Abstract
The circadian clock acts at the genomic level to coordinate internal behavioral and physiologic rhythms via the CLOCK-BMAL transcriptional heterodimer. Although the nuclear receptors REV-ERBα and β have been proposed to form an accessory feedback loop that contributes to clock function1,2, their precise roles and importance remain unresolved. To establish their regulatory potential we generated comparative cistromes of both REV-ERB isoforms, which revealed shared recognition at over 50% of their total sites and extensive overlap with the master circadian regulator BMAL1. While Rev-erbα has been shown to directly regulate Bmal1 expression1,2, the cistromic analysis reveals a direct connection between Bmal1 and Rev-erbα and β regulatory circuits than previously suspected. Genes within the intersection of the BMAL1, REV-ERBα and REV-ERBβ cistromes are highly enriched for both clock and metabolic functions. As predicted by the cistromic analysis, dual depletion of Rev-erbα/β function by creating double-knockout mice (DKOs) profoundly disrupted circadian expression of core circadian clock and lipid homeostatic gene networks. As a result, DKOs show strikingly altered circadian wheel-running behavior and deregulated lipid metabolism. These data now ally Rev-erbα/β with Per, Cry and other components of the principal feedback loop that drives circadian expression and suggest a more integral mechanism for the coordination of circadian rhythm and metabolism.
The circadian clock is a transcriptional mechanism that coordinates both behavioral and physiological processes such as the sleep-wake cycle and food intake. The existing model for the mammalian core molecular clock involves a transcriptional negative-feedback loop in which the transactivation of E-box-containing genes by CLOCK and BMAL1 is inhibited by the expression of Per1 and Cry1, themselves under transcriptional control of E-boxes 3. As functional redundancy is common for core clock components, deletion of multiple paralogs in mice is often required to uncover relevant phenotypes such as perturbations in circadian gene expression, metabolism and wheelrunning behavior 4,5,6,7.
As REV-ERBα, β and RORα, β, γ bind to a common response element (the RORE), their intrinsic repressive and inducive activities, respectively, are believed to establish the rhythmic expression of target genes such as Bmal1. However, the partial penetrance and mild period phenotype of Rev-erbα−/− mice 1 (essentially intact circadian wheel-running behavior) and the modest rhythmic phenotype upon partial Rev-erbβ depletion of Rev-erbα−/− cultured cells 2, suggested they are not required for principal core clock function. Rather, REV-ERBs are proposed to form an accessory feedback loop that stabilizes the clock and plays a role in receiving input signals or transmitting output pathways 2,8.
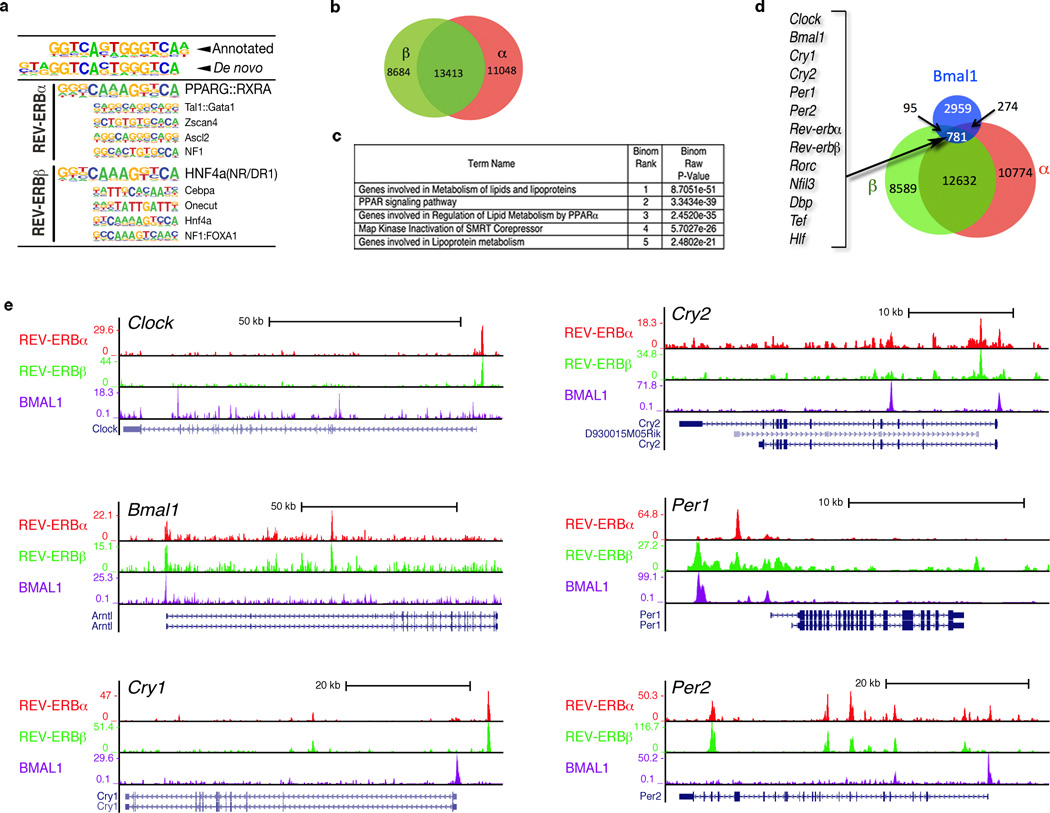
To clarify the regulatory potential of REV-ERBα and REV-ERBβ in circadian regulation, we generated isoform-specific antibodies (Supplementary Materials and Methods and Supplementary Fig. 1) and determined their genome-wide binding sites (cistromes) in the liver at Zeitgeber Time (ZT) 8, the peak of their protein expression (data not shown and 9). De novo motif analysis (Fig. 1a) revealed that in vivo, in addition to the classic REV-ERB DR2 motifs, other nuclear receptor binding sites (particularly DR1) are predominant in the peaks bound by both REV-ERBα and REV-ERBβ10. Though not surprising, extensive overlap was observed between the REV-ERB cistromes, with commonly bound peaks accounting for 54.8% and 60.7% of the total REV-ERBα and REV-ERBβ peaks, respectively (Fig. 1b). The somewhat limited overlap of our REV-ERBα cistrome with that published from Feng et al. 11 (< 50%) can most likely be attributed to differences in the antibody specificities (Supplementary Fig. 2, 3). Pathway analyses of our REV-ERBα and β overlapping peaks 12 revealed an enrichment in lipid metabolism genes (Fig. 1c), consistent with the hyperlipidemic phenotype previously observed in Rev-erbα null animals 13. Notably, loci encoding circadian clock genes (Clk, Bmal1, Cry1/2, Per1/2; see Fig. 1d) were also enriched in the REV-ERBα/β cistromic overlap, suggesting that the coordinated actions of both REV-ERBs are directly linked with clock function. A comparison of the REV-ERBα/β cistrome with published BMAL1 binding sites 14 revealed that 28% of BMAL1 peaks (at ZT6 and ZT10) were shared with the REV-ERBα/β (ZT8) cistrome and 68% of these peaks (781) were occupied by all three transcription factors. Clear binding sites for each of these transcription factors were found on “core clock” gene loci as well as on many clock controlled target genes (Rev-erb α/β, ROR, Dbp, Hlf, Tef, Nfil3; see Fig. 1d,e). In addition to circadian annotated loci, the BMAL1/REV-ERBα/REV-ERBβ triple intersection is highly enriched for genes in the receptor tyrosine kinase signaling pathway as well as those known for energy homeostasis (Supplementary Table 1). Their confluence at hundreds of clock and clock output genes suggest that beyond a simple ‘binary relationship’, Rev-erbα/β and Bmal1 cooperate to coordinately regulate clock and clock output genes.
Figure 1. Cistromic analyses of REV-ERBα and REV-ERBβ in liver.
a, De novo HOMER motif analysis of in vivo REV-ERBα and REV-ERBβ binding. b, Venn diagram depicting the unique and common REV- ERBα and REV-ERBβ bound peaks. c, Commonly bound REV-ERBα and REV-ERBβ peaks are enriched for genes involved in lipid metabolism and associated with PPARs. d, REV-ERBα, REV-ERBβ and BMAL1binding at canonical circadian clock genes. Left axis indicates tag counts. e, BMAL1 cistrome significantly overlaps with REV-ERBα and REV-ERBβ. Examples of Clock related genes in overlap are listed and selected peaks shown in Supplementary Figure 4.
To test the above proposal, we used Cre-Lox recombination to generate three genetically modified mouse lines in C57BL/6 backgrounds harboring either global, tissue-specific, or conditional knockouts of REV-ERBα and β loci. Global Rev-erbα−/− and β−/− single knockout mice were generated using a CMV-Cre transgenic allele (detailed in Supplementary Materials and Methods). Homozygous deletion of Rev-erbβ did not cause overt gross abnormalities, lethality or infertility, while Rev-erbα−/− mice, though viable, show frequent postnatal lethality (before 2 weeks of age) (Supplementary Table 2), with survivors exhibiting diminished fertility in both sexes. In contrast Rev-erbα−/− mice on a mixed 129/Sv and C57BL/6 background were reported to have reduced fertility but no postnatal lethality 15.
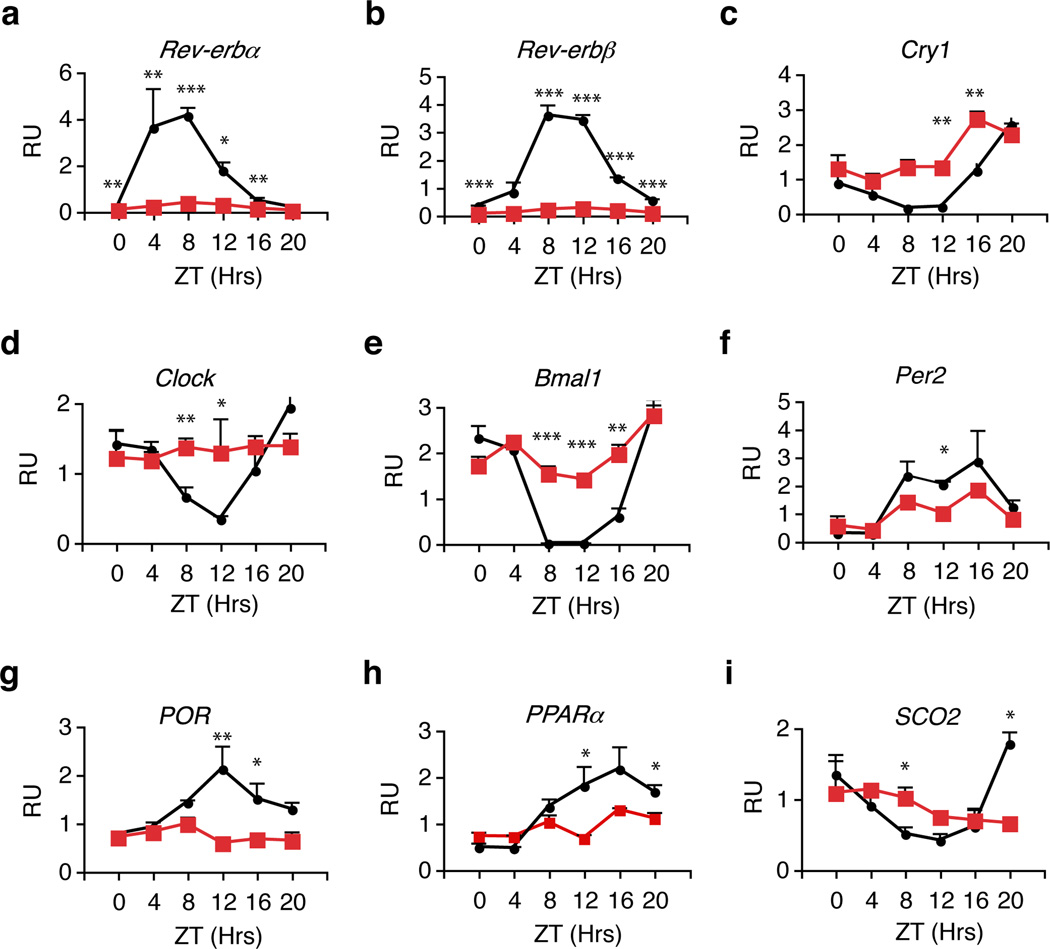
Liver-specific Rev-erbα−/− Rev-erbβ−/− mice were generated to assess combinatorial REV-ERB signaling in hepatic circadian gene expression via an albumin-Cre transgene 16. These liver-specific double knockout mice (L-DKO) were born at the expected Mendelian ratio, effectively bypassing the frequent lethality of the Rev-erbα−/− global deletion. Depletion of both Rev-erbα and Rev-erbβ (Fig. 2a-b) disrupted the circadian, and often total expression, of many hepatic core clock genes (Fig. 2c-f) and presumed output genes (Fig 2, g-i).
Figure 2. Circadian gene expression of many canonical core clock genes and output genes are disrupted in Livers of Rev-erbαlox/lox Rev-erbβlox/lox Albumin-Cre (L-DKO) mice.
The expression levels of a, Rev-erbα, b, Rev-erbβ, c-f, canonical core clock genes (Cry1, Clock, Bmal1 and Per2) g-i, presumed output genes (PoR, PPARα and Sco2) in livers from L-DKO (Albumin-Cre positive, red labels) and wildtype (Albumin-Cre negative, black labels) mice. Livers (n=3) were harvested at each indicated ZT under 12-hour light:dark cycle. QPCR was performed in technical triplicates. Relative Unit (RU) normalized with 36B4. Error bars indicate standard error of the mean, statistical significance determined by Student t-test (* p<0.05, ** p<0.01, *** p<0.001).
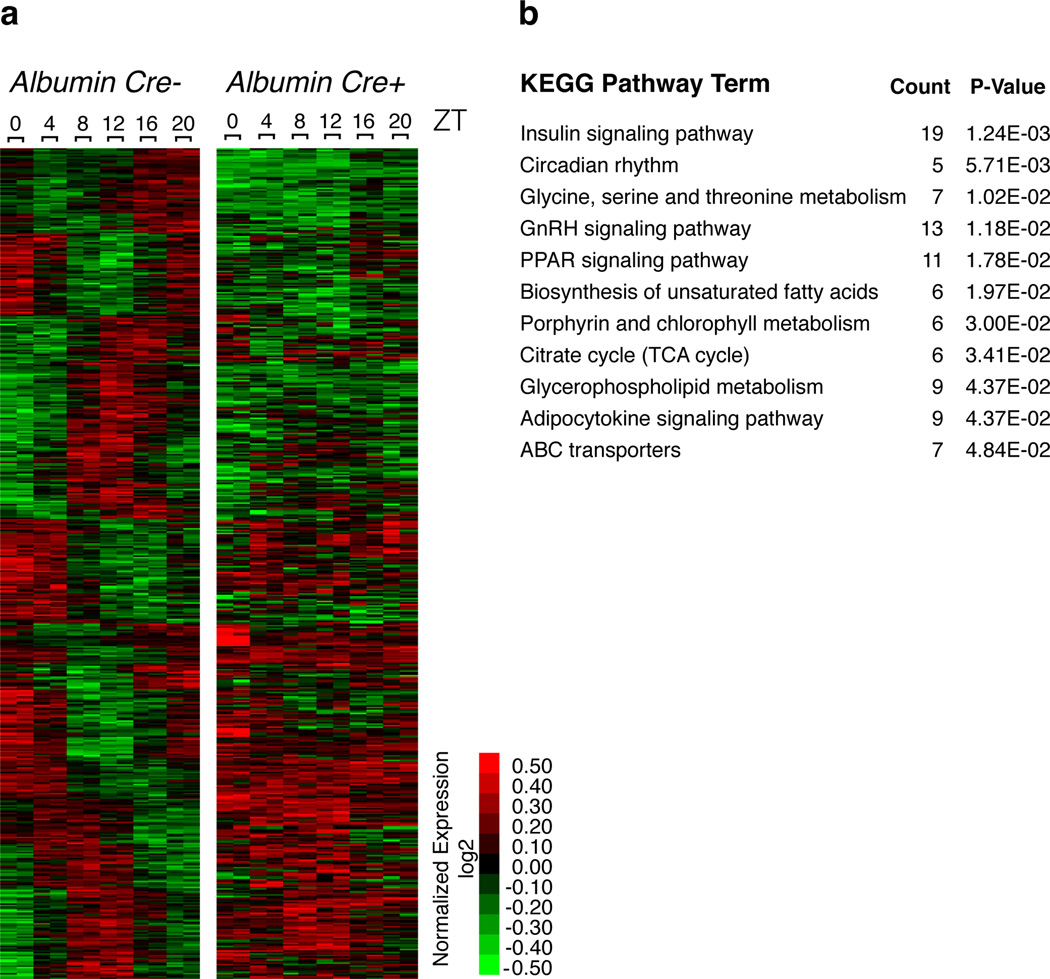
Microarray analysis of gene expression over a 24 hr time course (presented as Zeitgeber Time) revealed massive differences between the wildtype and L-DKO liver. Using CircWave v3.3 17 software, we determined that of the ~900 circadianly expressed genes in wildtype liver, the rhythmicity of more than 90% was perturbed in the L-DKO (Fig. 3a). The severity of the circadian disruption was comparable to that described for mice deficient in core clock components 18,19, emphasizing the functional impact associated with the extensive intersection between Rev-erbα/β and Bmal1 cistromes. Furthermore, the gene ontology of the Rev-erbα/β-dependent circadian transcriptome (those genes that lose rhythm in L-DKO; Fig. 3b) also mirrors that of their cistromes (Fig. 1c and Supplementary Table 1). This is of interest as the disrupted oscillation of these genes in the liver occurred in vivo in the presence of otherwise wildtype entraining signals and fully intact extra-hepatic clockwork (Supplementary Fig. 5). Notably, residual rhythmic expression of some genes were maintained in the L-DKO mice, indicating that they may be controlled by systemic cues, as suggested by Kornmann et al. 20.
Figure 3. Broad disruption of circadian transcriptome in the absence of Rev-erbα and Rev-erbβ.
a, Heatmap of genes with circadian expression in wild-type (left panel) and L-DKO (right panel) livers. 1227 unique accession numbers were selected based on fdr < 0.05. b, Genes expressed in a circadian manner that lose rhythm are highly associated with circadian and energy homeostasis functions as assessed by KEGG Pathway analysis.
To functionally link the cistromic and transcriptomic data, we examined the gene expression levels of Rev-erb/Bmal1 common cistromic sites. A comparison of the liver expression levels across ZT8 to ZT16 revealed that 45% of the co-occupied genes were significantly perturbed in the liver-specific DKO compared with WT liver (Supplementary Table 3). This strong correlation between REV-ERB occupancy and perturbed gene expression corroborates a direct role for REV-ERBs in maintaining rhythmic expression patterns for many of the above genes.
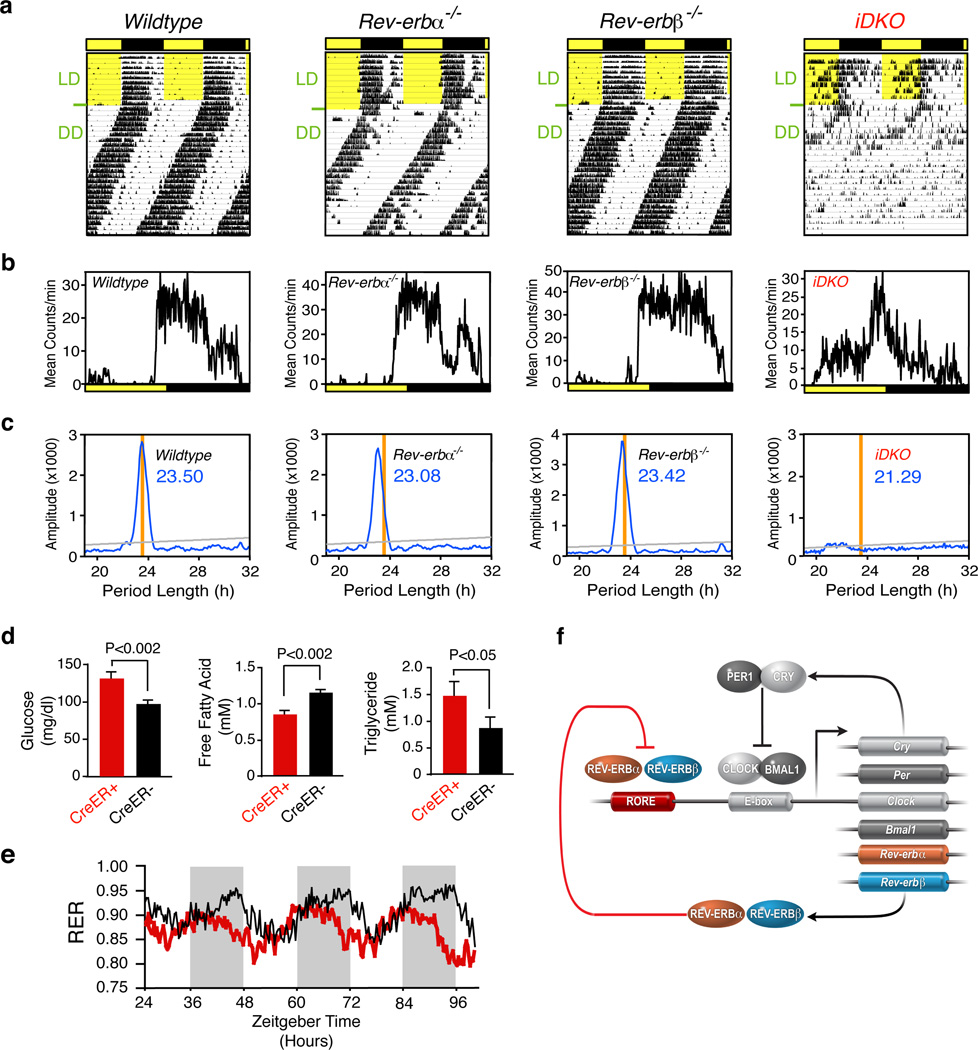
The benchmark assay for circadian dysfunction is wheel-running behavior, where the persistence or fragmentation of running rhythm is assessed in constant darkness after prior entrainment to a 12-hour light-dark cycle. Actograms recorded for Rev-erbα−/− mice in a pure C57Bl/6 background displayed a fully penetrant period shortening of 30 minutes (Fig. 4a,b), contrasting with the lack of fully penetrant phenotype reported in mixed background studies 1,15. No changes in the activity levels, activity consolidation, or phase angle of entrainment (activity onset relative to dark onset) were observed for Rev-erbα−/− mice. The Rev-erbβ−/− mice showed no change in any of the parameters of circadian activity rhythms (Fig. 4a,b), raising the possibility that REV-ERBβ may function outside the oscillator. Given the functional redundancy demonstrated for the established components of the core clock, we sought to address whether REV-ERBα and REV-ERBβ might cooperatively influence circadian rhythms in wheel running. Using the tamoxifen-activated CreER transgene, inducible double knockout (iDKO) mice were generated 21, allowing ubiquitous deletion of both genes in adulthood (see Supplementary Materials and Methods). While the wheel-running activity of Rev-erbαlox/lox Rev-erbβlox/lox animals treated with tamoxifen was normal, the phenotype of mice bearing the CreER transgene was more severe than the additive effects of the individual knockouts (Fig. 4a-c). The iDKO mice showed reduced and severely fragmented activity, and advanced phase angle of entrainment, features also found in Bmal1−/− mice 22. Furthermore, as shown in the periodogram in Fig. 4c, in mice with detectable activity rhythms, the free running period length under constant darkness was shortened by as much as 2.5 hours (see also Supplementary Tables 4, 5). The severe activity defects in iDKO mice is unexpected from the properties of the single KOs and supports a strongly selected, novel and cooperative role for REV-ERBα and β in rhythmic behavior 4–7.
Figure 4. Loss of both Rev-erbα and Rev-erbβ results in disrupted circadian wheel-running behavior and metabolic shift.
a-c, Voluntary locomotor activity of wildtype, Rev-erbα−/−, Rev-erbβ−/−, and Rev-erbα−/−Rev-erbβ−/− (iDKO) mice. a, Actograms showing wheel-running activity in constant darkness after prior entrainment in light/dark. b, Activity profiles during light dark cycles. c, Chi-square periodogram of the initial 20 days in constant darkness. (n=5–9 for each mutant strain, n= 5–6 littermate controls). Representative actograms from individual mice are shown. d, Triglyceride (n=6), fasting glucose (n=6) and free fatty acid (n=6) levels in iDKO and wildtype mice. e, Respiratory exchange ratio (RER) for wildtype (black) and iDKO (red) mice (n=4). f, Model depicting the activating (Clock/BMAL1) and repressive (REV-ERBα/REV-ERBβ) transcriptional complexes whose coordinate actions generate rhythmic gene expression.
Animals and humans with disrupted circadian rhythms have been shown to develop metabolic disorders including hyperlipidemia and hyperglycemia, suggesting a link between proper clock function and metabolism 23. To investigate if REV-ERB activity is similarly required for normal metabolic regulation, we compared metabolic parameters of tamoxifen-treated control and iDKO animals. Treated iDKO mice displayed increased circulating glucose and triglyceride levels, and a reduction in the level of free fatty acids compared to control littermates (Fig. 4d). The reduced fatty acids may reflect a more oxidative metabolism of the double knockout mice as exemplified by reduced RERs. We also note a distinct RER phase shift in constant dark conditions (Fig. 4e) indicating dysregulation of overall body metabolism. While previous studies have linked REV-ERBα with lipid homeostasis 13,24,25, the presence of both REV-ERBα and β on multiple key lipid and bile acid regulatory genes including LXR, FXR, ApoCIII, Cyp7a1, SHP, Insig2, and SREBP now provides a molecular mechanism for rhythmic cholesterol and bile acid metabolism. The current cistromic studies elevate REV-ERBβ to equal prominence with REV-ERBα in the transcriptional regulation of these pathways, and identify additional direct gene targets (Supplementary Fig. 6). The severity of the metabolic phenotype observed in the iDKO mice compared to the REV-ERBα KO mice is consistent with the cistromic analyses of the REV-ERBs.
Defining the genetic mechanisms that comprise the circadian clock is key to understanding how genomic rhythms are transformed into behavioral and metabolic physiology. In this study, using a combination of genome-wide cistromic profiling, mRNA expression analysis, and three new targeted gene knockout mouse models, we unequivocally demonstrate a more critical and central role for Rev-erbα/β circadian clock regulation than previously suspected. The genomic analysis confirms Bmal1 as a direct REV-ERBα/β target, while at the genome-wide level we find a predominance of both core clock and circadian output genes targeted by REV-ERBs, suggesting a more integral role for this family in the core clock than previously considered. These findings suggest that a dynamic balance between BMAL1 and REV-ERBα/β cistromes is used to regulate both core clock and clock output genes by co-localizing opposing epigenetic regulators to shared genomic targets. In vivo, using targeted and inducible double knockout mice, we demonstrate that Rev-erbα and β together function as integral drivers of the circadian clock, rather than simply as stabilizers of an output, thereby redefining the established paradigm for these receptors. The similarity of the DKO circadian phenotype to compound core clock mutants (Per1−/−Per2−/−, Cry1−/−Cry2−/−) 5,6,7 in severity, penetrance and strong period shortening, is more reflective of a pacemaker rather than a stabilizer of rhythm. Together, this leads to a model of mutual direct regulation, with BMAL1 controlling one loop and REV-ERBα/β a second loop of the core clock, with both loops using cistromic convergence to coordinate key clock and metabolic functions (Fig. 4f). The adaptive feature of the circadian clock enables its control of sleep-wake cycles, physiologic rhythms, energy homeostasis and behavior. In contrast, disruption of rhythm spawns a range of problems from jet lag, to more profound sleep disorders, obesity, metabolic disease, immune function and cancer 26,27,28,29. As partnered regulators, the recent development of both potent REV-ERB agonists (that enhance repression) 25 and REV-ERB antagonists (that relieve repression) 30 provide a new therapeutic approach to both reset disrupted rhythms and re-establish metabolic balance.
Methods Summary
For chromatin immunoprecipitation (ChIP), 5 month-old male C57BL/6J mice were euthanized by CO2 asphyxiation at ZT 8. Livers were removed and pooled for ChIP, processing and sequencing. Tamoxifen-induction of Cre Recombinase activity was accomplished by daily intraperitoneal injection of 2 mg tamoxifen (Sigma) in 100μl corn oil (Sigma) for 7 days. 3 month-old tamoxifen-treated animals were subjected to wheel running assays for 7 days after the end of treatment. For gene expression analysis, 5–6 month-old males were euthanized by cervical dislocation at indicated ZT points for rapid dissection and snap-freezing of the tissues. During the dark cycle, procedures were performed under red light. Detailed materials and methods are provided in the Supplementary Information.
Supplementary Material
Acknowledgments
Acknowledgements
We thank Samantha Kaufman, Jacqueline Alvarez, Ester Banayo, Henry Juguilon, Sandra Jacinto and Hiep Le for technical assistance; and Lita Ong and Sally Ganley for administrative assistance. We also thank Liming Pei for discussion. R.M.E is an Investigator of the Howard Hughes Medical Institute at The Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. H.C. is a recipient of National Research Service Award (T32-HL007770). This work was supported by National Institutes of Health Grants (DK062434, DK057978, DK090962, DK091618 and HL10278), National Health and Medical Research Council of Australia Project Grants (NHMRC 512354 and 632886), the Helmsley Charitable Trust, the Glenn Foundation and the Howard Hughes Medical Institute.
Footnotes
Author Contributions
X.Z., G.D.B., R.T.Y., M.D. and C.L. performed and/or analyzed the results from ChIP-Seq. R.T.Y, M.D., M.T.L. and C.K.G. performed and/or analyzed the results from microarray experiment. M.H., L.D. and S.P. performed and/or analyzed the wheel-running assay and the real time luciferase assay. J.A. performed gene targeting. H.C. and L.C. performed all experiments. H.C. and R.M.E. designed all experiments, analyzed all results and H.C., R.T.Y, M.D., A.A., S.P. and R.M.E. wrote the manuscript.
Author Information
Microarray and ChIP-Seq data sets have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE34020.
The authors declare no competing financial interests.
References
- 1.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 2.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 4.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 6.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 7.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 9.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 11.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomez P, et al. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor. Development. 2000;127:1489–1498. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- 16.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 17.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 18.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatanaka F, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 22.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, et al. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raspé E, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 25.Kumar N, et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151:3015–3025. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs JE, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojetin D, Wang Y, Kamenecka TM, Burris TP. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol. 2011;6:131–134. doi: 10.1021/cb1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.