Abstract
We detected Rickettsia felis DNA in Ctenocephalides felis and Bartonella quintana DNA in 3 Pulex irritans fleas taken from a pet Cercopithecus cephus monkey in Gabon, sub-Saharan Africa. This is the first report of B. quintana in the human flea.
Keywords: Bartonella quintana, fleas, monkey, primates, Rickettsia felis, Gabon, dispatch
Bartonellae are gram-negative bacteria that cause several human diseases and are transmitted by various arthropods, such as lice, ticks, and fleas (1). Bartonella quintana is a worldwide fastidious bacterium that infects humans and belongs to the alpha subgroup of the Proteobacteria. Recent reports suggest that humans are the natural reservoir of B. quintana and that the human body louse is the vector (1). However, we have recently reported molecular detection of several Bartonella species, including B. quintana, in Ctenocephalides felis fleas from France, which suggests that fleas may be important vectors of human disease (2). Fleas are found worldwide on mammals and are vectors of several major zoonoses, including plague caused by Yersinia pestis (3), murine typhus caused by Rickettsia typhi, and fleaborne spotted fever caused by R. felis (3). More than 2,000 species of fleas exist worldwide. While some species are highly host specific, others are more catholic and will feed on numerous hosts, especially in the absence of their preferred host (3). Several flea species, including Pulex irritans, C. canis, C. felis, Ceratophyllus gallinae, Ceratophyllus columbae, and Archaeopsylla erinacei, may infest humans. In this study, we collected P. irritans (human fleas) and C. felis fleas on a pet monkey in Gabon and report for the first time the molecular detection of B. quintana in P. irritans.
The Study
Four fleas collected from a pet monkey (Cercopithecus cephus) in Franceville, Gabon, were stored in 70% alcohol and sent to the World Health Organization (WHO) Collaborative Center for Rickettsial Reference and Research in Marseille, France, where molecular studies were performed in April 2005. Fleas were rinsed with distilled water for 10 min and dried on sterile filter paper in a laminar flow hood. Preliminary entomologic identification was performed by using reference taxonomic keys as previously reported (4).
Fleas were crushed individually in sterile Eppendorf tubes with the tip of a sterile pipette. DNA was then extracted by using the QIAamp Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer's instructions. Rickettsial DNA was detected by polymerase chain reaction (PCR) with primers targeting the citrate synthase gene (gltA) as previously described (4). R. montanensis DNA was used as positive control, and negative controls consisted of laboratory uninfected flea DNA. Bartonella DNA was detected by PCR with 3 sets of primers targeting the intergenic spacer (ITS) gene, and the B. quintana spacers 336 and 894 as previously described (4,5). B. elizabethae DNA was used as positive control and uninfected fleas as negative controls. Additionally, fleas were identified at the species level after amplification and sequencing of a portion of the 18S rDNA gene as previously described (4). PCR products were purified, and DNA sequencing was carried out by using the d-Rhodamine Terminator cycle sequencing ready reaction kit with Amplitaq Polymerase FS (Perkin-Elmer, Coignieres, France) as described by the manufacturer. For all PCR products, sequences from both DNA strands were determined twice. Sequencing products were resolved by using an ABI 3100 automated sequencer (Perkin-Elmer). Sequence analysis was performed by using the software package ABI Prism DNA Sequencing Analysis Software version 3.0 (Perkin-Elmer). All obtained sequences were compared with those available in GenBank by using the nucleotide-nucleotide BLAST (blastn) program (http://www.ncbi.nlm.nih.gov/BLAST/).
Using morphologic taxonomic keys, 3 fleas were identified as P. irritans and 1 flea as Ctenocephalides felis. These findings were unambiguously confirmed when a 331-bp fragment of the 18S rDNA gene showed 99.7% and 99.4% homology with previous sequences of C. felis (GenBank accession no. AF423914) and P. irritans (GenBank accession no. AF423915). When gltA primers were used, R. felis (GenBank accession no. AF516333, 100% homology) was detected in the C. felis, whereas the P. irritans as well as negative controls were negative. Using the ITS primers for Bartonella spp., we detected PCR products in the 3 P. irritans fleas, whereas the C. felis flea and negative controls were negative. By sequencing the ITS gene–amplified fragments from these 3 fleas, we identified B. quintana (GenBank accession no. AF368396, 100% homology). Two PCR procedures targeting specific B. quintana spacers previously described (5) were carried out to confirm the results. By using these primers, B. quintana type 1 sequence was obtained for the spacer 336 (GenBank accession no. AY660705, 100% homology) and B. quintana type 2 sequence for the spacer 894 (GenBank accession no. AY660713, 100% homology). Thus, according to current guidelines for B. quintana typing (5), we have amplified genotype 2 of B. quintana.
Conclusions
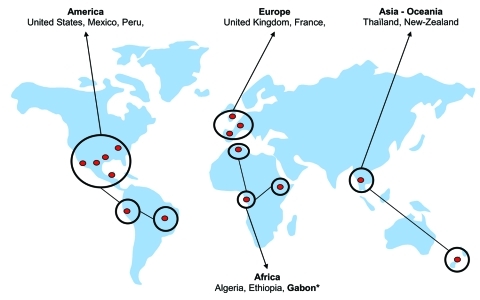
We present here the first molecular detection of R. felis in sub-Saharan Africa, Gabon (Figure). To date, 4 species of fleas have been associated worldwide with R. felis including C. felis (3,6), C. canis (6), P. irritans (3), and Archeopsylla erinacei (4). Thus, the amplification of R. felis in the C. felis flea from the monkey was not surprising but suggests that nonhuman primates may be infected as well as humans and may represent a reservoir of R. felis. The role of mammals, including rodents, hedgehogs, cats, dogs, and monkeys, in the life cycle and circulation of R. felis remains unclear and warrants further epidemiologic studies.
Figure.
Detection of Rickettsia felis in fleas worldwide. *This study.
We report for the first time that the human flea P. irritans can be infected with B. quintana. Apart from the body louse, the natural vector of B. quintana in humans, we have previously detected B. quintana in C. felis fleas with a prevalence of 4.5% in a series of 309 fleas collected in various regions of France (2). Thus, our results confirm that B. quintana may be found in the human flea and may explain 2 clinical reports of chronic adenopathy attributed to B. quintana infection for which the only epidemiologic risk factor identified was the presence of fleas (7,8). Few reports of detection of other bartonellae in fleas have been made (Table). Recently, the rodent flea Ctenophthalmus nobilis has been found to be a competent vector of at least 2 Bartonella species, B. grahamii, which has previously been associated with human infection, and B. taylorii (9). In contrast, no evidence of either horizontal or vertical transmission was seen in bank voles (Clethrionomys glareolus) injected with B. taylorii maintained in an arthropod-free environment, which suggests that fleas may be essential for transmitting some Bartonella spp (9). In the study of Stevenson et al, Bartonella spp. were detected in 38 Oropsylla hirsuta and 3 Oropsylla tuberculatus cynomuris prairie dog fleas in United States (10). In addition, new Bartonella genotypes, whose medical importance is not yet known, were detected in Pulex fleas in Peru (11), in 5 C. felis collected from cats, and in a Nosopsyllus fasciatus collected from a Rattus surifer specimen in Thailand (6).
Table. Bartonella species detected in fleas worldwide.
| Species | Species of Bartonella detected | Country/animal (ref.) |
|---|---|---|
| Ctenocephalides felis | B. clarridgeiae | France/cat (2,14) Thailand/cat (6) New Zealand/cat (15) |
| B. koehlerae | France/cat (2) | |
| B. quintana | France/cat (2) | |
| Ctenophthalmus nobilis | B. grahamii | United Kingdom/bank vole (9) |
| B. taylorii | United Kingdom/bank vole (9) | |
| Nosopsyllus fasciatus | Bartonella spp. | Thailand/rodent (6) |
| Oropsylla hirsuta | Bartonella spp. | United States/prairie dog (10) |
| O. tuberculatus cynomuris | Bartonella spp. | United States/prairie dog (10) |
| Pulex spp. | Bartonella spp. | Peru/human (11) |
| Pulex irritans | B. quintana | Gabon/monkey (this study) |
Although detection of Bartonella DNA is often reported from several sources, including fleas, mammals, and human samples, isolation of bartonellae by culture remains infrequent (12). Culture media and procedures used for Bartonella spp. have been highly variable and have questionable sensitivity (12). A novel chemically modified liquid medium that will support the growth of several Bartonella spp. has been recently developed and may provide an advantage over conventional blood agar culture for the isolation of Bartonella spp (13). The prevalence of B. quintana as well as other bartonellae in human fleas remains unknown, and this subject needs to be addressed to better define possible sources of Bartonella infections in humans.
Acknowledgments
We thank Paul Newton for English corrections.
Centre International de Recherches Médicales is supported by the Government of Gabon, Total-Fina-Elf Gabon, and the Ministère de la Coopération Française.
Biography
Dr Rolain conducts research at the Unité des Rickettsies, the national reference center for rickettsiosis and WHO collaborative center. The laboratory is primarily involved in the study of emerging and reemerging bacteria and arthropodborne diseases.
Footnotes
Suggested citation for this article: Rolain JM, Bourry O, Davoust B, Raoult D. Bartonella quintana, sub-Saharan Africa. Emerg Infect Dis [serial on the Internet]. 2005 Nov [date cited]. http://dx.doi.org/10.3201/eid1111.050861
References
- 1.Alsmark CM, Frank AC, Karlberg EO, Legault BA, Ardell DH, Canback B, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci U S A. 2004;101:9716–21. 10.1073/pnas.0305659101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rolain JM, Franc M, Davoust B, Raoult D. Molecular detection of Bartonella quintana, B. koehlerae, B. henselae, B. clarridgeiae, Rickettsia felis and Wolbachia pipientis in cat fleas, France. Emerg Infect Dis. 2003;9:338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad AF, Radulovic S, Higgins JA, Noden BH, Troyer JM. Flea-borne rickettsioses : ecologic considerations. Emerg Infect Dis. 1997;3:319–27. 10.3201/eid0303.970308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitam I, Parola P, Dittmar de la Cruz K, Matsumoto K, Baziz B, Rolain JM, et al. First molecular detection of Rickettsia felis in fleas from Algeria. Am J Trop Med Hyg. 2005. In press. [PubMed] [Google Scholar]
- 5.Foucault C, La Scola B, Lindroos H, Andersson SG, Raoult D. Multispacer typing technique for sequence-based typing of Bartonella quintana. J Clin Microbiol. 2005;43:41–8. 10.1128/JCM.43.1.41-48.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parola P, Sanogo OY, Lerdthusnee K, Zeaiter Z, Chauvancy G, Gonzalez JP, et al. Identification of Rickettsia spp. and Bartonella spp. in fleas from the Thai-Myanmar border. Ann N Y Acad Sci. 2003;990:173–81. 10.1111/j.1749-6632.2003.tb07359.x [DOI] [PubMed] [Google Scholar]
- 7.Drancourt M, Moal V, Brunet P, Dussol B, Berland Y, Raoult D. Bartonella (Rochalimaea) quintana infection in a seronegative hemodialyzed patient. J Clin Microbiol. 1996;34:1158–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raoult D, Drancourt M, Carta A, Gastaut JA. Bartonella (Rochalimaea) quintana isolation in patient with chronic adenopathy, lymphopenia, and a cat. Lancet. 1994;343:977. 10.1016/S0140-6736(94)90102-3 [DOI] [PubMed] [Google Scholar]
- 9.Bown KJ, Bennet M, Begon M. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg Infect Dis. 2004;10:684–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevenson HL, Bai Y, Kosoy MY, Montenieri JA, Lowell JL, Chu MC, et al. Detection of novel Bartonella strains and Yersinia pestis in prairie dogs and their fleas (Siphonaptera: Ceratophyllidae and Pulicidae) using multiplex polymerase chain reaction. J Med Entomol. 2003;40:329–37. 10.1603/0022-2585-40.3.329 [DOI] [PubMed] [Google Scholar]
- 11.Parola P, Shpynov S, Montoya M, Lopez M, Houpikian P, Zeaiter Z, et al. First molecular evidence of new Bartonella spp. in fleas and a tick from Peru. Am J Trop Med Hyg. 2002;67:135–6. [DOI] [PubMed] [Google Scholar]
- 12.La Scola B, Raoult D. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J Clin Microbiol. 1999;37:1899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggi RG, Duncan AW, Breitschwerdt EB. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J Clin Microbiol. 2005;43:2651–5. 10.1128/JCM.43.6.2651-2655.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurfield AN, Boulouis HJ, Chomel BB, Kasten RW, Heller R, Bouillin C, et al. Epidemiology of Bartonella infection in domestic cats in France. Vet Microbiol. 2001;80:185–98. 10.1016/S0378-1135(01)00304-2 [DOI] [PubMed] [Google Scholar]
- 15.Kelly PJ. A review of bacterial pathogens in Ctenocephalides felis in New Zealand. N Z Vet J. 2004;52:352–7. 10.1080/00480169.2004.36451 [DOI] [PubMed] [Google Scholar]