The ancient Greek word “φαγos” (phagos) stands for “eater” or “glutton” and “φαγɛĩν” (phagein) means “to eat”. Autophagy, also referred to as macroautophagy, was first discovered over 40 years ago as a process of cellular self-digestion, or “self-eating”, and was initially thought to be a nonspecific degradation process. It is now clear that autophagy is highly regulated, serving to remove damaged proteins and organelles from the cell as part of certain cellular stress responses. As such autophagy contributes to numerous physiological and pathophysiological processes1. Moreover, autophagy has recently emerged as a key regulating process in cancer formation, with evidence for either pro- or anti-tumorigenic effects reported2. Discovered almost 50 years ago as a T cell-derived mediator inhibiting the random migration of macrophages, today macrophage migration inhibitory factor (MIF) is known as a pleiotropic cytokine and non-classical chemokine with a variety of functions3. The MIF protein is structurally unique, not belonging to any of the established cytokine families. In spite of the gross architectural homology between the CXCL8 dimer and the MIF monomer, MIF also does not formally belong to any of the four classical chemokine sub-families4. Owing to its cytokine/chemokine activities, MIF is a pivotal inflammatory mediator that has been implicated in acute and chronic inflammatory diseases including septic shock, rheumatoid arthritis, and atherosclerosis3,5. Of note, MIF-blocking strategies have proven powerful in numerous mouse models of these diseases3,5. MIF was originally discovered as an inhibitor of random “macrophage” migration and as a promoter of the “phagocytotic” activity of these cells3. More recent data have established a critical role for MIF in inflammatory activation of macrophages, macrophage apoptosis, and inflammatory and atherogenic recruitment of the macrophage precursor cell, the monocyte3,4. Thus, the “macrophage” continues to be in the center of MIF biology, but it also has become clear that MIF functions extend way beyond this cell type.
Accordingly, it was found that MIF is not only expressed by T cells and macrophages, but also by other immune cell types, endothelial cells as well as various parenchymal cell types3. Moreover, various tumor cells express abundant MIF protein levels. While MIF is fairly ubiquitously expressed, its secretion, which follows a so-called non-conventional mechanism, is tightly controlled. Stimuli promoting the secretion of MIF encompass various inflammatory triggers such as bacterial endotoxin, bacterial exotoxins, or tumor necrosis factor (TNF)-α as well as pro-tumorigenic stimuli such as hypoxia, pro-oxidative stress, starvation, or growth stimulation3,6. MIF is not only produced by various tumor cells, but it has been well established to date that MIF is functionally involved in various cancers. MIF levels were found to be elevated in patients with cancers, and the levels of MIF were closely correlated with tumor aggressiveness and metastatic potential, overall suggesting that MIF contributes to disease severity and survival. Identified pro-tumorigenic effects of MIF encompass autocrine pro-oncogenic signaling processes through the CD74-ERK1/2 MAP kinase and CD74-PI3K/AKT pathways, but also a critical p53-dependent anti-apoptotic effect of MIF as well as pro-angiogenic effects6.
Breast cancer is a major malignancy in women with high mortality rates, i.e., 5-year survival rates in patients with advanced breast cancer of about 20%. MIF has been correlated with the development of breast cancer; however while elevated circulating levels of MIF have been unanimously associated with lower survival rates in breast cancer, enhanced intracellular expression of MIF in breast cells may even be beneficial7. MIF overexpression in breast cancer cells was most abundantly observed in non-invasive breast cancer cells, but not in invasive cells, which in turn expressed higher levels of the MIF receptor CD74, which is upregulated in numerous cancers including breast cancer8. Stimulation with exogenous MIF leads to a strong upregulation of MIF secretion and autocrine MIF promotes breast cancer cell proliferation and invasiveness7. MIF's chemokine-like functions are mediated through non-cognate binding to the chemokine receptors CXCR2 and CXCR4. However, while the MIF/CXCR4 axis has been implicated in glioblastoma growth, and metastatic invasion of drug-resistant colon cancer and rhabdomyosarcoma, so far no role in breast cancer has been reported9. Moreover, MIF intracellularly interacts with JAB1/CSN5, a quasi-oncogene that functions both as a component of E3 ligase-regulating COP9 signalosome complexes and as a coactivator of activator protein (AP)-1-driven transcription3. Although some of the interaction partners of MIF in breast cancer have been identified, the precise mechanism of how MIF contributes to breast cancer has remained unknown.
In their detailed mechanistic report, Wu et al.10 now have implicated steroid receptor coactivator-3 (SRC-3) as a major player in MIF-driven breast tumorigenesis. SRC-3 is a member of the p160 family of coactivators (SRC-1, SRC-2, SRC-3) and a well-known oncogene that has been associated with the development of breast cancer11,12. SRC-3 (also known as AIB1 for amplified in breast cancer 1) is amplified and overexpressed in 30%-60% of breast cancers and high levels of SRC-3 are associated with high levels of HER2/neu, EGFR, development of drug resistance and poor disease-free survival in patients with breast cancer. SRC-3 interacts with nuclear receptors and other transcriptional factors, including estrogen receptor, progesterone receptor, androgen receptor and NF-κB and is required for the expression of corresponding target genes. Moreover, SRC-3 functions are linked to pro-oncogenic extracellular stimuli11.
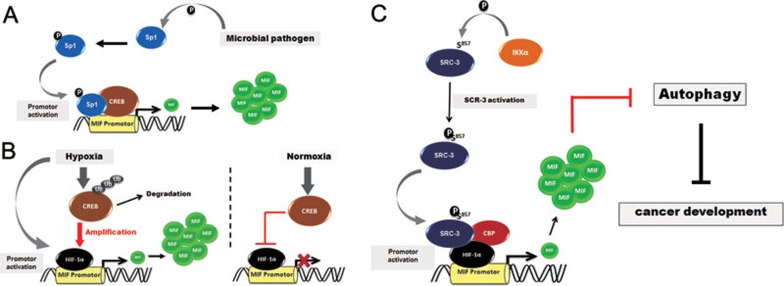
Transcriptional regulation of MIF was previously shown to be dependent on a hypoxia-responsive element (HRE) in the 5′UTR of the MIF gene and is further modulated by CREB expression (Figure 1A and 1B)13,14. Studies exploring the mechanism of MIF transcription so far have been restricted to fibroblasts, macrophages, and lung epithelial cells and the precise composition and mechanisms of the coactivator complexes driving and modulating MIF transcription have been unknown (MIF transcription is also modulated by upstream 5′ promoter polymorphisms, the -794 (CATT)5-8 microsatellite and the -173 G/C SNP, but here we are focusing on the non-polymorphic proximate promoter regions only). The current study by Wu et al.10 is important in that it describes for the first time that SRC-3 is part of the MIF promoter activation complex and serves to control the known HIF-1α/CREB/CBP effects (Figure 1C). Moreover, SRC-3-dependent MIF promoter activity is biologically relevant as loss of SRC-3 function reduces endogenous MIF expression at both mRNA and protein levels whereas gain of SRC-3 function activates the MIF promoter. Importantly, a phosphorylation of Ser857 on SRC-3 by IκB kinase-α (IKK-α) was identified and found to be required for the activation of the MIF promoter. Moreover, it was demonstrated that co-activation of the MIF promoter by HIF-1α is controlled by both SRC-3 and CBP (Figure 1C). Thus, SRC-3, in an IKK-dependent manner, was shown to regulate MIF expression by serving as a coactivator for HIF-1α (Figure 1). The study was performed in MCF-7 breast cancer cells and thus adds an important mechanistic piece as to how MIF expression is controlled in breast cancer and probably beyond.
Figure 1.
Schematic showing mechanisms controlling MIF transcription. (A) Control of the MIF promoter according to Roger et al.14. Microbial pathogens induce the phosphorylation, nuclear accumulation and DNA binding activity of Sp1, which acts together with cAMP response element binding protein (CREB) as positive regulators of human MIF gene expression. (B) Under Hypoxia, MIF gene expression is driven by hypoxia-inducible factor-1α (HIF-1α), and this process is enhanced by hypoxia-mediated CREB degradation, whereas under normoxia, this effect is suppressed by CREB13. (C) IKK-α-mediated phosphorylation of steroid receptor coactivator-3 (SRC-3) at Ser857 is responsible for the recruitment of CREB binding protein (CBP) and the co-activation of HIF-1α to induce the gene expression of MIF. Expressed MIF protein in turn inhibits autophagic cell death, favoring the proliferation and survival of breast tumor cells. Ub, ubiquitin.
MIF's role in cell survival, especially in tumor cells, has mainly been attributed to p53-dependent anti-apoptotic effects of MIF3. In addition to apoptosis, autophagic cell death is a major mechanism serving tumor suppressor functions2. Intriguingly, Wu et al. now interconnect the SRC-3/MIF axis and autophagy-driven processes of cell survival. Knockdown of SRC-3 or MIF promotes autophagy and the study shows that knockdown of SRC-3 or MIF reduces breast cancer cell viability through induction of autophagic cell death. Proof that cell death truly occurred through the autophagy pathway was convincingly demonstrated by various analyses. For example, the authors used MCF-7 cells stably expressing the autophagy marker LC3 fused with GFP (GFP-LC3) to visualize autophagosome formation in live cells and showed the formation of typical punctate GFP-LC3-containing structures, indicative of autophagic activity, when SRC-3 or MIF were knocked down by siRNA. In turn, treatment with exogenous recombinant MIF (rMIF) inhibited autophagic cell death induced by knockdown of SRC-3 or MIF. The critical role that cellular MIF levels exhibit in breast cancer behavior was underscored by a recent study by Schulz et al.15. The report showed that the HSP90 chaperone destabilizes MIF, promoting its degradation, and thereby inhibits breast tumor progression. Although Wu et al. did not specifically analyze MIF degradation processes in their SRC-3-expressing cell system, it may be concluded that MIF is able to inhibit autophagy and promote cell survival in an SRC-3-dependent manner. Thus, SRC-3 probably counteracts HSP90 in enhancing intracellular MIF levels in breast cancer cells. Overall, this underlines the importance of MIF regulation by SRC-3 and the tight connection of the SRC-3/MIF axis with autophagy regulation. Finally, Wu et al. offer in vivo evidence for the relevance of their findings, demonstrating that knockdown of MIF inhibits the tumorigenicity of MCF-7 cells in a tumor xenograft mouse model, although a role for autophagy-related processes was not specifically addressed in the in vivo model.
The report by Wu et al. should stimulate numerous follow-up studies. Since this is the first description of a link between MIF and autophagy, various mechanistic details constituting this novel connecting should be of interest to both the MIF and autophagy field. First, while Wu et al. have comprehensively characterized the upstream pathways encompassing SRC-3 activation by IKK-α and the composition and functioning of the MIF promoter complex, the “downstream” pathways connecting MIF with autophagic cell death in breast cancer cells remain unknown. For example, it is unclear whether intra- or extracellular MIF blocks the activation of the autophagy machinery. The observation by Wu et al. that exogenous rMIF was able to inhibit autophagic cell death induced by knockdown of SRC-3 or MIF would argue for an autocrine or paracrine role of MIF involving MIF secretion from SRC-3-activated breast cancer cells and activation of the same or neighboring cells by MIF by a receptor-mediated pathway. Interestingly, both CD74 and CXCR4 have already been associated with autophagy. On the other hand, the identified MIF interaction with JAB1/CSN5 might suggest a role for intracellular MIF ('intracrine action'), as CSN5 has previously been found to bind and activate the p160 family member SRC-13. The observed beneficial role of intracellular breast cancer cell-expressed MIF7 would argue against such a mechanism. Lastly, autophagy is connected to apoptosis. Depending on the cellular context and stimuli, autophagy may precede apoptosis and enhance or inhibit apoptosis, while in other cases, autophagy and apoptosis are mutually exclusive and function as backups for each other to ensure complete cell death. MIF has fairly unanimously been found to exert anti-apoptotic activities, yet the underlying mechanisms are not yet fully clear. Interestingly, the anti-apoptotic protein Bcl-2 has been shown to serve as a switch between the two cell death mechanisms and the MIF receptor CD74 has been found to be a potent activator of Bcl-2, at least in B cells3,4.
References
- Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernecke A, Bernhagen J, Weber C. Macrophage migration inhibitory factor in cardiovascular disease. Circulation. 2008;117:1594–1602. doi: 10.1161/CIRCULATIONAHA.107.729125. [DOI] [PubMed] [Google Scholar]
- Morand EF, Leech M, Bernhagen J. MIF: a new cytokine link between rheumatoid arthritis and atherosclerosis. Nat Rev Drug Discov. 2006;5:399–410. doi: 10.1038/nrd2029. [DOI] [PubMed] [Google Scholar]
- Mitchell RA. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell Signal. 2004;16:13–19. doi: 10.1016/j.cellsig.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Verjans E, Noetzel E, Bektas N, et al. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 2009;9:230. doi: 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese F, Clanchy FI. CD74: an emerging opportunity as a therapeutic target in cancer and autoimmune disease. Expert Opin Ther Targets. 2011;15:237–251. doi: 10.1517/14728222.2011.550879. [DOI] [PubMed] [Google Scholar]
- Dessein AF, Stechly L, Jonckheere N, et al. Autocrine induction of invasive and metastatic phenotypes by the MIF-CXCR4 axis in drug-resistant human colon cancer cells. Cancer Res. 2010;70:4644–4654. doi: 10.1158/0008-5472.CAN-09-3828. [DOI] [PubMed] [Google Scholar]
- Wu MY, Fu J, Xu J, O'Malley BW, Wu RC. Steroid receptor coactivator 3 regulates autophagy in breast cancer cells through macrophage migration inhibitory factor. Cell Res. 2012;22:1003–1021. doi: 10.1038/cr.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojis O, Rudraraju B, Gudi M, et al. The role of SRC-3 in human breast cancer. Nat Rev Clin Oncol. 2010;7:83–89. doi: 10.1038/nrclinonc.2009.219. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun. 2006;347:895–903. doi: 10.1016/j.bbrc.2006.06.148. [DOI] [PubMed] [Google Scholar]
- Roger T, Ding X, Chanson AL, Renner P, Calandra T. Regulation of constitutive and microbial pathogen-induced human macrophage migration inhibitory factor (MIF) gene expression. Eur J Immunol. 2007;37:3509–3521. doi: 10.1002/eji.200737357. [DOI] [PubMed] [Google Scholar]
- Schulz R, Marchenko ND, Holembowski L, et al. Inhibiting the HSP90 chaperone destabilizes macrophage migration inhibitory factor and thereby inhibits breast tumor progression. J Exp Med. 2012;209:275–289. doi: 10.1084/jem.20111117. [DOI] [PMC free article] [PubMed] [Google Scholar]