Abstract
Genetic recombination occurs during meiosis, the key developmental program of gametogenesis. Recombination in mammals has been recently linked to the activity of a histone H3 methyl-transferase, PRDM91–6, the product of the only known speciation gene in mammals7. PRDM9 is thought to determine the preferred recombination sites – recombination hotspots – through sequence-specific binding of its highly polymorphic multi-Zn-finger domain8. Nevertheless, Prdm9 knockout mice are proficient at initiating recombination 9. Here we map and analyze the genome-wide distribution of recombination initiation sites in Prdm9 knockout mice and in two mouse strains with different Prdm9 alleles and their F1 hybrid. We show that PRDM9 determines the positions of practically all hotspots in the mouse genome, with the remarkable exception of the pseudoautosomal region – the only area of the genome that undergoes recombination in 100% of cells10. Surprisingly, hotspots are still observed in Prdm9 knockout mice and as in wild-type, these hotspots are found at H3K4 trimethylation marks. However, in the absence of PRDM9, the majority of recombination is initiated at promoters and at other sites of PRDM9-independent H3K4 trimethylation. Such sites are rarely targeted in wild-type mice indicating an unexpected role of the PRDM9 protein in sequestering the recombination machinery away from gene promoter regions and other functional genomic elements.
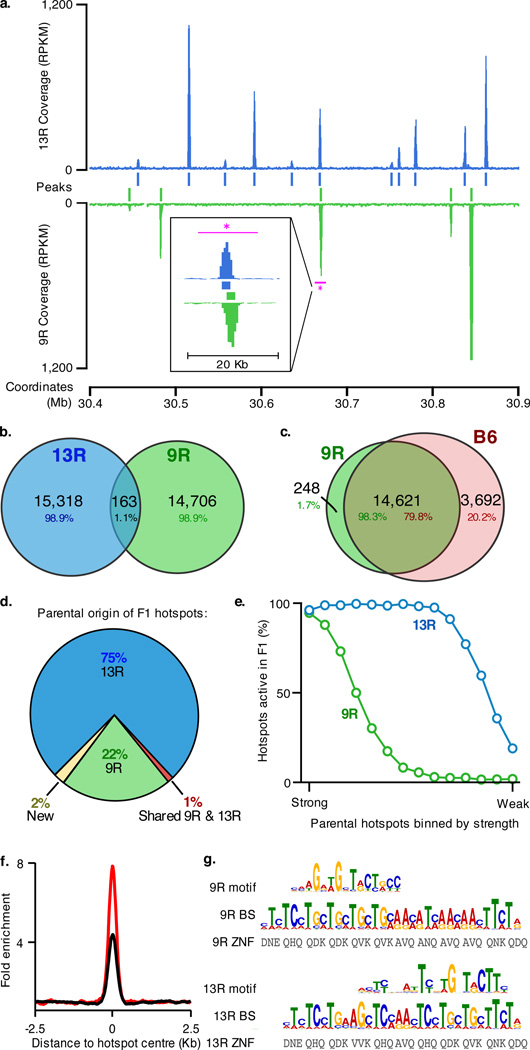
Homologous recombination is initiated by the introduction of DNA double stranded breaks (DSBs) by the SPO11 protein11, and we have previously shown that recombination sites can be mapped by sequencing the ends of breaks recovered by chromatin immunoprecipitation with antibodies to DMC1 protein bound to these ends (ChIP-Seq, SSDS)12,13. To directly assess the extent to which hotspot location depends on PRDM9 we have employed this approach to generate high-resolution maps of recombination initiation sites in two nearly identical (congenic) mouse strains with different Prdm9 alleles (strains 9R and 13R, Supplementary Fig. 1). We found that although the number of hotspots in 9R and 13R is similar (14,869 and 15,481, respectively) there is practically no overlap in the locations of hotspots (Fig. 1a,b). We next mapped recombination initiation sites in the most commonly used laboratory mouse strain C57Bl/6 (18,313 hotspots), which has the same Prdm9 allele as 9R. We found that more than 98% of 9R hotspots were present in C57Bl/6 (Fig. 1c). Taken together, these data constitute the first direct evidence that practically all recombination hotspots are PRDM9 dependent.
Figure 1.
DSB hotspots localize to different loci in 9R and 13R mice. a. DSB hotspots in a representative 0.5 Mb region on chromosome 3. The magenta bar indicates the region shown in the insert. This illustrates two adjacent, yet distinct hotspots in 9R and 13R. Data are smoothed using a 1 Kb sliding window (step 100 bp). Coverage is given as reads per Kb per million (RPKM) b. Overlap between 9R and 13R hotspots. Here and in subsequent panels, only overlaps in the central 400 bp of hotspots are counted. c. Overlap between 9R and C57Bl/6 (B6) hotspots. The excess of B6 hotspots is due to higher ChIP enrichment in this ChIP-Seq sample. B6 hotspots not found in 9R are a weak subset. d. Parental origin of DSB hotspots in the F1 (9R × 13R) mice. e. Stronger 9R and 13R hotspots are active in F1 progeny. The top 14,000 hotspots for each background were binned by strength and the overlap with F1 hotspots assessed for each bin. f. Distribution of hits to the 9R motif (black) and 13R motif (red) around their respective hotspots. Data are plotted in 200 nt steps. g. Alignment of 9R and 13R motifs to the predicted PRDM9 binding sites (see methods). ZNF, Zn finger contact residues; BS, predicted PRDM9 binding site.
We next assessed the number and distribution of recombination hotspots in mice heterozygous for functional Prdm9 alleles, as such mice would have potentially twice as many proficient PRDM9 binding sites. We found that in F1 hybrids derived from a 9R × 13R cross the number of detected hotspots (15,677) is comparable to those in the parental strains (see Supplementary Methods). Strikingly, although the vast majority of hotspots in the F1 mice coincided with hotspots in the parental lines, 75% corresponded to 13R hotspots, while only 22% corresponded to those of 9R (Fig. 1d). Hotspots from the reciprocal cross showed a similar bias (see Supplementary Methods). The 13R-derived hotspots in F1 were also significantly stronger than those of 9R origin (median of 575 and 280 reads, respectively; p = 10−182, Wilcoxon rank sum test), but the relative strengths of hotspots within parental pools were retained in the F1 background. Importantly, F1 hotspots composed a stronger than average subset of hotspots in both respective parental strains (Fig. 1e). In aggregate, these data indicate that the number of recombination hotspots does not double in the presence of two different Prdm9 alleles, but is rather maintained at a constant level with mostly the strongest of the potential hotspots being utilized. The dominance of the 13R allele could result from higher affinity of the 13R PRDM9 protein variant to its preferred DNA sequence however other scenarios are also possible.
The complete discordance between the 9R and 13R hotspot maps suggested that different DNA sequences might be recognized by the 9R and 13R PRDM9 protein variants. Indeed, we identified a distinct motif co-centred with hotspots in each strain. These motifs are present in nearly all hotspots (9R: 94% ; 13R: 97%) and enriched at hotspot centres 4 and 8-fold, respectively (Fig. 1f). The enrichment of better matches to motifs reaches 200-300-fold (Supplementary Fig. 2), and motif quality correlates with hotspot strength (Supplementary Fig. 3). We next predicted the putative DNA binding sites for the Zn-finger arrays of the 9R and 13R PRDM9 protein variants and found that each motif aligns with the predicted binding site for its respective Prdm9 allele but not with that of the other (Fig. 1g). Both human2 and mouse12 hotspot motifs were previously found to align to the C-terminal part of the PRDM9 Zn finger array (Supplementary Fig. 4). Our data suggest that DNA binding is not constrained to this region as the 9R motif aligns to N-terminal Zn fingers (Fig. 1g). Although these data are consistent with the sequence specificity of PRDM9 being a key determinant of hotspot positions in vivo, only a fraction of potential PRDM9 binding sites are utilized for hotspot formation. It is possible that sequence motifs do not capture some aspects of the binding site information such as di- or tri-nucleotide preferences of the PRDM9 protein. Since PRDM9 (or PRDM9-containing complex) must act within the constraints of the higher order chromatin organisation, it is also conceivable that the affinity of PRDM9 for its recognition sequence depends on the local chromatin environment.
In mammals, PRDM9 is the only histone H3 lysine 4 trimethyltransferase that has been reported to be specific to meiosis9. Since recombination hotspots are associated with meiosis-specific H3K4me3 marks12,14,15, the PRDM9 protein is likely responsible for their introduction8. If so, recombination hotspots in the strains with different Prdm9 alleles will correspond to H3K4me3 marks that are strain-specific. Indeed, we found that the vast majority of 9R and 13R hotspots overlap strain-specific H3K4me3 marks (9R: ≥83%; 13R: ≥89% - Supplementary Fig. 5a,b). Analysis of H3K4me3 in a Spo11−/− mouse revealed that H3K4me3 marks are present at 89% of potential hotspot loci despite the lack of SPO11-induced DSBs (Supplementary Fig. 5c), and in agreement with previous findings14,15. This indicates that hotspot-associated H3K4 trimethylation is not a consequence of DSB formation and/or repair, but a mark that is present prior to initiation of recombination.
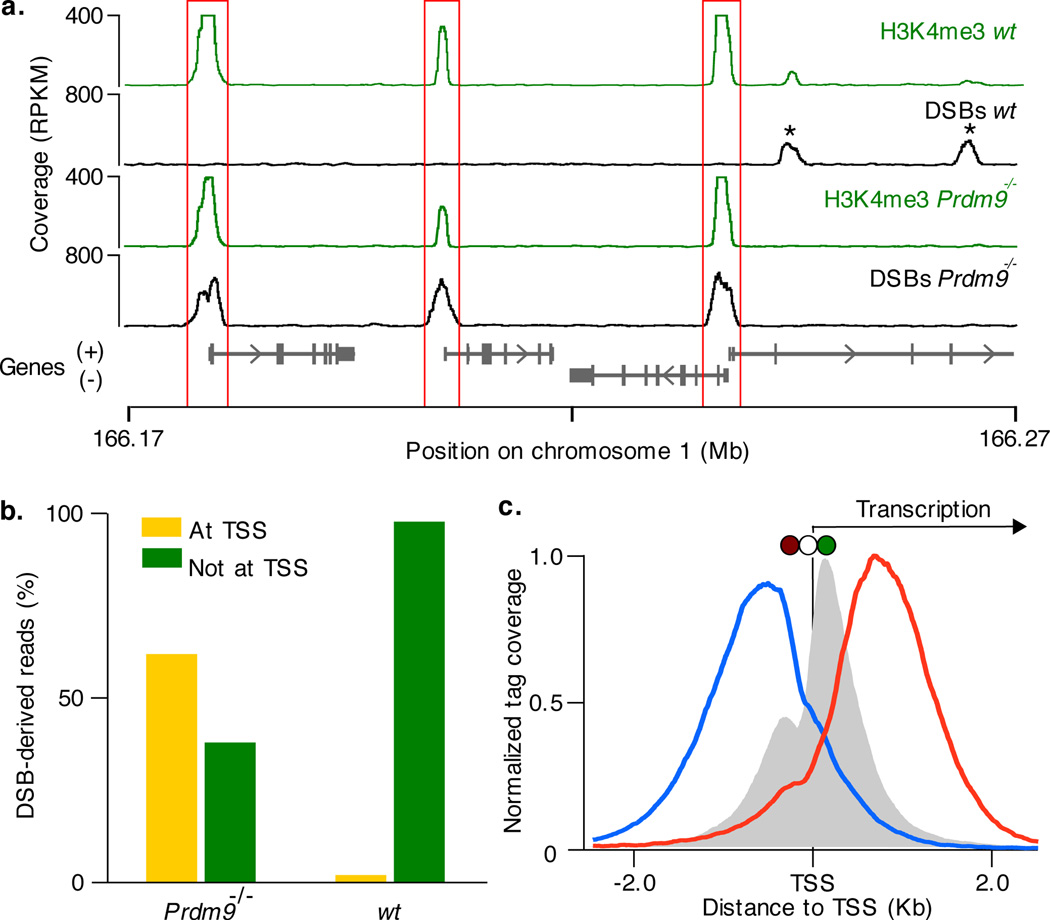
Despite the clear critical role of PRDM9 in the initiation of genetic recombination meiotic DSBs are present in Prdm9 knockout mice9. Remarkably, we found that initiation of recombination in these mice is not random or uniform, but is still clustered in hotspots. The vast majority (99%) of these hotspots do not overlap the hotspots detected in any of the wild type strains, nevertheless, 94% of Prdm9−/− hotspots still overlap H3K4me3 marks. Most of these marks (92%) are present in wild type testis and almost half are not specific to germline cells (Supplementary Fig. 5d). As H3K4me3 marks outnumber DSB hotspots, they do not appear to be sufficient for hotspot formation (Supplementary Fig. 6). H3K4me3 is a general mark of active transcription primarily associated with gene promoters16, enhancers17,18 and possibly other functional genomic elements. We found that in the absence of PRDM9 and PRDM9-introduced H3K4me3 marks, recombination hotspots are re-routed to these alternative H3K4me3 sites (Fig. 2a). Almost half (44%) of recombination hotspots in the Prdm9−/− mice localize to the promoters of annotated genes compared to just 3% in wild type. Unlike in wild type mice, promoter-associated hotspots in Prdm9−/− are relatively strong (Supplementary Fig. 7) and therefore, account for the majority of DSB-derived sequencing reads (62%; Fig. 2b). The H3K4me3 signal at promoters that overlap hotspots in Prdm9−/− is almost 100-fold stronger than at those that do not (median: 350 tags and 5 tags, respectively, P < 10−200, Wilcoxon test). Furthermore, promoter-overlapping hotspots are slightly enriched at genes expressed in early meiotic prophase (P = 10−138, one-sided binomial test; Supplementary Fig. 8). Prdm9−/− hotspots that are not associated with annotated promoters still likely represent DSBs formed at important genomic elements as they exhibit higher than expected sequence conservation (Supplementary Fig. 9). Indeed, 40% of these hotspots overlap histone crotonylation sites, a recently discovered marker of promoters and enhancers19 (see Supplementary Methods). The distribution of hotspots around transcription start sites (TSSs) in Prdm9−/− mice is reminiscent of that in S. cerevisiae where most hotspots are found at promoters20,21. However, while DSBs at yeast promoters form in the upstream, nucleosome-depleted region20,21, hotspots in Prdm9−/− mice form preferentially at the most frequently H3K4 trimethylated nucleosome at the +1 position, just downstream of the TSS (Fig. 2c).
Figure 2.
PRDM9 redirects DSBs away from functional genomic elements. a. In Prdm9−/− mice DSBs are formed at functional genomic elements marked by H3K4me3 (such as promoters, red boxes). These sites are refractory to DSB formation in wild type mice, where breaks are redirected to PRDM9-dependent H3K4me3 marks (*). b. Most DSBs occur in regions around TSSs in Prdm9−/− mice but not in wild type mice. c. The centre of Prdm9−/− hotspots at TSS coincides precisely with the H3K4me3 mark on the +1 nucleosome (red circle: −1 nucleosome; white circle: nucleosome free region; green circle: +1 nucleosome). Mean H3K4me3 coverage is grey, mean forward strand ssDNA coverage is blue and reverse strand coverage is red. The intersection point of the forward and reverse strand coverage is the mean hotspot centre (Supplementary Fig. 11).
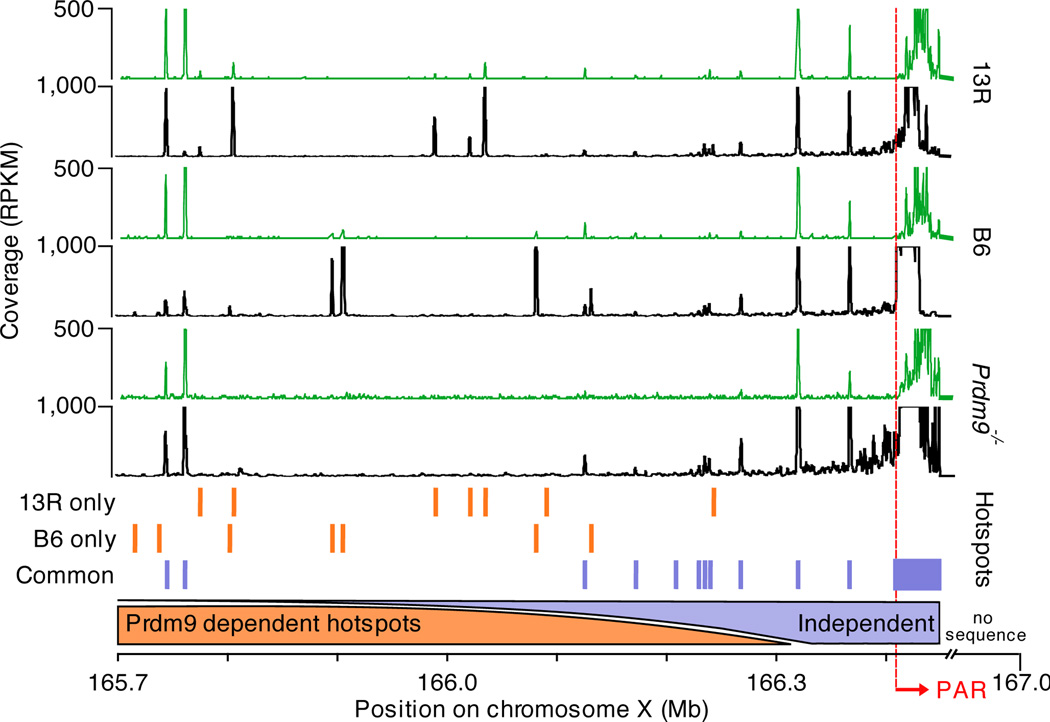
Unique in the genome is a short region of homology at the end of X and Y chromosome called the pseudo-autosomal region (PAR) that undergoes recombination in every meiosis10. We have recently demonstrated that the only sequenced region of the mouse PAR, adjacent to the interstitial PAR boundary, contains a large cluster of overlapping DSB hotspots spanning at least 40 Kb12. We now find that this hotspot cluster is still present in Prdm9−/− mice, and is shared between the strains with different Prdm9 alleles (Fig. 3). Akin to the hotspot cluster, the H3K4me3 signal in the PAR is far broader and stronger than any other H3K4me3 marks in the genome, but it is shifted slightly off the hotspot cluster, to within the PAR (Fig. 3). Recent studies have demonstrated that the structural organization of the PAR differs significantly from that of the rest of the chromosome22. It is conceivable that this region features constitutively accessible chromatin that allows initiation of recombination without participation of the PRDM9 protein. Initiation of homologous recombination at some of the hotspots in the PAR-flanking region is also defined by a PRDM9-independent mechanism. In this region, up to ~900 Kb upstream of PAR, there is an extraordinary concentration of hotspots shared between Prdm9−/− mice and wild type strains with different Prdm9 alleles (C57Bl/6, 9R and 13R; Fig. 3). As expected, the H3K4me3 marks at these shared hotspots are also strain independent. The density of PRDM9-independent hotspots in the PAR-flanking region appears to increase with the proximity to the PAR (Fig. 3), consistent with a specific chromatin organization or particular epigenetic mark characteristic to the PAR propagating into the adjacent area. Yet, PRDM9-dependent hotspots are also present in the PAR-flanking area, therefore the DNA in this region is still accessible to the PRDM9 protein.
Figure 3.
PRDM9-independent hotspots in the PAR and flanking region. Black tracks represent DMC1 tag coverage, green tracks depict H3K4me3 coverage. Orange bars illustrate hotspots unique to 13R or C57Bl/6 (B6) mice. Purple bars illustrate hotspots shared in 13R, C57Bl/6 and Prdm9−/− mice.
Why is the initiation of recombination in the PAR different from the rest of the genome? To ensure proper sex chromosome segregation during meiosis, at least one crossover must be formed inside the PAR10. Out of thousands of PRDM9-dependent hotspots only 200–400 are utilized in each meiosis and only 20–25 result in crossovers. Considering the short length of the PAR, a large fraction of cells would fail to introduce a crossover within this area simply by chance. It is possible that the mandatory crossover requirement is more efficiently fulfilled through an alternative mechanism of initiation, unique to the PAR. PRDM9-independent initiation of homologous recombination in the PAR would also facilitate the rapid evolution of both PRDM923 and the PAR24 by allowing them to evolve independently of each other. It is also conceivable that PRDM9-dependent and -independent recombination pathways utilize alternative isoforms of SPO11, as the number of meiotic DSBs in the PAR is diminished in mice expressing only the beta SPO11 isoform22.
Our study clearly demonstrates that the PRDM9 protein is not required for the initiation of genetic recombination per se, but rather, determines the positions (hotspots) where this initiation takes place. In the absence of PRDM9 the vast majority of DSB hotspots still coincide with H3K4me3 marks. Although a causal relationship is yet to be established, this suggests that H3K4me3 or/and mechanisms that induce H3K4me3 are necessary to provide a favourable chromatin environment for the initiation of meiotic recombination. Such an environment can be created with or without PRDM9, but only PRDM9-dependent H3K4me3 sites are associated with recombination hotspots in wild-type mice outside of the PAR. This indicates that PRDM9 is directly involved in recruitment of the recombination initiation machinery.
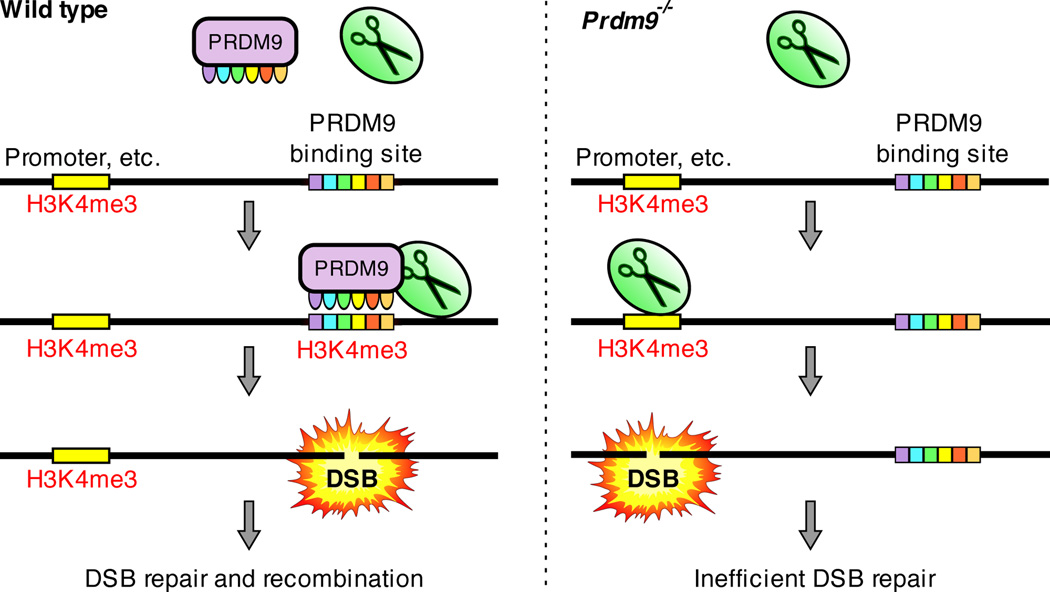
The sterility of Prdm9−/− mice suggests that the initiation of recombination at H3K4me3-marked functional elements may not be compatible with gametogenesis. Does recombination interfere with the transcription of essential meiotic genes? Since four chromatids are present before the first meiotic division it is unlikely that disruption of transcription on one of them will lead to a significant drop in overall transcription of any particular gene hit by a DSB. On the other hand, it is conceivable that the opposite is true and that transcription interferes with the repair of breaks introduced at promoters. The resulting aberrant recombination would account for the observation that in Prdm9−/− mice persistent DSBs are present and only partial synapsis is seen between homologous chromosomes9 (Supplementary Fig. 10). The Prdm9 gene exhibits complex epistatic interactions culminating in sterility of F1 hybrid males carrying a particular combination of functional Prdm9 alleles7. Unlike in Prdm9−/− mice, repair of meiotic DSBs in such males proceeds normally in a large fraction of cells with 40% of spermatocytes showing complete homologous synapsis7. While this suggests that distinct mechanisms are responsible for meiotic arrest in Prdm9−/− mice and in F1 hybrid males, we cannot exclude the possibility that an alternative role of PRDM9 in meiotic progression accounts for both sterility phenotypes. Although a function of PRDM9 in transcription has not been demonstrated, PRDM9 contains conserved domains implicated in transcription25,26 and could potentially be involved in executing the meiotic transcription program. It is also conceivable that PRDM9 is directly required for efficient progression of recombination. The potential importance of avoiding recombination at functional genomic elements is highlighted in the canid lineage, which has lost Prdm927,28. Though dogs retain recombination hotspots27, preliminary analyses suggest that unlike in Prdm9−/− mice, these hotspots do not appear to be enriched at promoters (Supplementary Information). A trivial explanation would be that dog Prdm9 resides in the unassembled parts of the genome, however, these data may also indicate that alternative mechanisms have evolved to target recombination to specialized genomic regions in species that lack Prdm9. Ultimately, we favour the hypothesis that PRDM9 prevents initiation of recombination in the areas of the genome where successful completion of recombination might be compromised by other nuclear processes (Fig. 4). The re-routing of DSBs from important genomic elements may also play a protective role against potential mutagenic effects of recombination.
Figure 4.
Proposed role of the PRDM9 protein. Left: In cells containing a functional copy of PRDM9 (Wild type) the DSB formation machinery (scissors) is directed to preferred DSB sites / PRDM9 binding sites. Right: In the absence of PRDM9, the DSB formation machinery opportunistically makes breaks at PRDM9-independent H3K4me3 marks such as those at promoters and enhancers. This results in inefficient DSB repair and meiotic arrest.
Methods Summary
SSDS13 using antibodies against DMC1 was used to identify DSB hotspots in the following mice: B10.S-H2t4/(9R)/J, B10.F-H2pb1/(13R)J, B10.S-H2t4/(9R)/J×B10.F-H2pb1/(13R)J (F1), C57Bl/6J and B6;129P2-Prdm9tm1Ymat/J. Chromatin immunoprecipitation using antibodies against H3K4me3 followed by Illumina sequencing was also performed on samples derived from each of these mice. DSB hotspots were defined by SISSRS29 and post-processed using a bespoke pipeline. Peaks in H3K4me3 data were called using MACS v1.3.730. Detailed methods are available in Supplementary Information.
Supplementary Material
Acknowledgements
We thank Shaila Sharmeen and Harold Smith for assistance with high-throughput sequencing. We also thank Michael Lichten and Peggy Hsieh for critical discussion of the manuscript. This research was supported by the NIDDK Intramural Research Program; by Basil O’Connor Starter Scholar Research Award Grant No. 5-FY07-667 from the March of Dimes Foundation (G.V.P.); NIH grant 1R01GM084104-01A1 from NIGMS (G.V.P.); New Investigator Start-up Grants FS71HU, R071HU and CS71HU from USUHS (G.V.P.).
Footnotes
Author Contributions:
K.B. and P.K. performed data analyses. F.S. performed all experiments. K.B. and G.V.P. wrote the manuscript. G.V.P. and R.D.C.O. designed and supervised the study. All authors contributed to experimental design, discussed the results and critiqued the manuscript.
Data deposition:
All datasets are fully described and available for download from the National Center for Biotechnology Information (US) GEO, accession number GSE35498.
References
- 1.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg IL, et al. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci U S A. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 8.Grey C, et al. Mouse PRDM9 DNA-Binding Specificity Determines Sites of Histone H3 Lysine 4 Trimethylation for Initiation of Meiotic Recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 10.Burgoyne PS. Genetic homology and crossing over in the X and Y chromosomes of Mammals. Hum Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- 11.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012 doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 22.Kauppi L, et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellott DW, Page DC. Reconstructing the evolution of vertebrate sex chromosomes. Cold Spring Harb Symp Quant Biol. 2009;74:345–353. doi: 10.1101/sqb.2009.74.048. [DOI] [PubMed] [Google Scholar]
- 25.Lim FL, Soulez M, Koczan D, Thiesen HJ, Knight JC. A KRAB-related domain and a novel transcription repression domain in proteins encoded by SSX genes that are disrupted in human sarcomas. Oncogene. 1998;17:2013–2018. doi: 10.1038/sj.onc.1202122. [DOI] [PubMed] [Google Scholar]
- 26.Margolin JF, et al. Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci U S A. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Axelsson E, et al. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz-Fuentes V, Di Rienzo A, Vilà C. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. 2011;6:e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.