Abstract
Viruses of thermophilic Archaea are unique in both their structures and genomic sequences. The most widespread and arguably best studied are the lemon-shaped fuselloviruses. The spindle-shaped virus morphology is unique to Archaea but widespread therein. The best studied fusellovirus is SSV1 from Beppu, Japan, which infects Sulfolobus solfataricus. Very little is known about the function of the genes in the SSV1 genome. Recently we have developed genetic tools to analyze these genes. In this study, we have deleted three SSV1 open reading frames (ORFs) ranging from completely conserved to poorly conserved: VP2, d244, and b129. Deletion of the universally conserved ORF b129, which encodes a predicted transcriptional regulator, results in loss of infectivity. Deletion of the poorly conserved predicted DNA-binding protein gene VP2 yields viable virus that is indistinguishable from wild-type. Deletion of the well-conserved ORF d244 that encodes a predicted nuclease yields viable virus. However, infection of S. solfataricus with virus lacking ORF d244 dramatically retards host growth, compared to the wild-type virus.
Keywords: DNA binding, nuclease, transcription factor
INTRODUCTION
Viruses of Archaea are very poorly understood with only about 50 known archaeal viruses relative to the ca. 5000 characterized viruses of bacteria, plants, and animals (Pina et al., 2011). The best studied of archaeal viruses are those infecting the thermoacidophiles, with an unprecedented new seven virus families introduced in the last few years to accommodate the astonishing morphological and sequence diversity present in these viruses (Pina et al., 2011).
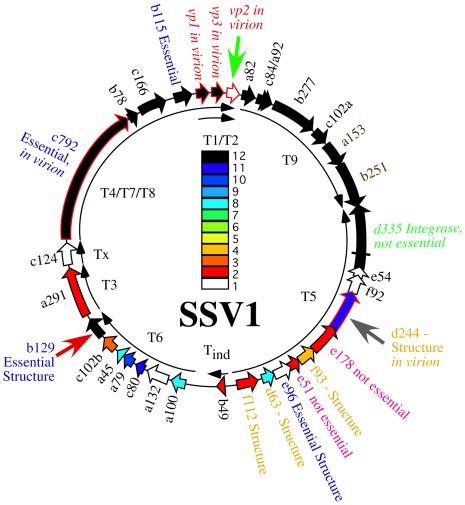
The Sulfolobus spindle-shaped viruses (SSVs) of the family Fuselloviridae were the first discovered and probably the best studied family of archaeal viruses. SSVs are found throughout the world in high temperature (> 70°C) and acidic (pH < 4) environments where their hosts, Sulfolobus solfataricus and its close relatives thrive (Wiedenheft et al., 2004; Held and Whitaker, 2009). The type virus, SSV1, encodes a positively supercoiled, 15.5 kbp circular dsDNA genome (NC_001338.1) that is enclosed within a lemon or spindle-shaped capsid (Yeats et al., 1982; Martin et al., 1984; Nadal et al., 1986). The genome encodes 34 open reading frames (ORFs; Palm et al., 1991), most of which have no recognizable homologs apart from other Fuselloviridae. The only SSV1 gene with clear homology to proteins outside the Fuselloviridae is the viral integrase, encoded by ORF d355. The main structural proteins purified from virus particles are the major and minor capsid proteins VP1 and VP3 and the putative DNA packaging protein VP2 (Reiter et al., 1987a). More recently, mass spectrometric analysis of SSV1 virions revealed two additional proteins, the products of ORFs c792 and d244 (Menon et al., 2008; Figure 1).
FIGURE 1.
Genome map of SSV1. Open reading frames are shown as block arrows and labeled as in Palm et al. (1991). Virus structural protein genes (Reiter et al., 1987a) and other proteins found in the virion (Menon et al., 2008) are outlined in red and labeled as “in virion.” Conservation of open reading frames in 12 canonical SSV genomes (SSV1, SSV2, SSV3, SSV4, SSV5, SSVRH, SSVK1, SSVL, SSVKM1, SSVKU1, SSVL2, and SSVGV1; Redder et al., 2009; Held and Whitaker, 2009; Stedman, unpublished) is listed with the color code in the middle of the genome with ORFs conserved in 12 genomes in black, ORFs conserved in 11 genomes in dark blue, etc. ORFs which did not tolerate insertion of the pBluescript plasmid are labeled as “Essential” in blue type. ORFs allowing insertion of the pBluescript plasmid without loss of virus function are labeled as “not essential” (Stedman et al., 1999). All ORFs whose products have been crystallized and structure determined are labeled as “Structure” (Lawrence et al., 2009; Menon et al., 2010). The gene for the SSV1-integrase is labeled in green and was shown to be not essential by deletion (Clore and Stedman, 2006). Transcripts are labeled as curved thin arrows (Reiter et al., 1987b; Fröls et al., 2007). ORFs targeted in this study are indicated with large arrows outside the genome map.
In the absence of homologous sequences, three complementary approaches have been used to try and determine the function of the proteins encoded in the SSV1 genome; structural genomics, comparative genomics, and genetics. Atomic resolution structures have been obtained by C. Martin Lawrence and his group for proteins encoded by SSV1 ORFs b129, f112, d63, e96, f93, and d244 or their homologs from other fuselloviruses. The products of ORFs b129, f112, and f93 resemble transcriptional regulators and d244 a novel nuclease (Lawrence et al., 2009; Menon et al., 2010). However, the function of these proteins in virus replication remains to be determined. Two of these ORFs, b129 and d244, are the targets of the current study.
In parallel, we and others have undertaken comparative genomic studies. Fifteen ORFs are completely conserved in 12 canonical SSV genomes (Stedman et al., 2003; Wiedenheft et al., 2004; Held and Whitaker, 2009; Redder et al., 2009; Stedman, unpublished; Figure 1). Most of the universally conserved genes are clustered in half of the genome with the notable exception of the VP2 gene, a target of this study. Conservation in the rest of the genome is lower. Nonetheless, there are very few completely unique genes in the SSV1 genome (Figure 1). It is highly probable that the conserved genes are required for virus function, but again this has not been confirmed.
We developed methods for gene disruption in order to determine the requirements for genes in the virus genome directly. About 10 years ago, we showed that four SSV1 ORFs did not tolerate insertion of the 3.2 kbp pBluescript plasmid and allow virus function. Twelve other SSV1 ORFs appeared, indirectly, to not tolerate insertion. However, two ORFs, e178 and e51, were able to tolerate insertion of the entire pBluescript plasmid (Stedman et al., 1999). This result allowed the development of viral shuttle vectors and the beginnings of Sulfolobus genetics (Jonuscheit et al., 2003). Insertion of the pBluescript plasmid and up to ca. 5 kbp of exogenous DNA in these ORFs does not appear to have a noticeable effect on virus function (Stedman et al., 1999; Jonuscheit et al., 2003; Clore and Stedman, 2006; Albers et al., 2006).
However, insertion of large DNA fragments into the SSV1 genome is not straightforward and the possible insertion locations are limited. Therefore, Long Inverse PCR (LIPCR) using high-fidelity highly processive DNA polymerases (e.g., Phusion®) was developed to specifically change the SSV1 genome at single nucleotide resolution. LIPCR was used to delete precisely the SSV1 viral integrase gene. Surprisingly, this “integrase-less” SSV1 was functional (Clore and Stedman, 2006). However, consistent with its conservation, the virus lacking the integrase gene is at a competitive disadvantage relative to integrase-containing viruses (Clore and Stedman, 2006). All of the SSV1 ORFs that can be deleted or tolerate insertion without abrogating virus function are in the “early” transcript, T5, that is induced soon after UV-irradiation of SSV-infected cultures (Reiter et al., 1987b; Fröls et al., 2007).
Three ORFs in the SSV1 genome were targeted for gene disruption in this study. The VP2 gene (NP_039802.1) was chosen for disruption because it is only present in SSV1 and the very distantly related SSV6 (Held and Whitaker, 2009; Redder et al., 2009), and is in the middle of the most highly conserved part of fusellovirus genomes (Figure 1). VP2 has DNA-binding activity (Reiter et al., 1987a; Iverson and Stedman, unpublished) that is presumably required for DNA packaging. ORF b129 (NP_039795.1) was chosen because it is intolerant of insertional mutagenesis (Stedman et al., 1999), a high resolution structure is known (Lawrence et al., 2009) and the gene is completely conserved in all SSVs (Figure 1). Finally, ORF d244 (NP_039781.1) was chosen for gene disruption because a high-resolution structure of its homolog from SSVRH is known (Menon et al., 2008) and it is conserved in most SSV genomes with the exception of SSVK1.
MATERIALS AND METHODS
CULTURE CONDITIONS
Sulfolobus solfataricus strains, Table 1, were grown aerobically at 76°C on plates or in liquid media containing yeast extract and sucrose as carbon and energy sources (YS Media), both as in Jonuscheit et al. (2003). Escherichia coli strains were grown in LB medium at 37°C as suggested by the manufacturer (Novagen).
Table 1.
Strains and plasmid vectors used in this work.
| Strain/vector | Description | Reference |
|---|---|---|
| S. solfataricus GΘ | MT4 Derivative | Cannio et al.(1998) |
| S. solfataricusS443 | Novel Sulfolobus isolate | Unpublished data |
| E.coli NovaBlue® | Expression strain | Novagen, Inc. |
| pAJC97 | SSV1 with TOPO PCR Blunt II | Clore and Stedman (2006) |
| pAJC97-ΔVP2 | pAJC97 lacking VP2 gene | This Work |
| pAJC97-Δd244 | pAJC97 lacking ORF d244 | This Work |
| pAJC97-Δb129 | pAJC97 lacking ORF b129 | This Work |
PURIFICATION OF DNA
Plasmid DNA used for LIPCR was purified from E. coli using the alkaline lysis method of Birnboim and Doly (1979). Plasmid DNA used to transform Sulfolobus was purified using the GeneJet Plasmid Purification Kit (Fermentas) following the manufacturer’s protocols. Total genomic DNA was isolated from S. solfataricus in late log phase growth (OD600 ~0.6) as in Stedman et al. (1999). Plasmid DNA was purified from a 50 mL culture of S. solfataricus transformed with SSV-Δd244 (late log, OD600 ~0.6) using the GeneJet plasmid purification kit (Fermentas) following the manufacturer’s protocols. This DNA was retransformed into E. coli (Novagen), purified therefrom and analyzed by restriction endonuclease digestion with EcoRI (Fermentas).
CONSTRUCTION OF SSV1 DELETION MUTANTS
Deletion mutants were constructed from the pAJC97 shuttle vector using LIPCR (Clore and Stedman, 2006). Primers were designed to overlap with the start and stop codon of the ORF to be deleted to keep the deletion in frame. Initially primers were designed using the archaea genome browser1. Primer melting temperatures were matched and then checked for potential primer dimer and secondary structure formation using online tools from IDT2. Table 2 contains a list of oligonucleotide sequences used. LIPCR was performed using Phusion® High-Fidelity DNA Polymerase (NEB/Finnzymes) at a final concentration of 0.005 U/μL. LIPCR cycling conditions as follows: initial denaturation at 98°C for 3 min; 35 cycles of 98°C for 15 s, annealing for 15 s, 72°C for 6 min, and a final extension at 72°C for 6 min. The annealing temperatures for deletion of VP2, ORF d244, and ORF b129 were 59, 53, and 66°C, respectively. DNA was precipitated directly from LIPCR reactions using sodium acetate at a final concentration of 0.3 M and 95% EtOH. This DNA was phosphorylated using T4 polynucleotide kinase according to the manufacturer’s protocols (Fermentas). DNA was ligated overnight (~20 h) at 16°C using 5 Weiss units of T4 DNA ligase (Fermentas). Ligated DNA was transformed into NovaBlue Singles chemically competent E. coli following the manufacturer’s protocol (Novagen). Plasmids were purified from single colonies and deletion constructs were identified by restriction endonuclease digestions. The deletion borders were confirmed by sequencing of the plasmids.
Table 2.
Oligonucleotides used in this work.
| Name | Sequence | Description |
|---|---|---|
| VP2 LIPCR F | 5′-CAC CGC AAG TAG GCC-3′ | Flanks VP2 gene for deletion |
| VP2 LIPCR R | 5′-CAC CCA CTT CAT ATC ACT CC-3′ | Flanks VP2 gene for deletion |
| d244 LIPCR F | 5′-ATC CAT TTA CCA TAA TCC ACC-3′ | Flanks ORF d244 for deletion |
| d244 LIPCR R | 5′-GGA AAA TGA TAT TCA ACT CAG AGG-3′ | Flanks ORF d244 for deletion |
| b129 LIPCR F | 5′-AGT TAG GCT CTT TTT AAA GTC TAC C-3′ | Flanks ORF b129 for deletion |
| b129 LIPCR R | 5′-TGA CTC CGT CAT CCT CTA AC-3′ | Flanks ORF b129 for deletion |
| VP2 Check F | 5′-ATT CAG ATT CTG WAT WCA GAA C-3′ | Amplifies VP2 gene and flanking sequences |
| VP2 Check R | 5′-TCS CCT AAC GCA CTC ATC-3′ | Amplifies VP2 gene and flanking sequences |
| d244 Check F | 5′-GGA ACT CCT CTC ATT AAC C-3′ | Amplifies ORF d244 and flanking sequences |
| d244 Check R | 5′-GAT CAT CAA CGA GTA TAT TGA CC-3′ | Amplifies ORF d244 and flanking sequences |
| b129 Check F | 5′-ATG AAG GCT GAG GAA ACA ATC GTG-3′ | Amplifies ORF b129 and flanking sequences |
| b129 Check R | 5′-TTA ATA TAG CTG CGA TGC AGT ATA GTT TAT TTG TGC-3′ | Amplifies ORF b129 and flanking sequences |
*Underlined sequence indicates ORF.
ELECTROPORATION OF SULFOLOBUS
Purified plasmid DNA was electroportated into Sulfolobus strain GΘ as in Schleper et al. (1992). Following electroporation (400Ω, 1.5 kV, 25 μF), cells were immediately resuspended in 1 mL of YS media at 75°C and incubated for 1 h at 75°C. The cells were then added to 50 mL of prewarmed YS media (75°C) and grown in liquid media as outlined below.
SCREEN FOR FUNCTIONAL INFECTIOUS VIRUS/HALO ASSAY
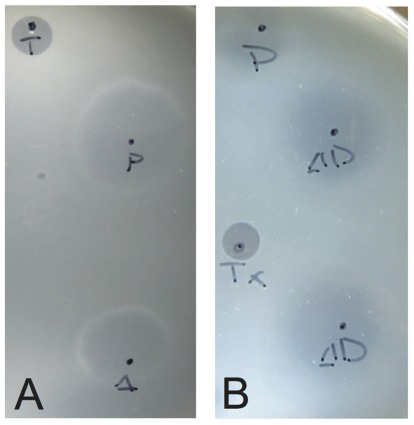
To confirm the presence of infectious virus, halo assays were performed in duplicate 48 and 72 h post-electroporation (Stedman et al., 2003). Uninfected Sulfolobus GΘ cells were diluted to an OD600 nm = ~0.3 and allowed to grow until the OD600 nm reached ~0.35 (about 2.5 h). Half of a milliliter of this uninfected culture was added to 5 mL YS media containing 0.2% wt/vol Gelrite® as a softlayer and poured onto prewarmed YS plates. Two microliters of supernatant from electroporated cultures was spotted onto the lawns and plates were incubated at 75°C for up to 3 days. A halo of host growth inhibition, typically observed 48–72 h after incubation, indicated the presence of an infectious virus (Figure 2).
FIGURE 2.
Typical growth inhibition of S. solfataricus on plates due to infectious virus. Lawns of S. solfataricus strain GΘ were prepared as in Stedman et al. (2003). Two microliters of supernatant from cultures transformed with either (A) SSV-ΔVP2 or (B) SSV-Δd244 were placed on the lawns where indicated. Δ indicates where SSV-ΔVP2 was spotted, ΔD where SSV-Δd244 was spotted. P indicates SSV-WT spotted as a positive control. T or Tx indicates 2 μL of 0.01% Triton X-100 spotted as a control for lawn growth.
GROWTH CURVES
Portions of halos of growth inhibition from infected S. solfataricus GΘ cells were removed from plates with a sterile pipette tip and inoculated into liquid YS media. The culture was grown to an OD600 nm of ~0.6. One milliliter of this culture was diluted in 100 mL YS media to an OD600 ~0.050. Cultures were placed in a shaking incubator at 75°C and the OD600 nm was measured every 24 h. After 96 h, 1 mL of culture was diluted into 100 mL fresh YS media and returned to 75°C. One milliliter of culture was removed 72 h after each dilution, cells removed by centrifugation (14000 rpm for 5 min in a microcentrifuge) and the supernatant was screened for virus using the halo assay above.
TRANSMISSION ELECTRON MICROSCOPY
Supernatant from infected cultures was collected by centrifugation at 14,000 rpm for 5 min in a microcentrifuge. Five microliters of supernatant was absorbed onto a 400 mesh carbon/formvar grid (Ted Pella) for 2 min and negatively stained with 2% uranyl acetate for 20 sec. Grids were viewed on a JEOL 100CX TEM operated at 100 keV and images captured with a Gatan imager.
RESULTS
SSV1 IS INFECTIOUS WITHOUT THE VP2 GENE
The VP2 protein was purified from SSV1 virus particles and reported to be a DNA-binding protein (Reiter et al., 1987a). Surprisingly, a gene for VP2 was not found in SSV2 (Stedman et al., 2003) or SSVRH or SSVK1 (Wiedenheft et al., 2004). Moreover, a homolog is not present in the S. solfataricus or S. islandicus genomes (She et al., 2001; Reno et al., 2009; Guo et al., 2011). However, a very distant relative of SSV1, SSV6, which also contains an atypical putative tail fiber protein, has a VP2 gene (Redder et al., 2009). Thus, it is not clear whether SSV1 can function without a VP2 gene.
Therefore, we made an in-frame deletion of the majority of the VP2 gene by LIPCR in the context of the pAJC97 SSV1 shuttle vector (Clore and Stedman, 2006), leaving the first four codons and the last four codons (including the stop codon) of the ORF intact (see Table 1). The putative promoter for the T9 “early” transcript was also left intact. The construct containing the deletion, pAJC97-ΔVP2, is hereafter referred to as SSV-ΔVP2.
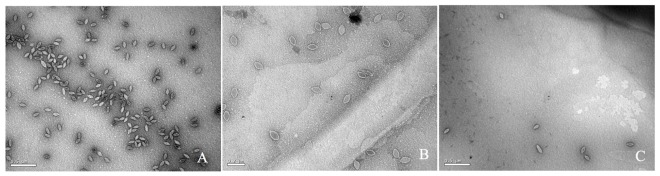
To determine if the SSV-ΔVP2 was able to make infectious virus, the shuttle vector was electroporated into S. solfataricus strain GΘ. Two days after electroporation, the supernatant from the transformed strains caused inhibition of growth of uninfected S. solfataricus strain GΘ on plates (Figure 2) that was indistinguishable from growth inhibition caused by the virus containing the VP2 gene. Similar growth inhibition was also observed on lawns of uninfected S. solfataricus strain S443, a new S. solfataricus isolate from Lassen Volcanic National Park that is a host for all tested SSVs (Ceballos et al., in preparation). Moreover, the supernatant contained SSV-like particles when observed by transmission electron microscopy (Figure 3).
FIGURE 3.
Transmission electron micrographs of SSV particles. Supernatants from cultures of S. solfataricus strain GΘ transformed with (A) pAJC97, (B) SSV-ΔVP2, (C) SSV-Δd244, were negatively stained with uranyl acetate and observed with a JEOL 100CX transmission electron microscope. Bar represents 0.2 μm (B) or 0.5 μm (A,C).
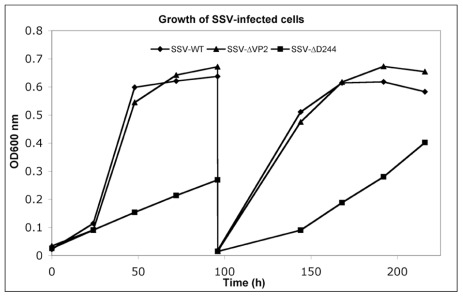
Infection by wild-type SSV1 and shuttle vectors does not drastically slow growth of cells in liquid culture for unknown reasons (Martin et al., 1984; Schleper et al., 1992; Stedman et al., 1999). The same is true of SSV-ΔVP2 (Figure 4). Infection with SSV-ΔVP2 was confirmed via PCR amplification (data not shown)
FIGURE 4.
Typical growth inhibition in liquid culture of virus constructs. Cultures of S. solfataricus GΘ infected with wild-type SSV1, diamonds, SSV-ΔVP2, triangles and SSV-Δd244, squares, were diluted in YS media to equal starting OD600 nm and incubated at 75°C. At the indicated times, samples were removed and the OD600 nm was determined and the presence of virus was confirmed in each culture via halo assay. After 96 h, 1 mL of cells were diluted 1:100 in fresh YS media and returned to 75°C.
SSV1 CONSTRUCTS LACKING THE CONSERVED ORF b129 DO NOT APPEAR TO MAKE INFECTIVE VIRUSES
The b129 ORF in SSV1 is universally conserved in all fuselloviruses (Redder et al., 2009). Moreover shuttle vectors with pBluescript inserted into ORF b129 did not produce infective virus when electroporated into Sulfolobus (Stedman et al., 1999). However, a similar insertion mutant in the equally conserved SSV1 viral integrase appears to be non-functional (Stedman et al., 1999), but an in-frame deletion was functional (Clore and Stedman, 2006). A structure for the b129 ORF is also known (Lawrence et al., 2009) and it contains two Zn-finger putative DNA-binding motifs.
The b129 ORF was deleted with LIPCR. The deletion of the b129 ORF left the first four and last two codons of the ORF intact and maintained the predicted T3 promoter (Reiter et al., 1987b). This construct is referred to as SSV-Δb129. Unlike the SSV-ΔVP2 construct, supernatants from Sulfolobus cells electroporated with SSV1-Δb129 did not cause zones of growth inhibition when spotted on lawns of uninfected S. solfataricus strain GΘ. A total of nine independent transformations were performed in which the wild-type virus consistently caused growth inhibition but SSV-Δb129 did not. Moreover, no halos of growth inhibition were formed on lawns of S. solfataricus strain S443. It is not currently known at which step of virus replication the SSV-Δb129 is deficient.
SSV1 LACKING ORF d244 IS INFECTIOUS BUT HAS A NOVEL PHENOTYPE
SSV1 ORF d244 is in the UV-inducible transcript T5, upstream of the viral integrase gene (Figure 1). The entire pBluescript plasmid can be inserted into the ORF directly upstream of ORF d244 without abrogating SSV1 function (Stedman et al., 1999). ORF d244 is well conserved in other Fusellovirus genomes with the exception of SSVK1 (Wiedenheft et al., 2004; Redder et al., 2009). The X-ray crystal structure of the homolog of SSV1 ORF d244, SSVRH ORF d212 has been solved and it is predicted to be a nuclease (Menon et al., 2010). Moreover, the product of ORF d244 has been reported to be in purified SSV1 particles (Menon et al., 2008).
The SSV1 d244 ORF was deleted with LIPCR. The deletion of the d244 ORF left the first two and last three codons of the ORF intact as well as maintained the ORF to avoid polar effects. This construct is referred to as SSV-Δd244.
To determine if SSV-Δd244 was able to make infectious virus, the shuttle vector was electroporated into S. solfataricus strain GΘ. Two days after electroporation, the supernatant from the transformed strains caused inhibition of growth of uninfected S. solfataricus strain GΘ on plates (Figure 2) and also inhibited growth of S. solfataricus strain S443 (data not shown). The supernatant contained SSV-like particles when observed by transmission electron microscopy (Figure 3).
Infection by wild-type SSV1, shuttle vectors and SSV-ΔVP2 does not slow growth of cells in liquid culture (Martin et al., 1984; Schleper et al., 1992; Stedman et al., 1999; see above). However, infection by SSV-Δd244 drastically slows growth of S. solfataricus strains GΘ and S443 in liquid culture (Figure 4). Infection with SSV-Δd244 was confirmed via PCR. Moreover, restriction endonuclease digestion of viral DNA recovered from transformed S. solfataricus cells and retransformed into E. coli revealed no obvious alterations of the SSV-Δd244 construct (data not shown).
DISCUSSION
THE PUTATIVE DNA PACKAGING PROTEIN VP2 IS NOT REQUIRED FOR SSV1 FUNCTION
The deletion of VP2 from SSV1 results in a functional virus that is indistinguishable from the wild-type virus (Figures 2–4). Based on the lack of conservation of VP2 this result is not completely unexpected. However, almost all viruses contain a genome packaging protein. There is no clear sequence homolog of VP2 in the host genome, but there are a number of small DNA-binding proteins, such as Sso7d or Cren7 that may be able to functionally substitute for VP2 in SSV1 genome packaging (Choli et al., 1988; Guo et al., 2008). This will be tested with mass spectrometry of SSV-ΔVP2 particles. Alternatively, the VP2 protein may be involved in maintenance of the positive supercoiling of the SSV1 viral genome (Nadal et al., 1986). It would be interesting to know if the topology of the viral DNA is affected by the absence of VP2. It is predicted that positive supercoiling should increase the thermal stability of the DNA, so SSV-ΔVP2 may be less thermally stable than the wild-type virus.
The VP2 gene may be more prevalent than previously thought. VP2-like sequences have been reported from metagenomic studies, one in an acid mine drainage metagenome (Andersson and Banfield, 2008) and the other from Boiling Springs Lake in California (Diemer and Stedman, unpublished). These VP2 genes may be in the context of a SSV6 or ASV-like genome (Redder et al., 2009).
THE PRODUCT OF ORF b129 APPEARS TO BE ESSENTIAL FOR SSV1 INFECTIVITY
Homologs of SSV1 ORF b129 are present in all known SSVs (Redder et al., 2009). The b129 ORF also does not tolerate insertion of the pBluescript plasmid (Stedman et al., 1999). Thus, it is not surprising that deletion of ORF b129 leads to an incompletely replicating virus. However, the SSV1 integrase, a gene also conserved in all fuselloviruses, did not appear to tolerate insertion of pBluescript (Stedman et al., 1999), but could be deleted with LIPCR without abrogating virus function (Clore and Stedman, 2006). This indicates that either polar effects are important, which seems unlikely since the SSV1 integrase is at the end of the T5 transcript, or that insufficient replicate transformations were performed in the earlier study.
Nine replicate transformations of S. solfataricus with SSV-Δb129 did not generate functional virus. However, we cannot absolutely determine that SSV1 ORF b129 is essential for virus function without complementation experiments, which are underway. The reasons for the apparent necessity of SSV1 ORF b129 are unclear, but the structure of the b129 ORF product, a predicted transcriptional regulator (Lawrence et al., 2009) and induction of the T6 transcript containing ORF b129 after UV-irradiation (Reiter et al., 1987b; Fröls et al., 2007) provides clues to its function.
The assay used herein for virus infection, ability to cause a zone of growth inhibition on a lawn of uninfected cells, is for virus spread and infectivity. There are many other aspects of virus replication that could be affected by disruption of ORF b129. An attractive hypothesis is that the b129 protein activates transcription of virus structural genes encoded by the “late” transcripts T7/8/9, T1, and T2 (Reiter et al., 1987b; Fröls et al., 2007; see Figure 1). This would be one of very few archaeal transcriptional activators characterized to date and the only the second archaeal viral transcriptional activator (Kessler et al., 2006). Thus, the SSV-Δb129 construct may be able to replicate its genome, integrate into the host, and have genome replication induced by UV-irradiation or some subset of these activities. Experiments to test these hypotheses are underway.
TRANSFECTION WITH SSV-Δd244 PRODUCES VIRUS AND RETARDS HOST CELL GROWTH
The SSV1 d244 ORF is well-conserved in fuselloviruses with the exception of SSVK1 (Wiedenheft et al., 2004; Redder et al., 2009). However, SSV1 lacking ORF d244 clearly makes infectious virus particles (Figures 2 and 3). Moreover, the zones of clearing produced by supernatants of cells transfected with SSV-Δd244 are clearer than those produced by either the wild-type or SSV-ΔVP2 viruses (Figure 3; unpublished data). They are reminiscent of zones of clearing produced by SSVK1 (data not shown). Unlike wild-type virus and SSV-ΔVP2, transfection by SSV-Δd244 leads to drastically reduced host growth (Figure 4). The reasons for this growth inhibition are unclear. Similar growth phenotypes have been observed in SSVK1 infections (Stedman et al., in preparation). SSVK1 consistently produces more virus than similar cultures of the wild-type virus, so this may account for the growth defect (unpublished data). Whether SSV-Δd244 consistently produces more virus than the wild-type or SSV-ΔVP2 is currently unknown.
The structure of the product of SSV1 ORF d244 is a predicted nuclease (Menon et al., 2010), similar to Holiday junction resolvase enzymes. Why the lack of a resolvase leads to slower host growth is unclear. Possibly SSV1 ORF d244 is involved in the specificity of SSV1 integration. SSV-K1 is known to integrate into multiple positions in the host genome (Wiedenheft et al., 2004), which may contribute to its higher copy number. Whether SSV-Δd244 integrates into multiple positions in the host genome is under investigation. On the other hand, there may be a defect in SSV-Δd244 replication or resolution of SSV replication intermediates that leads to accumulation of aberrant DNA, which, in turn, leads to slower host growth.
After multiple transfers of Sulfolobus cultures transfected with SSV-Δd244 into fresh media, growth rates recover to near wild-type rates (unpublished data). The virus is still present in these cultures by PCR and is able to inhibit Sulfolobus growth on plates (unpublished data) so the virus is not lost or apparently rearranged (see Results). Whether there are other genetic changes in the virus or host under these conditions remains to be determined. One attractive possibility is changes to the CRISPR repeat structures that are proposed to be important for acquired immunity in Sulfolobus (Held and Whitaker, 2009).
SUMMARY AND OUTLOOK
Comparative and structural genomics has identified a number of targets for gene disruption in the SSV1 genome. Here precise gene disruptions of the poorly conserved VP2 gene, and the well-conserved ORFs b129 and d244 are described. Deletions in VP2 may allow insights into DNA packaging in the SSV1 genome. Deletion of ORF b129 may allow the identification of the second archaeal virus transcriptional activator. Deletion of ORF d244 may allow insight into copy number regulation in SSVs, previously thought to be regulated by ORF d63 (Lawrence et al., 2009). Clearly, there are many more genes to be analyzed in the SSV1 genome and more insights that can be gained by combining comparative genomics, structural biology, and genetics. In the future, biochemical work will be added to this suite of techniques to gain fundamental understanding of this fascinating, unique, and ubiquitous archaeal virus family.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by Portland State University, NASA award NNX07AT63A. Subaward G258-08-W1951 and NSF grant MCB-0702020. The authors would like to thank Adam Clore for his advice on Long Inverse PCR and SSV comparative genomics.
Footnotes
REFERENCES
- Albers S. V., Jonuscheit M., Dinkelaker S., Urich T., Kletzin A., Tampé R., Driessen A. J., Schleper C. (2006). Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. F., Banfield J. F. (2008). Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320 1047–1050 [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7 1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannio R., Contursi P., Rossi M., Bartolucci S. (1998). An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180 3237–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choli T., Henning P., Wittmann-Liebold B., Reinhardt R. (1988). Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the Archaebacterium, Sulfolobus solfataricus. Biochim. Biophys. Acta 950 193–203 [DOI] [PubMed] [Google Scholar]
- Clore A. J., Stedman K. M. (2006). The SSV1 viral integrase is not essential. Virology 361 103–111 [DOI] [PubMed] [Google Scholar]
- Fröls S., Gordon P. M., Panlilio M. A., Schleper C., Sensen C. W. (2007). Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365 48–59 [DOI] [PubMed] [Google Scholar]
- Guo L., Brügger K., Liu C., Shah S. A., Zheng H., Zhu Y., Wang S., Lillestøl R. K., Chen L., Frank J., Prangishvili D., Paulin L., She Q., Huang L., Garrett R. A. (2011). Genome analyses of Icelandic strains of Sulfolobus islandicus model organisms for genetic and virus-host interaction studies. J. Bacteriol. 193 1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Feng Y., Zhang Z., Yao H., Luo Y., Wang J., Huang L. (2008). Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among crenarchaea. Nucleic Acids Res. 36 1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held N. L., Whitaker R. J. (2009). Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 11 457–466 [DOI] [PubMed] [Google Scholar]
- Jonuscheit M., Martusewitsch E., Stedman K. M., Schleper C. (2003). A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 48 1241–1252 [DOI] [PubMed] [Google Scholar]
- Kessler A., Sezonov G., Guijarro J. I., Desnoues N., Rose T., Delepierre M., Bell S. D., Prangishvili D. (2006). A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res. 34 4837–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. M., Menon S., Eilers B. J., Bothner B., Khayat R., Douglas T., Young M. J. (2009). Structural and functional studies of archaeal viruses. J. Biol. Chem. 284 12599–12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Yeats S., Janekovic D., Reiter W. D., Aicher W., Zillig W. (1984). SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 3 2165–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S. K., Eilers B. J., Young M. J., Lawrence C. M. (2010). The crystal structure of D212 from Sulfolobus spindle-shaped virus ragged hills reveals a new member of the PD-(D/E)XK nuclease superfamily. J. Virol. 84 5890–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S. K., Maaty W. S., Corn G. J., Kwok S. C., Eilers B. J., Kraft P., Gillitzer E., Young M. J., Bothner B., Lawrence C. M. (2008). Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376 270–278 [DOI] [PubMed] [Google Scholar]
- Nadal M., Mirambeau G., Forterre P., Reiter W. D., Duguet M. (1986). Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 321 256–258 [Google Scholar]
- Palm P., Schleper C., Grampp B., Yeats S., McWilliam P., Reiter W. D., Zillig W. (1991). Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185 242–250 [DOI] [PubMed] [Google Scholar]
- Pina M., Bize A., Forterre P., Prangishvili D. (2011). The archeoviruses. FEMS Microbiol. Rev. 35 1035–1054 [DOI] [PubMed] [Google Scholar]
- Redder P., Peng X., Brügger K., Shah S. A., Roesch F., Greve B., She Q., Schleper C., Forterre P., Garrett R. A., Prangishvili D. (2009). Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ. Microbiol. 11 2849–2862 [DOI] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Henschen A., Lottspeich F., Zillig W. D. (1987a). Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 206 144–153 [Google Scholar]
- Reiter W. D., Palm P., Yeats S., Zillig W. (1987b). Gene-expression in Archaebacteria - Physical mapping of constitutive and UV-inducible transcripts from the Sulfolobus virus-like particle SSV1. Mol. Gen. Genet. 209 270–275 [DOI] [PubMed] [Google Scholar]
- Reno M. L., Held N. L., Fields C. J., Burke P. V., Whitaker R. J. (2009). Biogeography of the Sulfolobus pan-genome. Proc. Natl. Acad. Sci. U.S.A. 106 8605–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C., Kubo K., Zillig W. (1992). The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: Demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U.S.A. 89 7645–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., Erauso G., Fletcher C., Gordon P. M., Heikamp-de Jong I., Jeffries A. C., Kozera C. J., Medina N., Peng X., Thi-Ngoc H. P., Redder P., Schenk M. E., Theriault C., Tolstrup N., Charlebois R. L., Doolittle W. F., Duguet M., Gaasterland T., Garrett R. A., Ragan M. A., Sensen C. W., Van der Oost J. (2001). The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 98 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman K. M., Schleper C., Rumpf E., Zillig W. (1999). Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics 152 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman K. M., She Q., Phan H., Arnold H. P., Holz I., Garrett R. A., Zillig W. (2003). Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154 295–302 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B., Stedman K. M., Roberto F., Willits D., Gleske A. K., Zoeller L., Snyder J., Douglas T., Young M. (2004). Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae genomes. J. Virol. 78 7438–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats S., McWilliam P., Zillig W. (1982). A plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J. 1 1035–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]