of I. persulcatus with Babesia microti pathogenic for immunocompromised humans has been shown by polymerase chain reaction (PCR) with genus- and species-specific primers (21), but nucleotide sequences of the specific PCR products remain unknown.
The second tick species, D. reticulatus, inhabits meadows and pastures (2), as well as near suburban areas from Europe to central Asia, but not taiga and dry steppes (22). D. reticulatus is well known as the vector of a canine pathogen, Babesia canis canis (23); Rickettsia spp (13), Francisella tularensis, and Coxiella burnetii were also found in this tick species (1). Borrelia spp. was also detected in different Dermacentor species, including D. reticulatus, by means of PCR (9) and an indirect immunofluorescence assay (24). Infection of D. reticulatus with Anaplasma/Ehrlichia and Bartonella species was previously unknown in spite of the detection of bacterial DNA in other Dermacentor species (17,25). Little or nothing was known of the genetic variability of the tickborne pathogens in ixodid ticks from Western Siberia (2); consequently, the aim of the present study was to study prevalence and genetic diversity of Borrelia, Anaplasma/Ehrlichia, Bartonella, and Babesia among I. persulcatus and D. reticulatus ticks in Western Siberia, Russia.
Materials and Methods
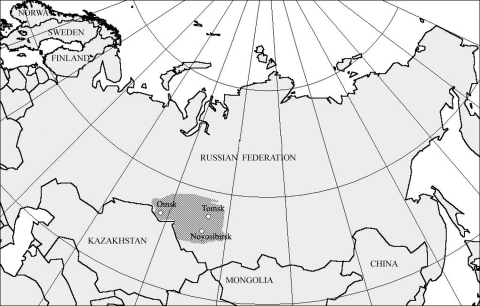
Unfed adult I. persulcatus ticks were collected by flagging of lower vegetation in different suburban places of mixed aspen-birch and pine forests of Novosibirsk (55°N, 83°E) (115 ticks) and Tomsk regions (56°N, 85°E) (12 ticks) (Figure 1) in May and June of 2003 and 2004. Questing imago of D. reticulatus ticks were collected by flagging in different locations of river valley and forest-steppe zones of Novosibirsk (72 ticks) and Omsk regions (55°N, 73°E) (15 ticks) from May to June of 2003 and 2004 (Figure 1). Nucleic acids were isolated by lysis of 127 individual I. persulcatus and 87 D. reticulatus ticks in guanidine thiocyanate followed by deproteinization with phenol-chloroform and precipitation with isopropanol.
Figure 1.
Areas where ticks were collected. Omsk, Tomsk, and Novosibirsk regions are shaded.
PCR Assay
To prevent contamination, we performed DNA isolation, PCR master mix assembly, and amplifications in separate rooms. Aerosol-free pipette tips were also used at each stage. We included negative control reactions with bidistillated water in each experiment at both steps of nested PCR. All reactions were performed in 20 μL reaction mixture containing 67 mmol/L Tris-HCl (pH 8.9), 16.6 mmol/L (NH4)2SO4, 2 mmol/L MgCl2, 0.01% Tween-20, 200 μmol/L each dNTP, 5% glycerol, 0.5 μmol/L specific primers, 2 U Taq DNA polymerase (Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences), and 2 μL tested DNA. Amplification was performed in a Tercic Thermal Cycler (DNA Technology, Moscow, Russia). For the inner reactions, 2 μL of the outer PCR products were added into the reaction mixture. PCR fragments were visualized under UV irradiation after electrophoresis in agarose gels containing ethidium bromide. To control DNA isolation from ticks, we performed PCR on an aliquot of purified DNA with the following universal primers targeted to 18S rRNA gene: forward 5´-AACCTGGTTGATCCTGCCAGTAGTCAT-3´ and reverse 5´-GAATGATCCTTCCGCAGGTTCACCTAC-3´ (26).
To detect tickborne infectious agents, we used nested PCR with specific primers both previously described (6,27,28) and designed in our study. Multiple sequence alignment of nucleotide sequences available in GenBank (http://www.ncbi.nlm.nih.gov) for each tickborne pathogen was performed by using ClustalW (http://www.ebi.ac.uk/clustalw/) (29). The desired specificity of selected primers and absence of cross-reactions were confirmed by BlastN homology search (http://www.ncbi.nlm.nih.gov/BLAST/). Formation of intra- and intermolecular primer dimers was reduced by using OLIGOS (http://www.basic.northwestern.edu/biotools/oligocalc.html).
PCR Detection of Borrelia-specific DNA
B. burgdorferi sensu lato DNA was detected by means of nested PCR with primers specific to conservative regions of 5S and 23S rRNA genes to amplify variable intergenic spacer (27). Primers for outer PCR were designed by aligning 5 GenBank nucleotide sequences of rRNA gene clusters for 3 species belonging to B. burgdorferi sensu lato complex, including B. burgdorferi sensu stricto, B. garinii, and Borrelia lusitaniae. Other criteria for primer design included the absence of possible cross-reactions with other genera. Outer reactions were performed with NC1 (5´-CCTGTTATCATTCCGAACACAG-3´) and NC2 (5´-TACTCCATTCGGTAATCTTGGG-3´) primers (35 cycles of 60 s at 94°C, 30 s at 58°C, and 30 s at 72°C). Inner reactions were carried out as previously described (27). DNA isolated from B. burgdorferi sensu stricto (strain B31), B. afzelii (strain Ip-21), and B. garinii (strains T6, 2, and 12) was used as positive control. Molecular typing of the PCR-positive samples was performed by using sequencing and restriction fragment length polymorphism analysis (27) by hydrolysis of the PCR products with the Tru9I restriction endonuclease (SibEnzyme, Novosibirsk, Russia) (isoschizomer MseI) with subsequent electrophoresis in 15% polyacrylamide gel.
PCR Detection of Anaplasma- and Ehrlichia-specific DNA
For Anaplasma/Ehrlichia detection, specific primers were designed by comparing 15 nucleotide sequences of 16S rRNA gene from 12 species (A. phagocytophilum, A. bovis, A. platys, A. centrale, A. marginale, E. muris, E. ruminantium, E. ewingii, E. chaffeensis, E. canis, Wolbachia pipientis, Rickettsia rickettsii). Primers EHR1, EHR2, and EHR3 were identical to the 16S rRNA gene nucleotide sequences of Anaplasma/Ehrlichia species but differed from W. pipientis sequence and markedly distinguished from the sequence of R. rickettsia. Outer reactions were performed by using EHR1 (forward, 5´-GAACGAACGCTGGCGGCAAGC-3´) and EHR2 (reverse, 5´-AGTA(T/C)CG(A/G)ACCAGATAGCCGC-3´) primers and inner reactions with EHR3 (forward, 5´-TGCATAGGAATCTACCTAGTAG-3´) and EHR2 primers (35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C). DNA isolated from A. phagocytophilum and A. marginale from spleens of ill cattle were used as positive controls. For PCR-positive samples, nested reactions with A. phagocytophilum–specific primers HGE1 (forward, 5´-CGGATTATTCTTTATAGCTTGC-3´) and HGE2 (reverse, 5´-CTTACCGAACCGCCTACATG-3´) were carried out in the same conditions.
PCR Detection of Bartonella-specific DNA
For Bartonella DNA detection, nested PCR with primers corresponding to groEL gene (6) was used. For outer reactions, primers BH1 (forward, 5´-GAAGAAACAACTTCTGACTATG-3´) and BH4 (reverse, 5´-CGCACAACCTTCACAGGATC-3´) were designed by aligning 31 nucleotide sequences of 13 Bartonella species, including Bartonella henselae, Bartonella quintana, Bartonella alsatica, Bartonella birtlesii, Bartonella bacilliformis, Bartonella capreoli, Bartonella doshiae, Bartonella grahamii, Bartonella koehlerae, Bartonella schoenbuchensis, Bartonella taylorii, Bartonella tribocorum, and Bartonella vinsonii. The outer reactions were carried out in 45 cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C. Both primer structures (HSP1, HSP2, and HSP4) and PCR conditions (45 cycles of 30s at 94°C, 30 s at 58°C, and 45 s at 72°C) for inner seminested reactions in the presence of 3 primers were identical to those previously described (6). Full-length genomic DNA samples isolated from B. henselae and B. quintana were used as positive controls.
PCR Detection of Babesia-specific DNA
Babesia DNA was detected by means of nested PCR with primers specific to the 18S rRNA gene. Specific primer PiroA had been previously described (28). Other Piroplasmida-specific primers were designed by comparing the 18S rRNA gene nucleotide sequences of 11 species (B. canis canis, Babesia canis vogeli, Babesia canis rossi, Babesia odocoilei, Babesia divergens, Babesia caballi, Babesia gibsoni, B. microti, Theileria parva, T. equi, Plasmodium falciparum). The chosen primers corresponded to the sequences of most Piroplasmida species (including those listed above) but significantly differed from those of P. falciparum. Outer reactions were performed with BS1 (forward, 5´-GACGGTAGGGTATTGGCCT-3´) and PiroC (reverse, 5´-CCAACAAAATAGAACCAAAGTCCTAC-3´) primers (36 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min at 72°C) and inner reactions with PiroA (forward, 5´-ATTACCCAATCCTGACACAGGG-3´) according to Armstrong et al. (28) (with the single nucleotide transition A→T at position 2 from the 5´ end of the primer) and PiroC primers (36 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C). For subsequent sequencing, the 1,304-bp fragment was synthesized in PCR with BS1 and BS2 (reverse, 5´-ATTCACCGGATCACTCGATC-3´) primers (40 cycles of 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C).
B. canis canis DNA isolated from blood samples of a dog with clinical signs of babesiosis confirmed by microscopic examination (GenBank accession no. AY527064) and B. microti DNA from Clethrionomys rutilus blood (AY943958) (V. Rar, unpub. data) were used as positive controls.
Sequencing of PCR Products
The PCR products were purified after gel electrophoresis in 1.5%–2% agarose gels with GFX Columns (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's instructions. Nucleotide sequences of the PCR products were determined by using BigDye Terminator Cycle Sequencing Kit and the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) at the DNA Sequencing Centre of the Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia. For initial species identification, the nested PCR products were sequenced in 1 direction. Detailed confirmation for each genetic group was performed by sequencing with forward and reverse outer or inner primers as needed.
Nucleotide sequences of PCR products determined in this study were analyzed by BlastN and aligned with ClustalW (29). Phylogenetic analysis was performed with MEGA 3.0 software (30). We used the unweighted pair-group method with arithmetic mean (UPGMA) and neighbor-joining algorithms with the Kimura 2-parameter model to generate the distance matrix as well as maximum parsimony and minimal evolution with a heuristic search. Bootstrap analysis was performed with 1,000 replications. GenBank accession numbers for the sequences used in the phylogenetic analysis are shown in Figures 2–5.
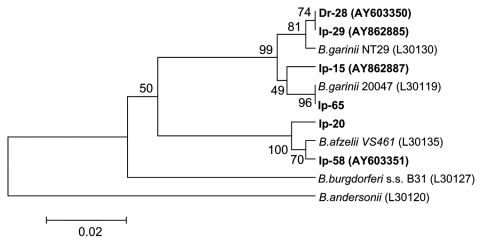
Figure 2.
Phylogenetic tree based on the Borrelia burgdorferi sensu lato 5S-23S rRNA intergenic spacer fragment sequences. Scale bar indicates an evolutionary distance of 0.02 nucleotides per position in the sequence. Borrelia andersonii was used as outgroup. Numbers above the branches indicate bootstrap support indexes. Samples isolated from Ixodes persulcatus (Ip) and Dermacentor reticulatus (Dr) in this research are in boldface.
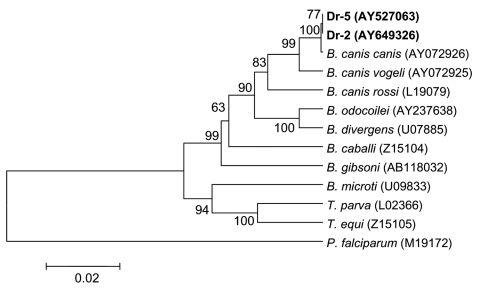
Figure 5.
Phylogenetic tree based on the Babesia 18S rRNA gene fragment sequences. Scale bar indicates an evolutionary distance of 0.02 nucleotides per position in the sequence. Plasmodium falciparum was used as outgroup. Numbers above the branches indicate bootstrap support indexes. Samples from Dermacentor reticulatus (Dr-2 and Dr-5) from this study are in boldface.
The nucleotide sequences determined in this study were deposited in GenBank under the following accession numbers: B. garinii, AY603350, AY862887, AY862885; B. afzelii, AY603351; A. phagocytophilum, AY587607; E. muris, AY587608; B. henselae, AY453166–AY453170; B. canis canis, AY527063, AY649326.
Results
Infection of I. persulcatus and D. reticulatus with 3 bacterial and 1 protozoan tickborne pathogens in Western Siberia, Russia were studied by nested PCR with genus-specific primers. To control DNA suitability for PCR analysis, we amplified the 18S rRNA gene in 125 of the 127 I. persulcatus samples tested and in 84 of the 87 D. reticulatus ticks studied. Therefore, the 5 samples in which we were unable to amplify tick DNA were excluded from further analysis. Both tick species contained Borrelia and Bartonella DNA, whereas Anaplasma/Ehrlichia DNA was detected only in I. persulcatus, and Babesia DNA was detected only in D. reticulatus ticks (Table).
Table. Prevalence of tickborne infectious agents in ticks in Western Siberia, Russia, 2003–2004.
| Pathogen | Prevalence (% ± SD)* |
|
|---|---|---|
| Ixodes persulcatus | Dermacentor reticulatus | |
| Borrelia spp. | 37.6 ± 4.3 | 3.6 ± 2.0 |
| Borrelia garinii NT29 | 18.4 ± 3.5 | 3.6 ± 2.0 |
| B. garinii 20047 | 8.8 ± 2.5 | 0 |
| Borrelia afzelii | 8.8 ± 2.5 | 0 |
| Mixed B. garinii NT29 + B. afzelii | 1.6 ± 1.1 | 0 |
| Anaplasma/Ehrlichia spp. | 11.2 ± 2.8 | 0 |
| Ehrlichia muris | 8.8 ± 2.5 | 0 |
| Anaplasma phagocytophilum | 2.4 ± 1.4 | 0 |
| Bartonella spp. | 37.6 ± 4.3 | 21.4 ± 4.5 |
| Babesia spp. | 0 | 3.6 ± 2.0 |
| Babesia canis canis | 0 | 3.6 ± 2.0 |
*SD, standard deviation.
In 37.6% ± 4.3% (standard deviation) of samples isolated from I. persulcatus and in 3.6% ± 2.0% of samples from D. reticulatus, DNA of B. burgdorferi sensu lato complex was found (Table). The nucleotide sequences of the 5S-23S intergenic spacer (216–237 bp) determined in this study were compared to those of other B. burgdorferi sensu lato sequences. The sequences from I. persulcatus ticks were placed in 2 clades of monophyletic origin, which corresponded to B. garinii and B. afzelii with excellent bootstrap support (99% and 100%, respectively), whereas samples from D. reticulatus were more closely related to B. garinii (Figure 2). Thirty-four PCR-positive samples contained DNA of B. garinii (23 samples of B. garinii group NT29 and 11 of B. garinii group 20047), and 11 samples contained B. afzelii DNA (Table). For 2 PCR-positive samples from I. persulcatus, the hydrolysis of the PCR products with the Tru9I restriction endonuclease resulted in 6 fragments of 108, 68, 57, 50, 38, and 20 bp that corresponded to a mixture of patterns C and D (27) and, consequently, 2 species, B. garinii group NT29 and B. afzelii (Table). Among samples obtained from D. reticulatus ticks, 3 contained B. garinii group NT29 DNA, but no other variants were found.
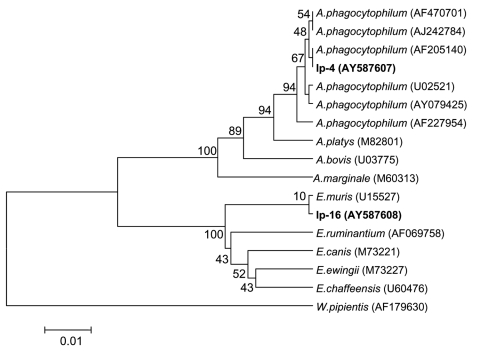
Anaplasma/Ehrlichia DNA was found in 14 I. persulcatus ticks but not in D. reticulatus ticks from different areas of Novosibirsk region. PCR with primers specific to A. phagocytophilum 16S rRNA gene showed the human pathogen DNA in 3 samples, Ip-4, Ip-45, and Ip-68, collected from different areas of Novosibirsk region. The nucleotide sequences of 629 bp of all these samples were identical to each other (GenBank accession no. AY587607) and to the known A. phagocytophilum sequence (AF205140). Nucleotide sequences from 11 other DNA samples were identical to each other (GenBank accession no. AY587608) and differed from E. muris DNA sequence (U15527) at the single position 91 (C→T). In a phylogenetic tree created by the UPGMA method, both A. phagocytophilum and E. muris sequences evidently formed the distinctive clusters (Figure 3).
Figure 3.
Phylogenetic tree based on the Anaplasma/Ehrlichia 16S rRNA gene fragment sequences. Scale bar indicates an evolutionary distance of 0.01 nucleotides per position in the sequence. Wolbachia pipientis was used as outgroup. Numbers above the branches indicate bootstrap support indexes. Samples from Ixodes persulcatus (Ip-4 and Ip-16) from this study are in boldface.
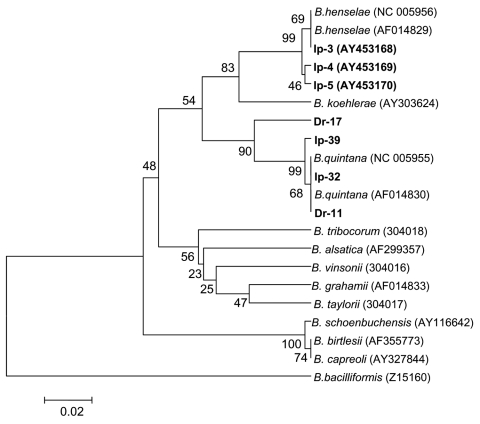
Bartonella DNA was detected by using nested PCR with primers that corresponded to the groEL gene in 47 I. persulcatus and 18 D. reticulatus ticks (Table). Comparative analysis of the groEL gene fragment nucleotide sequences of 190 bp showed 2 species, B. henselae and B. quintana, in both tick species. Part of the data is shown in Figure 4. The evidently separated 2 clades, B. henselae and B. quintana, were monophyletic with good statistical support (99% and 90%, respectively).
Figure 4.
Phylogenetic tree based on the Bartonella groEL gene fragment sequences. Scale bar indicates an evolutionary distance of 0.02 nucleotides per position in the sequence. Bartonella bacilliformis was used as outgroup. Numbers above the branches indicate bootstrap support indexes. Samples from Ixodes persulcatus (Ip) and Dermacentor reticulatus (Dr) from this study are in boldface.
Babesia DNA was found in 3 D. reticulatus ticks (Dr-2, Dr-4, Dr-5) by nested PCR and was not detected among I. persulcatus studied (Table). The nucleotide sequences of the Babesia 18S rRNA gene fragment of 1,203 bp determined in this study were similar to each other and to the single known full-length B. canis canis nucleotide sequence (GenBank accession no. AY072926). In the phylogenetic tree, nucleotide sequences from Dr-2 and Dr-5 as well as the B. canis canis sequence formed a distinctive cluster that was separated from other B. canis subspecies with excellent bootstrap support (Figure 5). Direct sequencing of the PCR fragment from the tick Dr-4 showed a mixture of nucleotide sequences with 2 undetermined bases at positions 609 and 610. Diluting DNA 10 times allowed us to determine 2 nucleotide sequences. The first was identical to those from Dr-2 and the second to a B. canis canis sequence found in canine blood from Croatia (AY072926).
UPGMA analysis produced phylogenetic trees (Figures 2–5) that were almost identical to the neighbor-joining trees and results of phylogenetic analysis with maximal parsimony and minimal evolution approaches (trees not shown).
Discussion
I. persulcatus is believed to maintain spirochetes transtadially and to transmit Borrelia to animals (31). Previously, the spirochetelike cells were isolated from I. persulcatus in Barbour-Stoenner-Kelly-H cultural medium (2) and were observed by indirect immunofluorescence assay (24). The nested PCR with subsequent sequencing showed that I. persulcatus contained both B. afzelii and B. garinii DNA (Table) as was previously shown (2,10,32,33). B. garinii appeared to be the prevalent species in I. persulcatus in Western Siberia (33). The B. garinii NT29 group is widely spread not only in Western Siberia but in the Russian Far East (GenBank accession no. AY429014, AY429015), Japan (34,35), and China (36). The nested PCR with subsequent sequencing allowed us to detect DNA of B. garinii group NT29 in 3.6% ± 2.0% of D. reticulatus ticks. Although Borrelia-specific DNA was detected in samples from D. reticulatus, numerous previous attempts to cultivate the living spirochetes were unsuccessful (2). Therefore, the ability of D. reticulatus to transmit Borrelia spp. remains unknown.
E. muris was the prevalent species among Anaplasma/Ehrlichia and was found in 8.8% ± 2.5% of I. persulcatus ticks in Western Siberia. This finding coincided with the E. muris prevalence (3%–13%) described in Baltic regions of Russia (12) and Siberia (16). The infection rate of I. persulcatus ticks with the human pathogen A. phagocytophilum (2.4% ± 1.4%) was significantly lower than the rate of infection with E. muris. In other regions, the infection rate of I. persulcatus with A. phagocytophilum varied from 1% to 4% in both China and Russia (14–16,37). The comparison of the Anaplasma 16S rRNA gene fragment nucleotide sequences (Figure 3) showed several genovariants of A. phagocytophilum. In I. persulcatus ticks, 4 types of sequences were found: 3 in China (GenBank accession nos. AY079425, AF205140, AF227954) and 1 in Korea (AF470701). All 3 nucleotide sequences of A. phagocytophilum determined in this study coincided with 1 genovariant from China (AF205140) found only in I. persulcatus (Figure 3) but not with the A. phagocytophilum isolated earlier in West Ural, Russia (GenBank accession no. AY094353) (38). No correlation was seen between genovariant and specific host or location.
Several tick species, such as deer ticks, I. persulcatus, I. ricinus, and I. pacificus, have been found to harbor Bartonella spp (17–20). Thus, PCR with primers specific to the 16S rRNA gene has shown Bartonella DNA in >70% of I. ricinus ticks in the Netherlands (18). Different Bartonella species, including B. henselae, have been detected in 19.2% of questing I. pacificus ticks in California by amplifying and sequencing the gltA gene fragment (17). More recently, B. henselae DNA has been found in 1.5% of I. ricinus ticks removed from humans in northwestern Italy (19) and in 38%–44% of I. persulcatus in Western Siberia (20). The high Bartonella infection rate of I. persulcatus in Western Siberia in 2003 and 2004 coincided with our observations from previous years (20). Moreover, both B. henselae and B. quintana were found not only in the 2 tick species studied (Figure 4) but also in Aedes mosquitos (O. Morozova, unpub. data). Only 2 human pathogens, B. henselae and B. quintana, were found in ixodid ticks in Siberia, despite sample collection for 4 years and phylogenetic analysis of all known Bartonella species (Figure 4).
We did not detect Babesia spp. in I. persulcatus. The only species of Babesia detected in D. reticulatus was B. canis canis, which causes babesiosis in dogs (7). D. reticulatus is the only known vector for B. canis canis (23,39). Comparison of the previously known Babesia canis canis 18S rRNA gene nucleotide sequences showed 3 genetic variants of B. canis canis in canine blood from Europe that differed at 2 variable positions 609 and 610 (26,40). Two of these variants were also seen in ticks in Novosibirsk. A new B. canis canis genetic variant that differed in a single nucleotide transition from those previously described was found. To our knowledge, this report is the first to identify nucleotide sequences of B. canis canis in ticks. B. microti was not found among tick samples studied, despite the presence of this human pathogen in small mammals in the same area (V. Rar, unpub. data).
When the 2 tick species were compared, I. persulcatus was more likely than D. reticulatus to be the host for tickborne bacterial infections examined in Western Siberia, Russia. The Borrelia, Anaplasma/Ehrlichia, and Bartonella infection rates for I. persulcatus exceeded those for D. reticulatus (Table). Moreover, Borrelia (10,33) and Bartonella (20) DNA from I. persulcatus could be easily detected in a single PCR, whereas nested PCR was required to detect DNA in samples from D. reticulatus. Neither Anaplasma nor Ehrlichia spp. were found in D. reticulatus. Conversely, Babesia spp. were detected only in D. reticulatus. The infection of unfed adult I. persulcatus and D. reticulatus ticks reflected transtadial transmission of tickborne infectious agents.
The experimentally observed and theoretically expected values of mixed infections of ticks with Borrelia, Ehrlichia, and Bartonella were statistically similar and consistent with independent distribution of these pathogens as previously reported (10). Thus, simultaneous coinfection with Borrelia, Anaplasma/Ehrlichia, and Bartonella found in 2.9% of I. persulcatus ticks slightly exceeded statistical probability of 1.8%. Further studies are required to establish the role of different tick species and biting arthropods as natural vectors of bacterial and protozoan agents.
Acknowledgments
We thank Natalya A. Titova for her help in DNA isolation from ticks and excellent technical assistance, Felipe Cabello for providing the B. burgdorferi sensu stricto strain B31, Edward I. Korenberg for providing the B. afzelii strain Ip-21, Dionysios Liveris for providing the A. phagocytophilum, Nikolay V. Rudakov for providing the A. marginale, and Michael Minnick for providing DNA isolated from B. henselae and B. quintana.
The study was supported in part by grant 02-01-113 from the Russian program "Vaccines of New Generation," grant N51 of the Program of the Integration in Basic Sciences of Siberian Branch of the Russian Academy of Sciences and grant "Fundamental Sciences to Medicine" of the Russian Academy of Sciences.
Biography
Dr Rar works in the Institute of Chemical Biology and Fundamental Medicine of the Siberian Branch of the Russian Academy of Sciences. Her research interests include molecular epidemiology, tickborne infections, and natural transmission cycles.
Footnotes
Suggested citation for this article: Rar VA, Fomenko NV, Dobrotvorsky AK, Livanova NN, Rudakova SA, Fedorov EG, et al. Tickborne pathogen detection, Western Siberia, Russia. Emerg Infect Dis [serial on the Internet]. 2005 Nov [date cited]. http://dx.doi.org/10.3201/eid1111.041195
References
- 1.Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. 2001;32:897–928. 10.1086/319347 [DOI] [PubMed] [Google Scholar]
- 2.Korenberg EI, Kovalevsky YV, Gorelova NB. Ecology of Borrelia burgdorferi sensu lato in Russia. In: Gray G, Kahl O, Lane RS, editors. Lyme borreliosis epidemiology and control. Oxford (UK): CAB International; 2002. p. 175–200. [Google Scholar]
- 3.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent' as subjective synonyms of Ehrlichia phagocytophilum. Int J Syst Evol Microbiol. 2001;51:2145–65. 10.1099/00207713-51-6-2145 [DOI] [PubMed] [Google Scholar]
- 4.Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacomo V, Kelly PJ, Raoult D. Natural history of Bartonella infections (an exception to Koch's postulate). Clin Diagn Lab Immunol. 2002;9:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeaiter Z, Fournier PE, Raoult D. Genomic variation of Bartonella henselae strains detected in lymph nodes of patients with cat scratch disease. J Clin Microbiol. 2002;40:1023–30. 10.1128/JCM.40.3.1023-1030.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homer MJ, Aguilar-Delfin I, Telford SR, Krause PJ, Persing DH. Babesiosis. Clin Microbiol Rev. 2000;13:451–69. 10.1128/CMR.13.3.451-469.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippova NA. The taiga tick Ixodes persulcatus (Acarina, Ixodidae). Morphology, systematics, ecology, medical importance. Leningrad (Russia): Nauka; 1985. [Google Scholar]
- 9.Hubbard MJ, Baker AS, Cann KJ. Distribution of Borrelia burgdorferi s.l. spirochaete DNA in British ticks (Argasidae and Ixodidae) since the 19th century, assessed by PCR. Med Vet Entomol. 1998;12:89–97. 10.1046/j.1365-2915.1998.00088.x [DOI] [PubMed] [Google Scholar]
- 10.Morozova OV, Dobrotvorsky AK, Livanova NN, Tkachev SE, Bakhvalova VN, Beklemishev AB, et al. PCR Detection of Borrelia burgdorferi sensu lato, tick-borne encephalitis virus and human granulocytic ehrlichiosis agent in Ixodes persulcatus ticks from Western Siberia, Russia. J Clin Microbiol. 2002;40:3802–4. 10.1128/JCM.40.10.3802-3804.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen B, Rikihisa Y, Mott J, Fuerst PA, Kawahara M, Suto C. Ehrlichia muris sp. nov., identified on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int J Syst Bacteriol. 1995;45:250–4. 10.1099/00207713-45-2-250 [DOI] [PubMed] [Google Scholar]
- 12.Alekseev AN, Dubinina HV, van de Pol I, Schouls LM. Identification of Ehrlichia spp. and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J Clin Microbiol. 2001;39:2237–42. 10.1128/JCM.39.6.2237-2242.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shpynov S, Fournier PE, Rudakov N, Tankibaev M, Tarasevich I, Raoult D. Detection of a Rickettsia closely related to Rickettsia aeschlimannii, "Rickettsia heilongjiangensis," Rickettsia sp. strain RpA4, and Ehrlichia muris in ticks collected in Russia and Kazakhstan. J Clin Microbiol. 2004;42:2221–3. 10.1128/JCM.42.5.2221-2223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao WC, Zhao QM, Zhang PH, Yang H, Wu XM, Wen BH, et al. Prevalence of Anaplasma phagocytophila and Borrelia burgdorferi in Ixodes persulcatus ticks from northeastern China. Am J Trop Med Hyg. 2003;68:547–50. [DOI] [PubMed] [Google Scholar]
- 15.Semenov AV, Alekseev AN, Dubinina EV, Kaufmann U, Jensen RM. Detection of the genotypic heterogeneity of Ixodes persulcatus Schulze (Acari: Ixodidae) of the north-west region of Russia and characteristics of distribution of tick-borne pathogens causing Lyme disease and Ehrlichia infections in various genotypes [article in Russian]. Med Parazitol (Mosk). 2001;3:11–5. [PubMed] [Google Scholar]
- 16.Shpynov SN, Rudakov NV, Iastrebov VK, Leonova GN, Khazova TG, Yegorova NV, et al. New evidence for the detection of Ehrlichia and Anaplasma in Ixodes ticks in Russia and Kazakhstan [article in Russian]. Med Parazitol (Mosk). 2004;2:10–4. [PubMed] [Google Scholar]
- 17.Chang CC, Chomel BB, Kasten RW, Romano V, Tietze N. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J Clin Microbiol. 2001;39:1121–6. 10.1128/JCM.39.4.1221-1226.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J Clin Microbiol. 1999;37:2215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanogo YO, Zeaiter Z, Caruso G, Merola F, Shpynov S, Brouqui P, et al. Bartonella henselae in Ixodes ricinus ticks (Acari: Ixodida) removed from humans, Belluno province, Italy. Emerg Infect Dis. 2003;9:329–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morozova OV, Cabello FC, Dobrotvorsky AK. Semi-nested PCR detection of Bartonella henselae in Ixodes persulcatus ticks from Western Siberia, Russia. Vector Borne Zoonotic Dis. 2004;4:306–9. 10.1089/vbz.2004.4.306 [DOI] [PubMed] [Google Scholar]
- 21.Alekseev AN, Semenov AV, Dubinina HV. Evidence of Babesia microti infection in multi-infected Ixodes persulcatus ticks in Russia. Exp Appl Acarol. 2003;29:345–53. 10.1023/A:1025841901909 [DOI] [PubMed] [Google Scholar]
- 22.Kulik IL, Vinokurova NS. Distribution range of the meadow tick Dermacentor pictus (Ixodidae) in the USSR [article in Russian]. Parazitologiia. 1983;17:207–13. [PubMed] [Google Scholar]
- 23.Uilenberg G, Franssen FF, Perie NM, Spanjer AA. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet Q. 1989;11:33–40. 10.1080/01652176.1989.9694194 [DOI] [PubMed] [Google Scholar]
- 24.Kahl O, Janetzki C, Gray JS, Stein J, Bauch RJ. Tick infection rates with Borrelia: Ixodes ricinus versus Haemaphysalis concinna and Dermacentor reticulatus in two locations in eastern Germany. Med Vet Entomol. 1992;6:363–6. 10.1111/j.1365-2915.1992.tb00634.x [DOI] [PubMed] [Google Scholar]
- 25.Holden K, Boothby J, Anand S, Massung R. Detection of Borrelia burgdorferi, Ehrlichia chaffeensis, and Anaplasma phagocytophilum in ticks (Acari: Ixodidae) from a coastal region of California. J Med Entomol. 2003;40:534–9. 10.1603/0022-2585-40.4.534 [DOI] [PubMed] [Google Scholar]
- 26.Caccio SM, Antunovi B, Moretti A, Mangili V, Marinculic A, Baric R, et al. Molecular characterisation of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Vet Parasitol. 2002;106:285–92. 10.1016/S0304-4017(02)00112-7 [DOI] [PubMed] [Google Scholar]
- 27.Postic D, Assous M, Grimont P, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–52. 10.1099/00207713-44-4-743 [DOI] [PubMed] [Google Scholar]
- 28.Armstrong PM, Katavolos P, Caporale DA, Smith RP, Spielman A, Telford SR. Diversity of Babesia infecting deer ticks (Ixodes dammini). Am J Trop Med Hyg. 1998;58:739–42. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, Nakao M. Transmission of the Lyme disease spirochete, Borrelia garinii, between infected and uninfected immature Ixodes persulcatus during cofeeding on mice. J Parasitol. 1997;83:547–50. 10.2307/3284432 [DOI] [PubMed] [Google Scholar]
- 32.Postic D, Korenberg E, Gorelova N, Kovalevski Y, Bellenger E, Baranton G. Borrelia burgdorferi sensu lato in Russia and neighbouring countries: high incidence of mixed isolates. Res Microbiol. 1997;148:691–702. 10.1016/S0923-2508(99)80068-0 [DOI] [PubMed] [Google Scholar]
- 33.Livanova N, Morozova O, Morozov I, Beklemishev A, Cabello F, Dobrotvorsky A. Characterization of Borrelia burgdorferi sensu lato from Novosibirsk Region (West Siberia, Russia) based on direct PCR. Eur J Epidemiol. 2003;18:1155–8. 10.1023/B:EJEP.0000006632.26884.59 [DOI] [PubMed] [Google Scholar]
- 34.Postic D, Belfaiza J, Isogai E, Saint Girons I, Grimont PA, Baranton G. A new genomic species in Borrelia burgdorferi sensu lato isolated from Japanese ticks. Res Microbiol. 1993;144:467–73. 10.1016/0923-2508(93)90054-6 [DOI] [PubMed] [Google Scholar]
- 35.Masuzawa T, Komikado T, Iwaki A, Suzuki H, Kaneda K, Yanagihara Y. Characterization of Borrelia sp. isolated from Ixodes tanuki, I. turdus, and I. columnae in Japan by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. FEMS Microbiol Lett. 1996;142:77–83. 10.1111/j.1574-6968.1996.tb08411.x [DOI] [PubMed] [Google Scholar]
- 36.Li M, Masuzawa T, Takada N, Ishiguro F, Fujita H, Iwaki A, et al. Lyme disease Borrelia species in northeastern China resemble those isolated from far eastern Russia and Japan. Appl Environ Microbiol. 1998;64:2705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao W, Zhao Q, Zhang P, Dumler J, Zhang X, Fang L, et al. Granulocytic ehrlichiae in Ixodes persulcatus ticks from an area in China where Lyme disease is endemic. J Clin Microbiol. 2000;38:4208–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telford S, Korenberg E, Goethert H, Kovalevskii U, Gorelova N, Spielman A. Detection of natural foci of babesiosis and granulocytic ehrlichioses in Russia [article in Russian]. Zh Mikrobiol Epidemiol Immunobiol. 2002;6:21–5. [PubMed] [Google Scholar]
- 39.Hauschild S, Schein E. The subspecies specificity of Babesia canis. Berl Munch Tierarztl Wochenschr. 1996;109:216–9. [PubMed] [Google Scholar]
- 40.Duh D, Tozon N, Petrovec M, Strasek K, Avsic-Zupanc T. Canine babesiosis in Slovenia: molecular evidence of Babesia canis canis and Babesia canis vogeli. Vet Res. 2004;35:363–8. 10.1051/vetres:2004018 [DOI] [PubMed] [Google Scholar]