Abstract
Subunits of the RNA processing exosome assemble into structurally distinct protein complexes that function in disparate cellular compartments and RNA metabolic pathways. Here, in a genetic, cell biological, and transcriptomic analysis, we examine the role of Dis3 – an essential polypeptide with endo- and 3’ to 5’ exo-ribonuclease activity – in cell cycle progression. We present several lines of evidence that perturbation of DIS3 affects microtubule (MT) localization and structure in Saccharomyces cerevisiae. Cells with a DIS3 mutant: (i) accumulate anaphase and pre-anaphase mitotic spindles; (ii) exhibit spindles that are mis-oriented and displaced from the bud neck; (iii) harbor elongated spindle-associated astral MTs; (iv) have an increased G1 astral MT length and number; and (v) are hypersensitive to MT poisons. Mutations in the core exosome genes RRP4 and MTR3 and the exosome cofactor gene MTR4 – but not other exosome subunit gene mutants – also elicit MT phenotypes. RNA deep sequencing analysis (RNA-seq) shows broad changes in the levels of cell cycle- and MT-related transcripts in mutant strains. Collectively, the data presented in this study suggests an evolutionarily conserved role for Dis3 in linking RNA metabolism, MTs, and cell cycle progression.
Keywords: Exozyme, exosome, mitotic spindle, mitosis, Dis3, Mtr3
INTRODUCTION
Exosome subunits are implicated in numerous aspects of RNA metabolism in the budding yeast Saccharomyces cerevisiae. These include the turnover, processing, or surveillance of messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and cryptic-unstable, stable-unannotated, upstream-non-coding and promoter-associated transcripts (Butler 2002; Kadaba et al. 2004; Houseley et al. 2006; Chekanova et al. 2007; Preker et al. 2008). Following the discovery of a 5-subunit exosome complex in budding yeast (Mitchell et al. 1997), it was proposed that the multiplicity of RNA substrates reflected the activities of its distinct subunits or subsets of subunits (Mitchell and Tollervey 2000). This idea has been revised and promulgated (Kiss and Andrulis 2011). Since this complex and its subunits are conserved from archaea to eukaryotes, defining its structure and functional mechanism(s) in budding yeast will ultimately facilitate understanding evolutionarily conserved RNA processing and turnover pathways.
The canonical yeast exosome complex is composed of nine subunits (Allmang et al. 1999b) and is commonly referred to as the “core exosome.” Six of the core subunits have RNase PH domains (Rrp41/Ski6, Rrp42, Rrp43, Rrp45, Rrp46, and Mtr3) and three are S1/KH RNA-binding domain proteins (Csl4, Rrp4, and Rrp40). A tenth polypeptide, Dis3 (also referred to as Rrp44 in yeast), is a homolog of the eubacterial RNase R/RNase II. Dis3 is present in all eukaryotes, but is not found in the exosome complexes of trypanosomes and archaebacteria (Estevez et al. 2003; Evguenieva-Hackenburg et al. 2003; Buttner et al. 2006). Therefore, sensu stricto, Dis3 is not an evolutionarily conserved component of the core exosome. Several proteins interact with the exosome in a compartment specific manner. One nuclear cofactor, Mtr4, a DEAD box RNA helicase (de la Cruz et al. 1998; Allmang et al. 1999a; van Hoof et al. 2000a; Torchet et al. 2002; Schilders et al. 2007), is also a component of TRAMP, a multiprotein complex implicated in post-transcriptional RNA polyadenylation (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; San Paolo et al. 2009). Another polypeptide, Rrp6, that associates with yeast nuclear-localized exosome subunits and complexes, is a homolog of eubacterial RNase D (Briggs et al. 1998; Allmang et al. 1999b). Although Rrp6 predominantly associates with the nuclear exosome complex, that association is not required for Rrp6 to complete at least some of its RNA processing functions in yeast (Callahan and Butler 2008; Callahan and Butler 2010). Rrp6 has also been shown to reside in a core exosome-independent complex (Graham et al. 2009b). Rrp47/C1D/Lrp1 is an additional nuclear cofactor that functionally and physically interacts with Rrp6 (Mitchell et al. 2003; Hieronymus et al. 2004; Houalla et al. 2006; Schilders et al. 2007).
It has been suggested that Dis3 is the only catalytically active RNase associated with the yeast exosome core (Liu et al. 2006; Dziembowski et al. 2007; Bonneau et al. 2009; Malet et al. 2010; Schaeffer and van Hoof 2011). However, a DIS3 mutation that ablates its 3’ to 5’ exoribonuclease activity in vitro is not lethal, even in the absence of RRP6 the other exosome component known to possess RNase activity (Dziembowski et al. 2007). Thus, either the Dis3 exoRNase active site mutation does not completely destroy its catalytic activity in vivo or the essential function lies in its endoribonuclease activity or elsewhere (Mamolen and Andrulis 2009; Schaeffer et al. 2009; Mamolen et al. 2010). In short, the essential role of Dis3 is unclear. In this regard, the dis3-54 allele was originally identified in a Schizosaccharomyces pombe genetic screen for mutants that are defective in sister chromatid disjunction. This mutant is hypersensitive to the nucleotide analog caffeine (Ohkura et al. 1988), a drug known to cause bypass of the G2/M checkpoint (Schlegel and Pardee 1986; Eastman 2004). Further, DIS3 is implicated in regulating the fission yeast cdc2 kinase (mammalian Cdk1; (Kinoshita et al. 1991)), a key protein controlling the G2/M transition (Stark and Taylor 2006). DIS3 is required for mitotic progression and has poorly-characterized links to phosphatase pathways (Kinoshita et al. 1991; Wilson et al. 1991; Shimanuki et al. 1993). Both the human and S. pombe Dis3 protein interact directly with the small GTPase Ran (Noguchi et al. 1996; Shiomi et al. 1998), a mitotic regulator that affects nucleocytoplasmic transport, spindle integrity, and chromatin-nuclear envelope relations (Dasso 2002; Li et al. 2003; Gruss and Vernos 2004; Goodman and Zheng 2006; Ciciarello et al. 2007). Dis3 can interact with GTP-, GDP-bound or nucleotide-free Ran and enhances the guanine nucleotide exchange activity of Rcc1, the Ran guanine nucleotide exchange factor (GEF; (Noguchi et al. 1996; Shiomi et al. 1998)). Consistent with this physical interaction, mutations of DIS3 and GSP1/CNR1 (yeast Ran) genes share several RNA processing phenotypes (Suzuki et al. 2001). Based upon this particular observation, it was proposed that Ran regulates the assembly or disassembly of the exosome complex (Suzuki et al. 2001), but this has not been addressed.
Here, we present our investigation of the genetic links between DIS3 and cell cycle progression in the budding yeast Saccharomyces cerevisiae. We began with mtr17-1, an allele of DIS3 that was isolated in a genetic selection for mutants which accumulate poly A+ in the nucleus and therefore could be defective in nucleocytoplasmic mRNA transport (mtr; (Kadowaki et al. 1994; Suzuki et al. 2001)). This DIS3 allele (its historic ties to the mtr screen inadvertently lost when renamed rrp44-1 (Bousquet-Antonelli et al. 2000; Torchet et al. 2002)) has a general RNA processing defect. We show that perturbation of DIS3 and a few other—but not all—exosome subunit genes, leads to widespread defects in microtubule (MT) structure and function, including defects in spatial and temporal organization of the mitotic spindle. As revealed by RNA deep sequencing (RNA-seq), these phenotypes may be an indirect consequence of disrupting the metabolism of critical cell cycle regulator mRNAs. Our genetic, cell biological, and RNA-seq data show that the tested mutants have distinct phenotypes and affect cell cycle-related transcripts differentially. Moreover, our study bolsters the idea that Dis3 has a conserved function in cell cycle progression.
MATERIALS AND METHODS
Strain construction and analysis
GFP-TUB1 integrants were made by digesting plasmid pAFS91 (Straight et al. 1997) with StuI and transforming yeast cells. Cells were plated on SC-Ura media, and Ura+ transformants were screened for GFP-tubulin protein expression and the presence of fluorescent spindles and spindle pole bodies. For all experiments, cells were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) because GFP-TUB1 expression was better in YPD than in synthetic complete media. For cell spotting experiments, cells were inoculated at an equal density, grown overnight, then 10-fold serially diluted five times and spotted onto plates. For the benomyl (10 µg/ml) and nocodazole (4 µg/ml) sensitivity experiments, plates were incubated at room temperature for 4–5 days. Pictures of plates were taken using the Gel Documentation system (Bio-Rad Laboratories; Hercules, CA).
Cell manipulation, counting, and analysis
Yeast cell transformations, genetic crosses, genotyping, and analysis and processing was performed using established lab protocols or those described (Burke et al. 2000). Protein extraction for western blotting was done by treating cell pellets with freezing on dry ice, vortexing with glass beads, and boiling in SDS sample buffer. Astral MT lengths were calculated as described previously (Shaw et al. 1997). Briefly, cell images were captured using a Zeiss microscope at a size resolution of 67.5 nm/pixel. Images were analyzed using Adobe Photoshop, with the measure tool set to pixels. Astral MT length was then measured in pixels from the center of the spindle pole body to the MT tip, and pixel measurements were then converted to micrometers using the conversion above. Preanaphase spindle angles were determined as described (Lee et al. 1999) and calculated as follows. Using Adobe Photoshop, the measure tool was used to trace the spindle. The measure tool was anchored at a site along the mother-bud axis and then another line was drawn to represent the mother-bud axis. Conveniently, the measure tool reports the angle at the anchor point.
Deep Sequencing
Yeast strains were grown in 50 mL of YPD in a 250 mL flask at 30°C and 250 rpm to a final OD600 of 0.4, and temperature shifted for 2 hours at 37°C. Cells were harvested in 50 mL conical tubes by centrifugation at 3,400 rpm for 4 min at RT, resuspended in 1.0 mL of water, centrifuged at 14,000 rpm for 2 min, and the supernatant was discarded by aspiration. Cells were frozen on dry ice and stored at −80°C. RNA was isolated from 20 OD units of each strain by a modified hot acid phenol method. Total RNA was purified with on-column DNase treatment using the Qiagen RNeasy kit. Purity (OD260/280 ratio of 2.2 to 2.3) was evaluated on a nano-drop spectrophotometer and integrity of RNA was confirmed with a bioanalyzer RNA 6000 nano chip. RNA libraries were generated using an Illumina poly-A select mRNA kit, and a unique 3 base barcode was included into each library. All libraries were simultaneously sequenced in a single lane of an Illumina GAIIx at the Microarray and Genomic Analysis Core Facility of the Huntsman Cancer Institute, University of Utah. 36 cycles of sequencing were performed, and each read resulted in 1.1 billion filter passed bases. The resulting data were split by barcodes using SplitBarcode (Christopher Maxwell, U. of Utah). The individual samples were groomed using the FASTQ groomer in the NGS: QC and Manipulation tab, and aligned to the sacCer2 reference genome using the Map with Bowtie for Illumina function of the NGS: Mapping tab of the Galaxy website http://main.g2.bx.psu.edu/ (Blankenberg et al. 2010a; Blankenberg et al. 2010b; Goecks et al. 2010). The aligned sequences are available in GEO accession number (GSE29018) and were then uploaded into Avadis NGS software (STRAND Scientific Intelligence Inc.), reads were normalized, then numerical values were exported and analyzed using Microsoft Excel and Access suites. The reads associated with individual genes and Saccharomyces Genome Database (SGD) Gene Ontology (GO) terms linked to mitosis, cell cycle progression, and MTs were examined both individually and as groups with genes 85% (compared to WT) and below counted as decreased, and genes 115% and higher counted as increased.
RESULTS
Saccharomyces cerevisiae DIS3, MTR3, and MTR4 mutant strains accumulate mitotic spindles
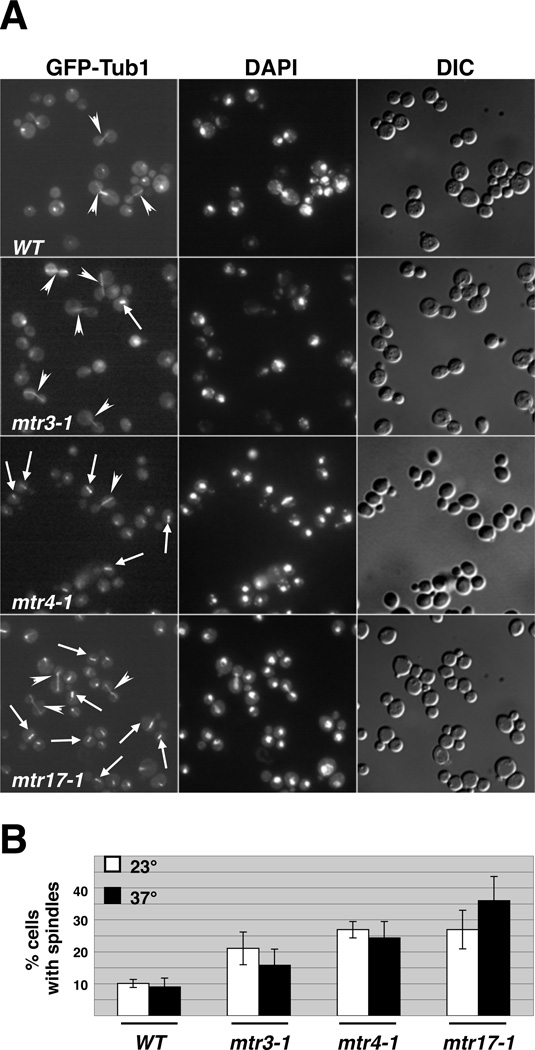
A genetic link between DIS3 and mitosis was observed in Schizosaccharomyces pombe (Ohkura et al. 1988; Kinoshita et al. 1991), and we sought to understand whether this relationship was evolutionarily conserved. We started with mtr17-1, a Saccharomyces cerevisiae DIS3 mutant that was isolated in a genetic selection for defects in mRNA export (Kadowaki et al. 1994; Suzuki et al. 2001). In order to analyze whether the mtr17-1 mutant had mitotic defects, we chromosomally integrated GFP-α-tubulin (GFP-TUB1) to visualize the spindle. Wild-type cells with GFP-TUB1 exhibited anaphase spindles that extended from the mother to daughter cells and had few pre-anaphase spindles (small, short spindles that extend across the nucleus in the mother cell). In contrast, mtr17-1 cells had an abundance of both pre-anaphase and anaphase spindles (Figure 1A), even at the permissive temperature. Quantitative analysis of three different GFP-TUB1 integrants demonstrated that the fraction of mtr17-1 cells with a visible spindle was 2.5- to 3.5-fold higher than in the wild-type strain (Figure 1B).
FIGURE 1.
Strains harboring mutations in DIS3, MTR3, and MTR4 accumulate mitotic spindles. (A) Examining microtubule architecture in exosome subunit and cofactor gene mutant cells. Wild-type or mutant cells expressing GFP-TUB1 were fixed and stained with DAPI to visualize the DNA. Arrows, pre-anaphase spindles; arrowheads, anaphase spindles. (B) Quantitative analysis of mitotic spindle accumulation phenotype in mtr mutants. Cells were scored for the presence of an anaphase or preanaphase spindle at either the permissive (23°) or restrictive (37°) temperatures. At the restrictive temperature, these cells arrest growth and accumulate polyA+ RNA in the nucleus (Kadowaki et al. 1994).
To determine whether this spindle accumulation phenotype was restricted to the mtr17-1 mutant, we expanded our analysis to include two other exosome associated mutants isolated in the mRNA transport screen, mtr3-1, a core exosome subunit, and mtr4-1, a nuclear exosome cofactor (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996). Visual examination (Figure 1A) and quantitative analysis (Figure 1B) of GFP-TUB1 in these mutant strains revealed a 2–3 fold increase in cells containing pre-anaphase or anaphase spindles at both the permissive and non-permissive temperatures. Surprisingly, spindle accumulation at the permissive temperature in these mutants is not accompanied by a slowing of cell growth either in liquid culture or on plates ((Kadowaki et al. 1994; Kadowaki et al. 1995); Figure 4A; data not shown). Mutation of exosome genes are known to affect the levels of mRNAs of unrelated pathways, and thus the temperature sensitivity and spindle-accumulation phenotypes are likely caused by different sets of perturbed mRNAs. Finally, for the sake of brevity, we henceforth refer to these three genes (DIS3, MTR3, and MTR4) and mutants as 3-3-4 genes.
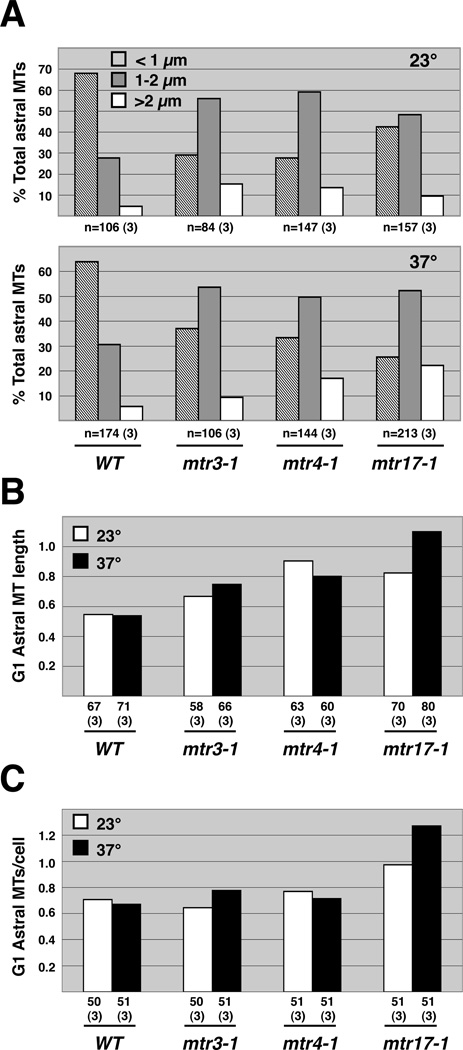
FIGURE 4.
Astral microtubules are elongated in mtr mutants. (A) Cells grown at either the permissive (top) or restrictive (bottom) temperature have longer astral MTs. (B) G1 astral MTs are elongated in mtr mutants. (C) The number of astral MTs in a G1 cell is increased in mtr17-1 mutants. Neither mtr3-1 nor mtr4-1 have this phenotype.
Spindle misorientation and mispositioning in 3-3-4 mutants is an apparent consequence of spindle assembly mistiming
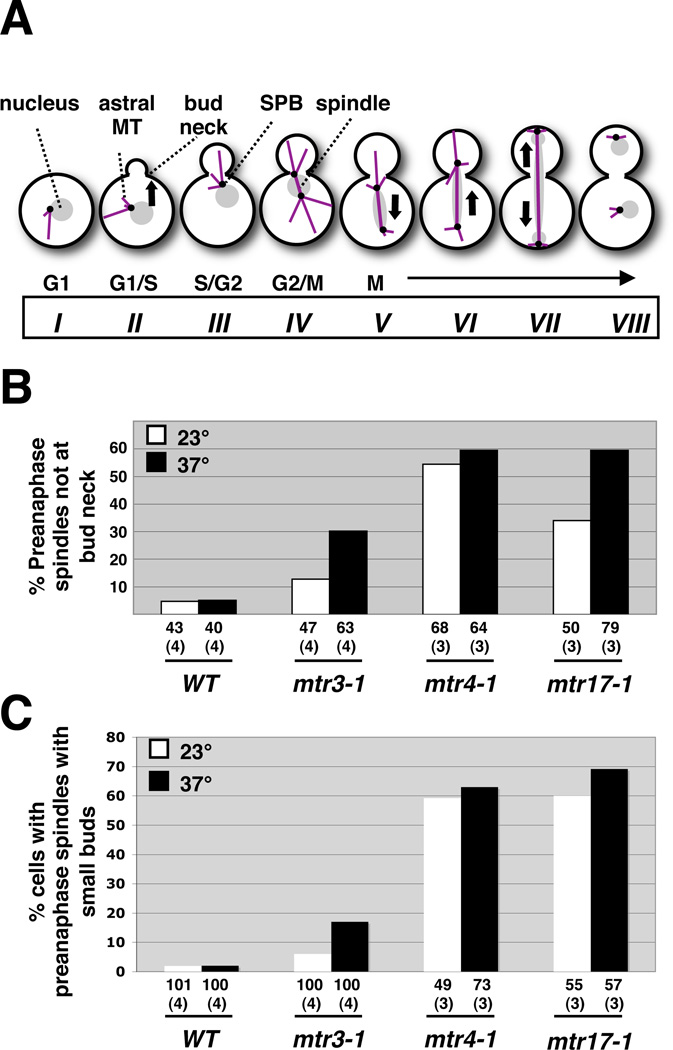
During asymmetric cell division in S. cerevisiae, it is critical that the division plane be orthogonal to the axis of the mitotic spindle. Yeast cells establish a constriction point during bud emergence, and thus, prior to mitosis, the cell has already marked the eventual cleavage site. In a normal yeast cell cycle, pre-anaphase spindles form at the mother-bud neck and orient along the mother-bud axis. Following this, the spindle elongates into the daughter to allow proper chromosome segregation (Figure 3A, stages V-IX; (Shaw et al. 1997; Smeets and Segal 2002; Kusch et al. 2003)). In abnormal spindle assembly, spindles can fail to align properly along the mother-bud axis and to attach at the mother-bud neck (Figure 2A; see below). To better understand the nature of the spindle accumulation phenotype in these 3-3-4 mutants, we characterized the orientation of preanaphase spindles by scoring the angle of the spindle relative to the mother-bud axis. These mutants harbored misoriented preanaphase spindles at both permissive and restrictive temperatures but increased in frequency at the restrictive condition (Figure 2B). As with the spindle accumulation phenotype, both mtr17-1 and mtr4-1 showed more pronounced orientation defects than mtr3-1 (Figure 2C).
FIGURE 3.
Preanaphase spindle assembly is mistimed in the DIS3, MTR3, and MTR4 mutants. (A) Schematic showing the coordination among nuclear migration, bud formation, and spindle formation, orientation, alignment, and disassembly in a normal cell cycle of Saccharomyces cerevisiae. The roman numerals signify the arbitrary stages. Note that the preanaphase spindle forms in stage IV, when the bud has almost achieved its full size. Adapted from (Shaw et al. 1997). (B) Preanaphase spindles are displaced from the bud neck in mtr mutants. In this analysis, the positioning of the spindle and the nucleus (DNA stain) was scored relative to the bud neck. There is a strong correlation between preanaphase spindle misorientation and mispositioning. (C) mtr mutants uncouple normal spindle assembly timing from bud growth. These mutants have a preanaphase spindle (observed in stage IV) in a cell that morphologically is at stage II or III. This phenotype is observed both at the permissive and restrictive temperature (right panel).
FIGURE 2.
Preanaphase spindles in mtr mutants are misoriented. (A) Germane features of the preanaphase spindle in budding yeast. The angle corresponds to the positioning of the preanaphase spindle along the mother bud axis. (B) and (C) mtr mutants accumulate misoriented preanaphase spindles. At both the permissive (top) and restrictive (bottom) temperatures, the frequency of properly oriented (0–30°) spindles decreases with a concomitant increase in the misoriented spindles. Note the 3- to 5-fold increase in misoriented spindles highlighted in (C).
In order for the preanaphase spindle to orient properly, it must be positioned at the bud neck through interactions between the astral MTs and the septal ring at the bud cell cortex (Figure 3A; (Palmer et al. 1992; Sullivan and Huffaker 1992; Shaw et al. 1997; Hwang et al. 2003)). We thus tested whether the 3-3-4 mutants had lost these interactions by scoring the preanaphase spindles as proximal to or detached from the bud neck (e.g., as shown in Figure 1A). We observed that up to 60% of the preanaphase spindles were mislocalized within the mother cell in these mutants (Figure 3B). Again, both mtr17-1 and mtr4-1 mutants exhibited a higher frequency of mispositioned preanaphase spindles than did mtr3-1, with more pronounced phenotypes at 37° for all the mutants.
Spindles that fail to orient along the mother-bud axis can elongate and segregate chromosomes in the mother cell (Sullivan and Huffaker 1992). We considered the possibility that 3-3-4 mutant cells were undergoing a process to correct premature, or mistimed, spindle assembly. Since the spindle does not assemble and elongate in a normal yeast cell cycle until the bud has enlarged significantly (Figure 3A, IV–VIII), we assayed this by examining the size of the bud (as an indicator of cell cycle stage) in these 3-3-4 mutants relative to the wild-type cells. Remarkably, the preanaphase spindles, normally found in cells with enlarged buds (Figure 3A, IV), are present in 3-3-4 cells with small buds (Figure 3A, II and III). This indicates that mutation of either DIS3 or MTR4 (and, to a lesser extent, MTR3) uncouples spindle formation from cell (and bud) growth and normal cell cycle cues.
General astral MT defects in 3-3-4 mutants
To orient along the mother-bud axis, the spindle first attaches at the bud neck via interactions between molecules linking astral MTs to the bud neck (Figure 3A; (Palmer et al. 1992; Sullivan and Huffaker 1992; Shaw et al. 1997; Hwang et al. 2003)). Hence, a general astral MT defect might cause the observed 3-3-4 spindle misoriention, mispositioning, and mistiming phenotypes. To explore this idea, we measured the lengths of the astral MTs (as described in (Shaw et al. 1997)) that were associated with preanaphase spindles at both permissive and restrictive temperatures (Figure 4A). The 3-3-4 mutants showed an increase in the number of long astral MTs relative to the wild-type control strain. Once again, mtr17-1 and mtr4-1 had more pronounced phenotypes than mtr3-1. Thus, these data show that spindle phenotypes are accompanied by changes in spindle-associated astral MTs.
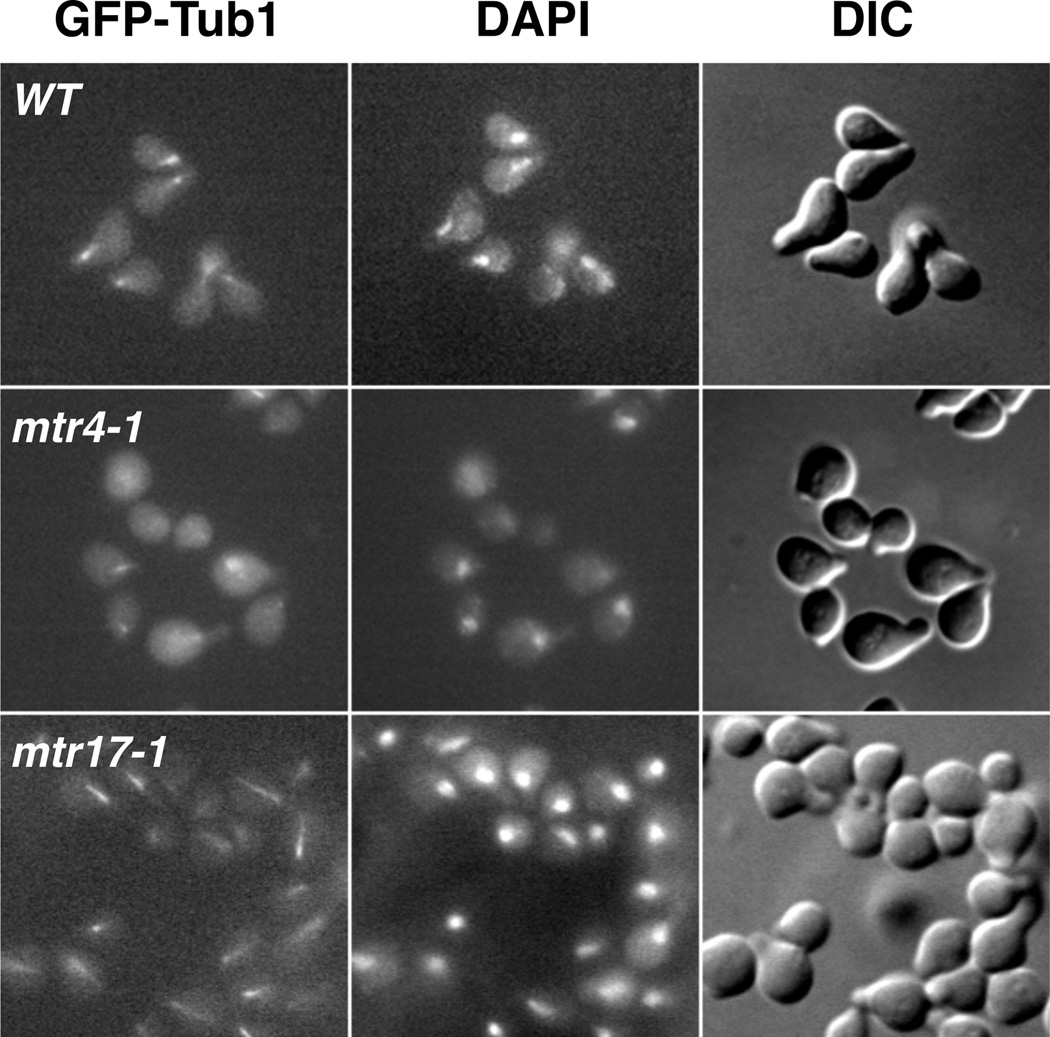
To determine whether there was a general defect in astral MT length control, we scored the length of astral MTs in G1, or unbudded, cells (Figure 3A, I). We observed long astral MTs in the 3-3-4 cells in G1 as well (Figure 4B). This was not, however, accompanied by a change in G1 astral MT number for mtr3-1 or mtr4-1. In contrast, we did observe up to a two-fold increase in G1 astral MTs in mtr17-1 (Figure 4C). Moreover, α-factor treated mtr17-1 cells exhibit elongated astral MTs, with an average length of 2.4 microns (141 astral MTs scored; Figure 5). In contrast, both the wild-type and mtr4-1 cells exhibited shorter MTs (~1.7 microns, 128 scored). These elongated astral MTs in mtr17-1 cells did not affect the length of the schmoo or compromise the rate or efficiency of release from the α-factor block (data not shown). Taken together, these genetic and cell biological data suggest that mtr17-1 has a more penetrant MT phenotype than mtr3-1 or mtr4-1.
FIGURE 5.
The mtr17-1 strain, but not mtr4-1, has extended cytoplasmic microtubules in response to α-factor. mtr3-1 cells could not be examined here because they are MATα and thus not responsive to α-factor treatment.
Deep sequencing analysis shows that 3-3-4 mutants exhibit changes in cell cycle-related mRNAs
As 3-3-4 genes have been linked to multiple steps of RNA metabolism, we reasoned that the MT and cell cycle phenotypes we observed might be an indirect consequence of changes in the levels of mRNAs related to theses processes. Rather than assay a few MT and cell cycle mRNAs by RT-PCR, we used RNA deep sequencing (RNA-seq) to take an unbiased look at the entire population of mRNAs within each strain. We then searched the RNA-seq data for genes with Saccharomyces Genome Database Gene Ontology (GO) terms related to mitosis and MTs and found 190 genes within 16 GO categories. The average changes in mRNA levels (compared to WT) of each GO category is presented in Table 1, while the individual transcript levels are listed in Tables S2.1a–S2.16a. To identify trends we set an arbitrary cut-off of 15% change in transcript level compared to the WT. We chose this cut-off level because the number of genes within each GO category is low and therefore setting too stringent of a cut off will eliminate both real and artifactual trends. Tables tallying the number of increased (≥ 115% of WT levels) and decreased (≤ 85%) genes within each GO category are shown (Tables S2.1b–S2.16b). Within a GO category, individual transcript levels varied from the wild type by as much as 6-fold. When combining our 14 mitosis-related GO categories, transcripts in mtr4-1 were essentially unchanged (102%) but were increased in mtr3-1 and mtr17-1 (120% and 147%, respectively) strains (Table 1). What is striking is that the average transcript level within the entire RNA-seq dataset is ~18% decreased for each of the 3-3-4 mutants, and yet the mitosis-related subset of genes is unchanged or increased. In addition, the average change in transcript level of the combined GO categories masks larger changes, which are evident when examining individual genes within a GO category.
Table 1.
Levels of mitosis related genes altered in mtr mutant strains
| Go Term (# of genes) | Set Avg WT |
Set Avg mtr3-1 |
Set Avg mtr4-1 |
Set Avg mtr17-1 |
|||
|---|---|---|---|---|---|---|---|
|
General mitosis Negative Regulation of mitotic exit (6) |
100 | 110.8 | 84.3 | 111.7 | |||
| Regulation of mitotic exit (16) | 100 | 153.1 | 112.8 | 158.7 | |||
| mitotic cell cycle spindle assembly checkpoint (20) | 100 | 94.6 | 88.5 | 121.3 | |||
| mitotic metaphase/anaphase transition (14) | 100 | 133.2 | 133.7 | 250.2 | |||
| regulation of mitotic metaphase- anaphase transition (4) | 100 | 141.3 | 84.6 | 160.8 | |||
| G1/S Checkpoint (1) | 100 | 71.3 | 116.0 | 106.9 | |||
| G2/M transition of mitotic cell cycle (29) | 100 | 99.4 | 87.8 | 124.9 | Set Avg mtr3-1 |
Set Avg mtr4-1 |
Set Avg mtr17-1 |
| regulation of exit from mitosis (20) | 100 | 145.6 | 112.6 | 157.5 | 118.7 | 102.5 | 149.0 |
|
Spindle orientation/position establishment of mitotic spindle orientation (12) |
100 | 121.0 | 97.0 | 162.2 | Set Avg mtr3-1 |
Set Avg mtr4-1 |
Set Avg mtr17-1 |
| mitotic cell cycle spindle orientation checkpoint (6) | 100 | 157.4 | 177.3 | 192.2 | 139.2 | 137.1 | 177.2 |
|
Bud associated Cell budding (8) |
100 | 100.9 | 59.7 | 91.9 | |||
| Septin Ring assembly (9) | 100 | 130.7 | 101.4 | 139.6 | Set Avg mtr3-1 |
Set Avg mtr4-1 |
Set Avg mtr17-1 |
| mitotic spindle elongation (23) | 100 | 109.0 | 89.6 | 166.3 | 113.5 | 83.5 | 132.6 |
|
Astral Microtubules Astral Microtubules (2) |
100 | 116.8 | 78.7 | 109.5 | |||
| selected Go-term average Whole Sample Average |
100 100 |
120.4 83.7 |
101.7 81.3 |
146.7 81.4 |
|||
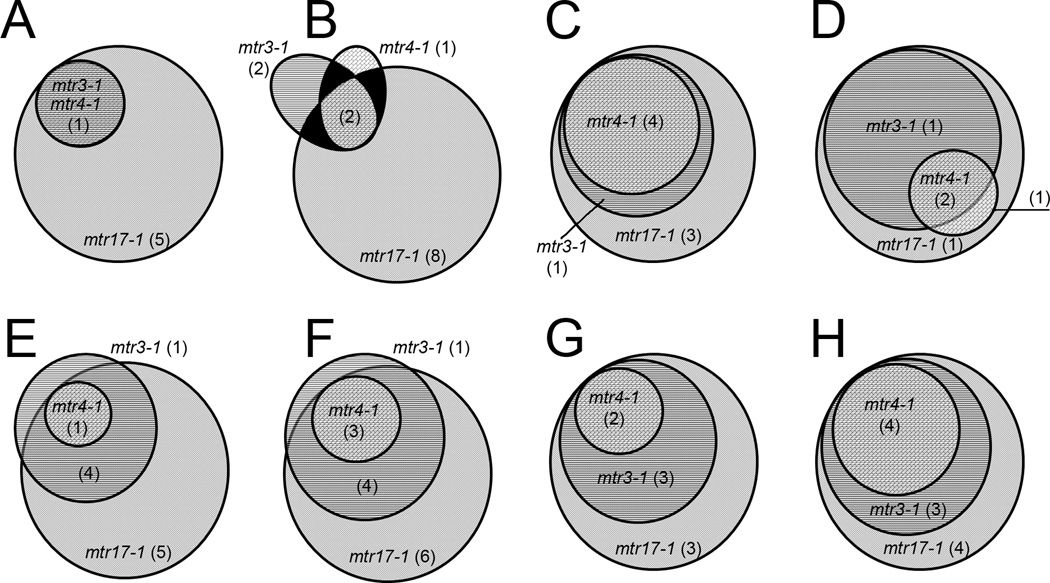
A pattern of differential mRNA levels emerged when Venn diagrams were prepared for all increased transcripts from mitosis-related GO categories containing more than 10 genes (Figure 6). In each of these GO categories, the mtr17-1 mutant always accounted for the largest number of increased genes, and with only three exceptions, all the genes increased in mtr3-1 and mtr4-1 were also increased in mtr17-1 (Figure 6). Also, nearly every mitosis transcript that increased in the mtr4-1 strain also increased in the mtr3-1 strain. Large changes (2 to 4 fold) in some transcripts associated with the spindle orientation checkpoint, suggests that Dis3 is required to properly survey those mRNAs (Table 1, Tables S2.8 and S2.9). We also observed that the levels of the tubulin transcripts (TUB1, TUB2, and TUB3) were essentially unchanged in mtr3-1 (three gene average of 103%) and mtr4-1 (91%) strains, but they were increased (137%) in the mtr17-1 strain (Table S2.2). Notably, nearly every gene associated with the tubulin complex assembly factor GO term was down-regulated in all 3-3-4 strains (Table S2.1). The consequences of this mis-regulation still needs to be determined. These RNA-seq data support the idea that the cell biological phenotypes (i.e., aberrant spindle positioning, orientation, elongation) may emerge from metabolic changes in mRNAs encoding important polypeptides in mitotic and cell cycle regulatory pathways.
FIGURE 6.
Hierarchical regulation of mitosis-related transcripts by mtr mutants. Venn diagrams showing the relationships, as observed by deep sequencing, of increased (≥115% of WT levels) genes in SGD GO terms associated with mitosis. After (A) tubulin associated, the diagrams are shown in cell cycle order beginning with (B) G2/M transition, (C) Mitotic cell cycle spindle assembly checkpoint, (D) Establishment of mitotic spindle orientation, (E) Mitotic metaphase/anaphase transition, (F) Mitotic spindle elongation, (G) Regulation of mitotic exit, and (H) Regulation of exit from mitosis. Only GO terms with ten or more genes are depicted.
Spindle phenotypes of 3-3-4 mutants: A consequence of a general MT assembly or structural defect?
In addition to investigating cell biological phenotypes related to mitosis, we also inquired whether 3-3-4 mutants had phenotypes associated with a general defect in MT structure and function. We plated cultures of mutant and wild-type yeast cells onto control media or media containing the MT destabilizing drug, benomyl (Figure 7). Each of the 3-3-4 mutants were hypersensitive to this drug in comparison to the wild-type strain. Lesser phenotypes were also observed with nocodazole (Table 2). Importantly, wild-type copies of these genes complemented the MT drug sensitivity, indicating that these and other MT phenotypes (Figures 1–4) are due to loss of Dis3, Mtr3, and Mtr4 function. By comparison, yeast cells overexpressing either DIS3 or MTR3 from the GAL1 promoter were not more sensitive than the wild-type strain to a range (5–25 µg/ml) of benomyl concentrations (data not shown). Consistent with our results, the S. pombe dis3-54 allele is hypersensitive to MT poisons (Murakami et al. 2007).
FIGURE 7.
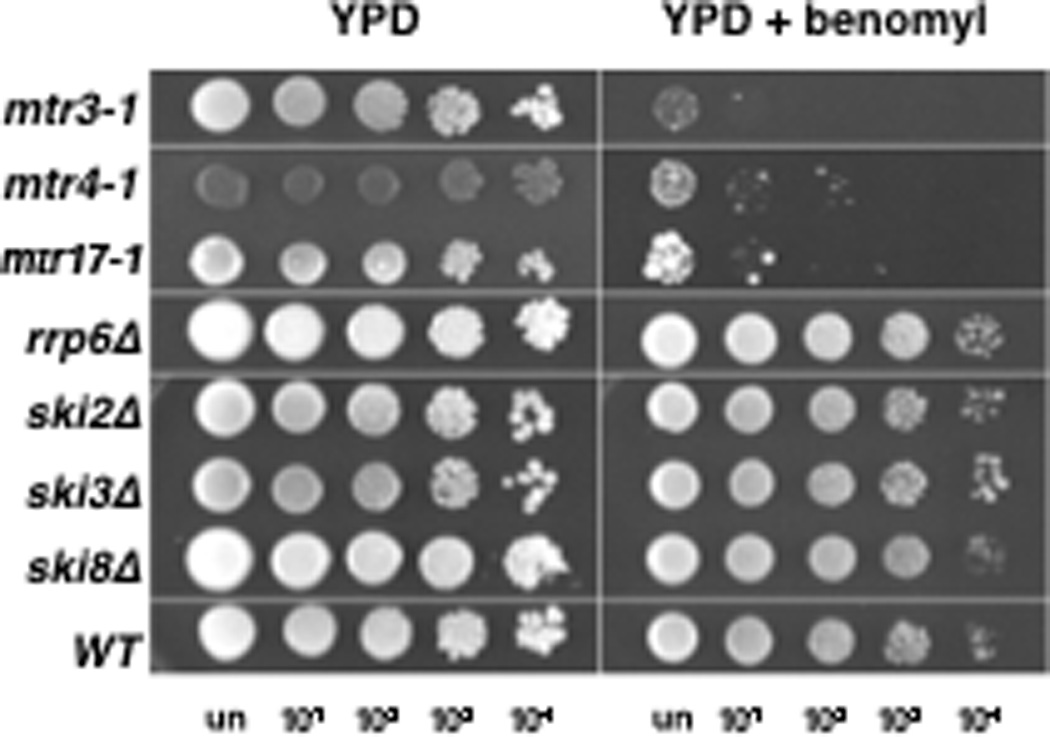
A general microtubule defect in mtr17-1, mtr3-1, and mtr4-1 mutants. Mutants are hypersensitive to the MT destabilizing drug benomyl. In contrast, null mutants of RRP6, SKI2, SKI3, and SKI8 are insensitive to the drug. Cells were serially diluted 10-fold and dilutions plated onto control media or media containing 10 µg/ml benomyl.
Table 2.
Growth of mtr and exosome gene mutant strains on MT poisons
| Drug | |||
|---|---|---|---|
| Strain | Ben | Noc | Gene Function |
| WT | + | + | NA |
| mtr1-1 | + | + | Ran GTPase GEF Rcc1 |
| mtr2-1 | − | ± | mRNA transport factor |
| mtr3-1 | − | ± | Core exosome subunit |
| mtr4-1 | − | ± | Nuclear exosome cofactor |
| mtr5-1 | ± | − | ND |
| mtr6-1 | ± | − | ND |
| mtr7-1 | ± | ± | Acetyl-CoA carboxylase |
| mtr8-1 | + | − | ND |
| mtr10-1 | ± | + | Nuclear import receptor |
| mtr11-1 | + | + | ND |
| mtr12-1 | + | + | ND |
| mtr13-1 | + | + | RNA binding protein |
| mtr14-1 | + | + | Core exosome subunit Rrp40 |
| mtr15-1 | + | + | ND |
| mtr16-1 | + | ± | ND |
| mtr17-1 | − | − | Mitotic regulator Dis3 |
| rrp4-1 | ± | ± | Core exosome subunit |
| ski6-100 | ± | + | Core exosome subunit |
| ski2Δ | + | + | Cytoplasmic exosome cofactor |
| ski3Δ | + | + | Cytoplasmic exosome cofactor |
| ski8Δ | + | + | Cytoplasmic exosome cofactor |
| rrp6Δ | + | + | Nuclear exosome subunit |
| rrp47Δ | + | ND | Nuclear exosome cofactor |
Ben, benomyl; Noc, nocodazole; ND, not determined; NA, not applicable.
Streak growth at 25°: good (+), intermediate (±), or poor (−).
By comparison, we observed that a strain lacking Rrp6 was not sensitive to benomyl. This result is different from that of a recent large scale study (Daniel et al. 2006), but that study did not quantify the rrp6Δ sensitivity to benomyl and was in a different strain background. Finally, mutations in the cytoplasmic exosome cofactor genes SKI2, SKI3, and SKI8 did not confer sensitivity to benomyl or nocodazole (Figure 7 and Table 2). We conclude that 3-3-4 mutants cause a general defect in MT structure and/or function. As a specificity test, we determined whether all mtr mutants, not just the 3-3-4 group, were sensitive to MT poisons (Table 2). Importantly, the mtr14-1 mutant, a previously undisclosed allele of the core exosome subunit RRP40, was insensitive to the drugs. Moreover, other than a strain harboring a mutation in the known mRNA export factor, MTR2, no other mtr strain exhibited hypersensitivity to benomyl. Collectively, these data show that MT drug sensitivity is specific to 3-3-4 mutants, and not a general characteristic of mtr or exosome core or cofactor gene mutants.
An RRP4 mutant allele exhibits MT and cell cycle phenotypes at permissive and restrictive temperatures
To examine whether these phenotypes were specific to 3-3-4 genes or are strain background specific, we analyzed a temperature-sensitive rrp4-1 strain that has clear defects in RNA processing and decay at 37° (Mitchell et al. 1996; de la Cruz et al. 1998; van Hoof et al. 2000a). Serial dilution experiments with this strain revealed some sensitivity to benomyl (Figure S1A) as well as to nocodazole (Table 2). This suggests that proper Rrp4 function, levels, or both is important to maintain MT integrity. In order to determine whether rrp4-1 had defects in spindle assembly and function, we chromosomally integrated GFP-TUB1 in this strain. Visual examination of GFP-TUB1 in rrp4-1 revealed an increase in the fraction of cells containing spindles at the permissive temperature (Figure S1B, bottom). At the restrictive temperature, many but not all rrp4-1 cells arrested with a large bud, a short spindle, and the nucleus at the bud neck. This phenotype is consistent with a cdc arrest and differs from the 3-3-4 mutants; we have not pursued this further. In contrast, wild-type cells did not exhibit these phenotypes (Figure S1B, top). We conclude that perturbation of RRP4 function affects cell cycle progression in a related yet distinct manner from 3-3-4 genes.
DISCUSSION
We present several lines of evidence from S. cerevisiae that extend the previous observations linking Dis3 to mitosis in S. pombe. We also demonstrate an important role for Dis3 in MT dynamics, and extend these observations to other physically- and functionally-interacting partners of Dis3.
Dis3 and core exosome subunit contributions to MT organization
Our genetic, cell biological, and RNA-seq data indicate that Dis3, Mtr3, and Mtr4 have related but distinct functionalities in MT organization. These three proteins may function as part of the core exosome, or as one or more exozymes – subunit complexes that are structurally and functionally independent from the core exosome (for a review see (Kiss and Andrulis 2011)). In this regard, Mtr4 is found in at least two different TRAMP complexes (TRAMP4 and TRAMP5) that survey both overlapping and distinct sets of RNAs (San Paolo et al. 2009). Thus, the mtr4-1 phenotypes could be due to disrupted TRAMP-dependent polyadenylation that is Dis3- and core-independent. As increased mitosis-related transcripts are most abundant in the mtr17-1 strain and often excluded from the mtr3-1 (a core exosome subunit) strain, our RNA-seq data suggest that, just like Rrp6 (Callahan and Butler 2008; Callahan and Butler 2010), Dis3 has RNA processing roles outside the context of the core exosome. An alternate explanation is that Dis3 serves as a master regulator of core exosome-mediated RNA surveillance, but also has RNA metabolic functions that are independent from the core exosome (Kiss and Andrulis, unpublished data). The data presented here are consistent with, and cannot distinguish between, both of these explanations.
More generally, since Mtr proteins are involved in mRNA transport (Kadowaki et al. 1994), the severe MT phenotypes of the 3-3-4 strains could arise indirectly from perturbed nucleocytoplasmic RNA mobilization. Alternatively, 3-3-4 phenotypes could arise from a mis-regulation of 3-3-4 physical interactions in one or more exozymes – thereby disrupting substrate recognition and metabolism. Finally, given that the levels of exosome subunits are under tight cellular control in Trypanosomes and both human and fly cells (Estevez et al. 2003; Graham et al. 2006; van Dijk et al. 2007; Kiss and Andrulis 2010), it is also possible that the mis-expression of 3-3-4 genes or mis-folding of their gene products could change their relational stability and stoichiometry. Additional biochemical studies are required to address these issue and determine whether these polypeptides have direct roles—whether overlapping or distinct—in regulating MT structure and function.
Mitotic defects as an indirect consequence of disrupted mRNA metabolism
Given the roles of Dis3 and other exosome subunits and cofactors in numerous distinct RNA metabolic pathways, it is reasonable that the observed MT phenotypes arise due to defects in these pathways. For example, the DIS3, MTR3, and MTR4 alleles used in this study were isolated in a genetic screen for mutant yeast that accumulate poly A+ RNA in the nucleus (Kadowaki et al. 1994). DIS3 and other core exosome subunits are known to function in rRNA processing (Mitchell et al. 1997; Allmang et al. 1999a; Allmang et al. 2000). A defect in rRNA processing has the potential to deplete ribosomes, thereby reducing translational tempo and/or output, thus leading to a diminished pool of proteins critical for temporally and spatially regulating MT assembly. In this regard, the yeast 60S ribosomal synthesis factor Rrp14 is required for proper mitotic spindle orientation (Oeffinger et al. 2007). Alternatively, defects in Dis3- or exosome-mediated 3’ processing of snRNAs, snoRNAs, or mRNAs (Allmang et al. 1999a; van Hoof et al. 2000a; van Hoof et al. 2000b) could broadly perturb gene expression or, more narrowly, a particular class of transcripts, and thus, indirectly affect the MTs. Finally, Dis3-mediated surveillance and turnover of defective tRNAs (Kadaba et al. 2004; Schneider et al. 2007) could also indirectly affect MTs by perturbing translation. Consistent with this possibility, the mtr17-1 strain does have a tRNA processing defect (Kadowaki et al. 1994).
Although specific defects in RNA transport, surveillance, and processing are possible explanations for the 3-3-4 phenotypes, we consider a general RNA metabolic defect to be a more parsimonious explanation. Indeed, both our RNA-seq data and published data showing that DIS3 and exosome genes have been implicated in turnover of nuclear and cytoplasmic mRNAs support this point of view (Bousquet-Antonelli et al. 2000; van Hoof et al. 2000a; Das et al. 2003). One possibility is that the observed MT phenotypes arise from the inappropriate stabilization of an mRNA(s) encoding MTs themselves or a MT assembly, nucleation, or maintenance factor(s). Alternatively, unregulated or aberrant Dis3- or exozyme-mediated mRNA turnover on a global scale could elicit the observed MT phenotypes.
Growing number of links between RNA metabolic proteins and cell cycle progression
There are several additional links between RNA metabolic proteins and mitosis that are relevant to this discussion. For example, human and fission yeast Dis3 interacts physically with Ran – a validated central player in MT dynamics and spindle assembly – in vitro and in vivo, Dis3 stimulates Ran GEF activity, and S. cerevisiae DIS3 and Ran gene mutants share RNA processing phenotypes (Noguchi et al. 1996; Shiomi et al. 1998; Suzuki et al. 2001). Although we did not explore how Dis3/Ran interaction is related to our spindle phenotypes, we are curious about and cannot outright dismiss a mechanistic contribution.
Our 3-3-4 data join a growing body of evidence linking ribonucleometabolic factors and pathways to MT function and cell cycle progression. Mutations in the budding and fission yeast cytoplasmic 5’ to 3’ exoribonuclease Xrn1 (Interthal et al. 1995; Szankasi and Smith 1996; Pathak et al. 2005), the dhp1+ gene encoding a nuclear 5’ to 3’ exoribonuclease (Shobuike et al. 2001), the nonessential rRNA polyadenylation factor Cid14 (Win et al. 2006), the cleavage and polyadenylation factors Pfs2 (Wang et al. 2005) and Glc7/PP1/Dis2 (Ohkura et al. 1988; Proudfoot 2004; Trinkle-Mulcahy and Lamond 2006), the RNase III gene, RNT1, and the nucleolar ribonucleoprotein endoribonuclease MRP (Cai et al. 2002; Gill et al. 2004; Gill et al. 2006) exhibit, with varying severity and scope, defects in mitotic entry, progression, or exit. We have also found that depletion of Rrp6 and Dis3 from Drosophila cells results in defects in mitotic chromosome dynamics and cell proliferation (Graham et al. 2009c; Kiss and Andrulis 2010). These findings, as a whole, suggest that proteins controlling RNA metabolism play critical roles in cell cycle progression.
CONCLUSIONS
This study describes the observation that mutation of budding yeast DIS3 and certain exosome subunit or cofactor genes affects MTs and the coupling of spindle assembly to cell morphogenesis. By examining the intact cell and a particular class of mRNAs, we have detected some unprecedented connections concerning Dis3, Rrp6, and exosome subunit localization, interactions, and function (this study and (Graham et al. 2006; Graham et al. 2009a)). The molecular mechanisms of how these RNases are important for cell cycle progression and spindle function in several eukaryotes suggests a common, conserved cellular mechanism that awaits elucidation.
Supplementary Material
FIGURE S1. Proper RRP4 function is required for growth in response to a MT poison and for cell cycle progression. (A) rrp4-1 cells are hypersensitive to benomyl. Spotting assays as described in Figure 5. (B) The rrp4-1 mutant cells accumulate spindles at 23° and have large buds and nuclei at their bud necks at 37°.
ACKNOWLEDGEMENTS
We would like to thank Roy Parker, Ambro van Hoof, Philip Mitchell and David Tollervey for yeast strains and T. Huffaker and members of the Andrulis lab for discussions and comments on the manuscript. We thank Christopher Maxwell for the barcode splitting software. This work was supported by Research Grant from the American Foundation for Aging Research (AFAR AA05105) and by a grant from the National Institutes of Health (R01 GM072820) to E.D.A.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases. Genes Dev. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010a;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Chapter 19. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 19 10. 2010b. pp. 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3' end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Burke DJ, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Press; 2000. [Google Scholar]
- Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP. The exosome: a macromolecular cage for controlled RNA degradation. Mol Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161:1029–1042. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. The Journal of biological chemistry. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, Alonso J, Brukhin V, Grossniklaus U, Ecker JR, Belostotsky DA. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci. 2007;64:1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Keyes BE, Ng YP, Freeman CO, Burke DJ. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. The Ran GTPase: theme and variations. Curr Biol. 2002;12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3' end formation of 5.8S rRNA in Saccharomyces cerevisiae. Embo J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Eastman A. Cell cycle checkpoints and their impact on anticancer therapeutic strategies. J Cell Biochem. 2004;91:223–231. doi: 10.1002/jcb.10699. [DOI] [PubMed] [Google Scholar]
- Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenburg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gill T, Aulds J, Schmitt ME. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B, Zheng Y. Mitotic spindle morphogenesis: Ran on the microtubule cytoskeleton and beyond. Biochem Soc Trans. 2006;34:716–721. doi: 10.1042/BST0340716. [DOI] [PubMed] [Google Scholar]
- Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome and importin-alpha3. Traffic. 2009a;10:499–513. doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome, and importin-alpha3. Traffic. 2009b;10:499–513. doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED. Core exosome-independent roles for Rrp6 in cell cycle progression. Mol Biol Cell. 2009c;20:2242–2253. doi: 10.1091/mbc.E08-08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast. 2006;23:439–454. doi: 10.1002/yea.1369. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Bellocq C, Bahler J, Bashkirov VI, Edelstein S, Heyer WD. A role of Sep1 (=Kem1, Xrn1) as a microtubule-associated protein in Saccharomyces cerevisiae. Embo J. 1995;14:1057–1066. doi: 10.1002/j.1460-2075.1995.tb07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Schneiter R, Hitomi M, Tartakoff AM. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Goebl M, Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol. 1991;11:5839–5847. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED. Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA. 2010;16:781–791. doi: 10.1261/rna.1906710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED. The exozyme model: a continuum of functionally distinct complexes. Rna. 2011;17:1–13. doi: 10.1261/rna.2364811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, Liakopoulos D, Barral Y. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 2003;13:562–569. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Cao K, Zheng Y. Ran in the spindle checkpoint: a new function for a versatile GTPase. Trends Cell Biol. 2003;13:553–557. doi: 10.1016/j.tcb.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, Saibil HR, Lorentzen E. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–942. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamolen M, Andrulis ED. Characterization of the Drosophila melanogaster Dis3 ribonuclease. Biochem Biophys Res Commun. 2009;390:529–534. doi: 10.1016/j.bbrc.2009.09.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamolen M, Smith A, Andrulis ED. Drosophila melanogaster Dis3 N-terminal domains are required for ribonuclease activities, nuclear localization and exosome interactions. Nucleic Acids Res. 2010;38:5507–5517. doi: 10.1093/nar/gkq295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3' processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'-->5' exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. The 3' end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. Musing on the structural organization of the exosome complex. Nat Struct Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease Activity of Dis3 Is Required for Mitotic Progression and Provides a Possible Link between Heterochromatin and Kinetochore Function. PLoS ONE. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, Nishimoto T. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. Embo J. 1996;15:5595–5605. [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Fatica A, Rout MP, Tollervey D. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 2007;35:1354–1366. doi: 10.1093/nar/gkl824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. Embo J. 1988;7:1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CC, Gonzales FA, Zanchin NI. Temperature-sensitive mutants of the exosome subunit Rrp43p show a deficiency in mRNA degradation and no longer interact with the exosome. Nucleic Acids Res. 2002;30:4186–4198. doi: 10.1093/nar/gkf545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R, Bogomolnaya LM, Guo J, Polymenis M. A role for KEM1 at the START of the cell cycle in Saccharomyces cerevisiae. Curr Genet. 2005;48:300–309. doi: 10.1007/s00294-005-0030-5. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000555. e1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1013180108. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R, Pardee AB. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Kinoshita N, Ohkura H, Yoshida T, Toda T, Yanagida M. Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol Biol Cell. 1993;4:303–313. doi: 10.1091/mbc.4.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. Human dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae dis3. J Biochem (Tokyo) 1998;123:883–890. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- Shobuike T, Tatebayashi K, Tani T, Sugano S, Ikeda H. The dhp1(+) gene, encoding a putative nuclear 5'-->3' exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucleic Acids Res. 2001;29:1326–1333. doi: 10.1093/nar/29.6.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets MF, Segal M. Spindle polarity in S. cerevisiae: MEN can tell. Cell Cycle. 2002;1:308–311. doi: 10.4161/cc.1.5.143. [DOI] [PubMed] [Google Scholar]
- Stark GR, Taylor WR. Control of the G2/M transition. Mol Biotechnol. 2006;32:227–248. doi: 10.1385/MB:32:3:227. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Noguchi E, Nakashima N, Oki M, Ohba T, Tartakoff A, Ohishi M, Nishimoto T. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3' processing of 7S-to-5.8S rRNA and in degradation of the excised 5'-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics. 2001;158:613–625. doi: 10.1093/genetics/158.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. Requirement of S. pombe exonuclease II, a homologue of S. cerevisiae Sep1, for normal mitotic growth and viability. Curr Genet. 1996;30:284–293. doi: 10.1007/s002940050134. [DOI] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3'-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Lamond AI. Mitotic phosphatases: no longer silent partners. Curr Opin Cell Biol. 2006;18:623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. Rna. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000a;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3'-to-5' degradation of mRNA. Mol Cell Biol. 2000b;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Asakawa K, Win TZ, Toda T, Norbury CJ. Inactivation of the pre-mRNA cleavage and polyadenylation factor Pfs2 in fission yeast causes lethal cell cycle defects. Mol Cell Biol. 2005;25:2288–2296. doi: 10.1128/MCB.25.6.2288-2296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Brenner AA, White TB, Engler MJ, Gaughran JP, Tatchell K. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol Cell Biol. 1991;11:3369–3373. doi: 10.1128/mcb.11.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. Proper RRP4 function is required for growth in response to a MT poison and for cell cycle progression. (A) rrp4-1 cells are hypersensitive to benomyl. Spotting assays as described in Figure 5. (B) The rrp4-1 mutant cells accumulate spindles at 23° and have large buds and nuclei at their bud necks at 37°.