Abstract
The BRAF inhibitors (BRAFi) induce anti-tumor responses in nearly 60% of patients with advanced V600BRAF-mutant melanomas but only 5% of patients with V600BRAF-mutant colorectal carcinomas. Earlier studies of how a subset of melanoma that initially responds to BRAFi but later acquires drug resistance pointed to the importance of receptor tyrosine kinases (RTKs) in drug escape. In a pair of recent reports, this RTK-mediated mechanism of acquired BRAFi resistance in melanoma is re-surfacing in the context of innate or primary BRAFi resistance in V600BRAF-mutant colorectal carcinomas, suggesting potential upfront therapeutic strategies to prevent BRAFi resistance.
V600BRAF mutations are found in >50% of melanomas, nearly 100% of hairy cell leukemias but smaller subsets of more common human malignancies (e.g., colorectal, thyroid)1. The in-human “druggability” of mutant BRAF has been best demonstrated in metastatic BRAF mutant melanomas using the novel small-molecule BRAF inhibitor (BRAFi) PLX4032/vemurafenib, producing survival benefits2. Early clinical results of BRAFi in colorectal carcinoma, however, were disappointing, with only 5% of patients (1 of 21 patients) experiencing a partial response and 19% of patients (4 of 21 patients) experiencing minor responses3. This difference in the clinical results (melanoma vs. colorectal carcinoma) may relate less to their ontological origins but more to alternative states of a dynamic and plastic survival signaling network.
The majority of BRAF mutant melanomas responds to BRAFi rapidly but acquires drug resistance within a median time of 6-7 months. The specific mechanisms of acquired BRAFi resistance are variegated but fall under two core pathways: 1) reactivation of RAF-MEK-ERK MAPK signaling, and 2) activation of MAPK-redundant signaling via the receptor tyrosine kinase (RTK)-PI3K-AKT pathway, which is parallel but interconnected to the MAPK pathway. MAPK reactivation can occur via NRAS activating mutations4, COT overexpression5, V600BRAF alternative splicing6, V600BRAF amplification7, and MEK1 activating mutation8,9. MAPK-redundant signaling via RTK overexpression has been shown to result in AKT activation and RAS-CRAF-MEK signaling, bypassing mutant BRAF4,10,11. The repertoire of RTK overexpressed appears restricted but shares a common pattern of PDGFRβ and EGFR overexpression, at least in melanoma cell lines with acquired resistance to vemurafenib4. It is unclear at present how this overexpression of a select number of wild-type RTKs contributes to the molecular details of survival pathway redundancy and cooperativity. Nevertheless, understanding how melanomas acquire BRAFi resistance via core pathways may shed key insights into mechanisms of innate BRAFi resistance in multiple malignancies. Hence, it came as not a complete surprise that a pair of papers published recently implicated RTKs in innate BRAFi resistance in colorectal cancer cell lines12,13. Both studies pointed to EGFR activation and downstream signaling as a key component to innate BRAFi resistance, at least in a majority of colorectal carcinoma (CRC) cell lines examined.
Corcoran et al.12 showed that BRAF mutant CRC cell lines, in contrast to BRAF mutant melanoma cell lines, displayed innate resistance to growth inhibition by vemurafenib. An important clue implicating RTK involvement in innate vemurafenib resistance of BRAF mutant CRC cell lines came from the observation that p-ERK recovery occurred soon (hours to days) after vemurafenib treatment, unlike the kinetics of p-ERK recovery in BRAF mutant melanoma cell lines. This relatively rapid recovery of p-ERK post vemurafenib treatment in CRC cell lines is akin to that in melanoma cell lines with acquired BRAFi resistance driven by RTK overexpresion10. Corcoran et al. then traced this propensity for early p-ERK recovery to vemurafenib treatment (24 h)-dependent enhancement of (activated) RAS-GTP levels and MEK activity, parallel to elevated RAS-GTP levels in melanoma cell lines with RTK-driven, acquired BRAFi resistance4. In phospho-RTK arrays, they determined that the p-EGFR level (among others such as p-c-MET and p-IGF1R levels) was elevated in CRC cell lines relative to those in melanoma cells. Vemurafenib treatment (24 h) did not significantly enhance the p-EGFR level (but did elevate the p-IGFR1 level). Elevated p-EGFR levels in BRAF mutant CRC cell lines were correlated with elevated total EGFR levels (i.e., overexpressed compared with BRAF mutant melanoma cell lines). Thus, several observations correlated with innate BRAFi resistance in CRC cell lines: RTK (mostly consistently EGFR) overexpression (at baseline); upregulation of activation-associated phosphorylation of RTKs (at baseline); and upregulation of RAS-GTP levels (in response to BRAFi treatment). Curiously, although EGFR is highly phosphorylated at baseline, the RAS-GTP levels only rose in response to vemurafenib treatment.
Corcoran et al. further showed that small-molecule EGFR inhibitors (EGFRi) could downregulate, partially or completely, the RAS-GTP level induced by vemurafenib treatment. The combination of vemurafenib (BRAFi) and gefitnib (EGFRi) could synergistically reduce p-ERK levels and the net growth inhibition of most but not all CRC cell lines studied, suggesting that survival in some CRC cell lines may also depend on other RTKs and downstream signaling (e.g., AKT). Consistently, the combination of vemurafenib and erlotinib (EGFRi) stabilized the growth of, but did not cause significant regression of, CRC xenografts. Simultaneous inhibition or genetic knockdown of multiple RTKs was not explored, leaving unresolved the issue of how multiple RTKs may potentially play cooperative survival roles at baseline or in response to kinase inhibitor therapy.
Prahallad et al.13 also compared CRC and melanoma cell lines and showed that EGFR expression is generally higher in CRC cell lines. Vemurafenib treatment (6 h) of the WiDr CRC cell line led to an induction in p-EGFR and p-AKT levels, concomitant with the expected suppression of p-MEK and p-ERK. MEK inhibition, by AZD6244 treatment, similarly led to the rebound phosphorylation of EGFR. Based on earlier literature showing that the ERK kinase phosphorylates Cdc25c, activating its phosphatase activity, and that Cdc25c can dephosphorylate EGFR, Prahallad et al. went on to show that Cdc25c knockdown mimicked vemurafenib treatment in inducing p-EGFR levels. As predicted, vemurafenib treatment of CRC cell line inhibited Cdc25c phosphorylation at a key threonine (Thr 48), which was previously demonstrated to be a key event for its phosphatase activity. Addition of an EGFRi (cetuximab or gefitnib) to the BRAFi vemurafenib treatment downregulated the baseline level of p-ERK and the BRAFi-induced p-AKT level (but not the baseline p-AKT level). Moreover, addition of an EGFRi sensitized CRC cell lines to growth inhibition by vemurafenib in vitro but did not induce tumor regression in vivo, again suggesting incomplete survival signaling blockade. Accordingly, it has been shown that the effect of vemurafenib in shrinking CRC tumor xenografts was enhanced by combining with an AKT inhibitor (MK-2206)14. Moreover, in this study, the addition of vemurafenib to erlotinib treatment also resulted in increased anti-tumor activity and improved survival in xenograft models. It should be pointed out that Prahallad et al. did not formally assess BRAFi and EGFRi synergy, nor did they examine the diversity of RTK overexpression/activity and its contribution to downstream survival signaling (e.g., AKT).
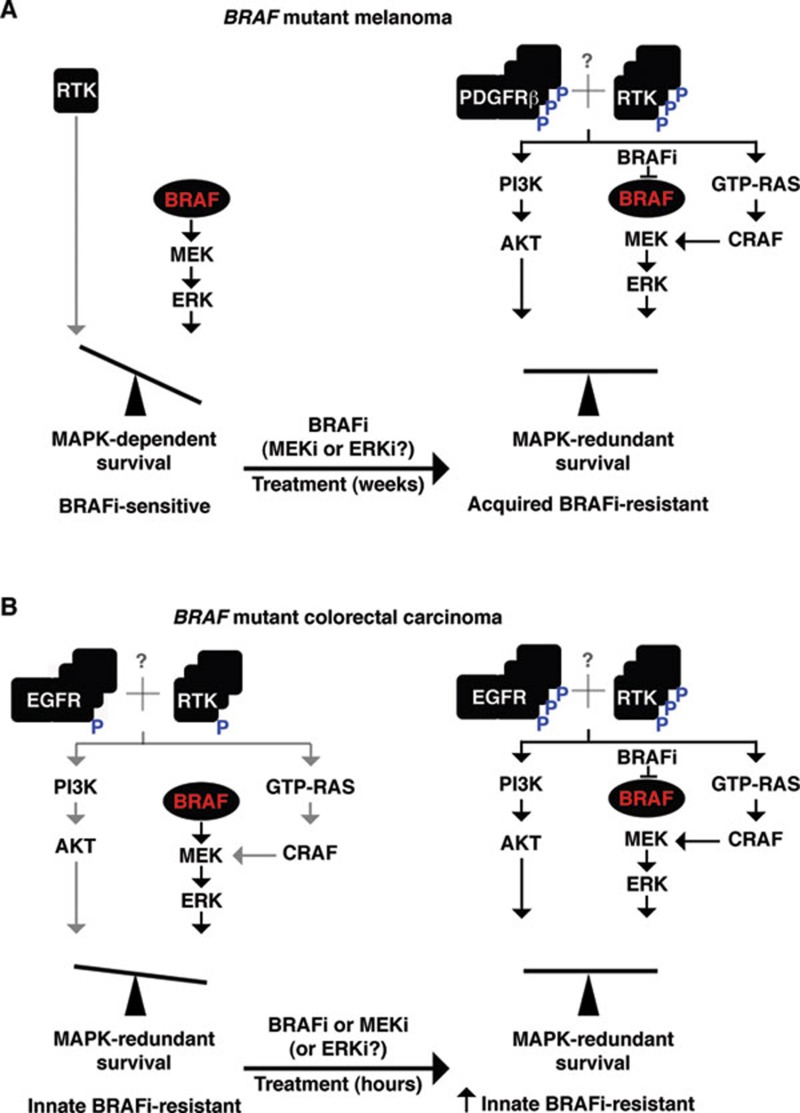
These works, along with prior studies4,10, highlight the importance of expression and activity level of RTKs as a key sensitivity determinant of BRAFi resistance in BRAF mutant cancer cell lines (Figure 1). An important question remains as to whether the diversity of RTK overexpression and/or upregulation participates in and contributes to the full BRAFi resistance phenotype. A recent study afforded us a systems-wide view of the RTKinome reprogramming in response to MEK inhibition in the so-called triple-negative breast cancer cell lines15. The balance of the MAPK vs. RTK network signaling may be dynamically influenced by kinase inhibitors targeting RAF or MEK. This daunting diversity of RTK expression/activity may corner us into abandoning a combination of RTK inhibitors (already approved for clinical usage) with a BRAF inhibitor. Instead, we might need to resort to downstream pathway inhibitors not yet approved for clinical usage (e.g., an inhibitor of MEK with an inhibitor of the PI3K-AKT-mTORC1/2 axis) before we have a chance to corner BRAF mutant cancers into death.
Figure 1.
Upregulation of receptor tyrosine kinase(s) (RTKs) as a key sensitivity determinant of BRAFi resistance in BRAF mutant cancer cell lines. (A) In BRAF mutant melanoma cell lines, RTKs are generally expressed at very low levels and contribute minimally to survival signaling, resulting in a strong addiction to mutant BRAF signaling and sensitivity to BRAFi. When BRAF mutant melanoma cell lines acquire BRAFi resistance, they upregulate the expression and activity of PDGFRb and other RTKs, resulting in reactivation of MEK-ERK as well as MAPK-redundant PI3K-AKT survival signaling. (B) In BRAF mutant colorectal carcinoma (CRC) cell lines, EGFR and other RTKs are upregulated by overexpression and some level of activation, resulting in MAPK-redundant survival signaling and conferring innate or primary BRAFi resistance. Treatment of CRC cell lines wth a BRAF or a MEK inhibitor can further activate EGFR and potentially other RTKs and stimulate GTP-RAS levels, consolidating innate BRAFi resistance. Red denotes mutated protein (e.g., BRAF); gray symbols denote weak signaling or interactions; multiplicity of protein symbols denotes overexpression; P in blue denotes activation-associated phosphorylation.
References
- Lo RS. Combinatorial therapies to overcome B-RAF inhibitor resistance in melanomas. Pharmacogenomics. 2012;13:125–128. doi: 10.2217/pgs.11.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz S, Desai J, Chan E, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors J Clin Oncol 201028(15s)suppl; abstr 3534. [Google Scholar]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. Acquired resistance to RAF inhibitors is mediated by splicing isoforms of BRAF (V600E) that dimerize in a RAS independent manner. Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies V600B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, et al. Preexisting MEK1 exon 3 mutations in V600BRAF melanomas do not confer resistance to BRAF inhibitors Cancer Discov 2012 April 1. doi: 10.1158/2159-8290.CD-12-0022 [DOI] [PMC free article] [PubMed]
- Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Yang H, Higgins B, Kolinsky K, et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res. 2012;72:779–789. doi: 10.1158/0008-5472.CAN-11-2941. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307, 321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]