Abstract
MicroRNAs (miRNAs) are critical post-transcriptional regulators and are derived from hairpin-shaped primary transcripts via a series of processing steps. However, how the production of individual miRNAs is regulated remains largely unknown. Similarly, loss or overexpression of the key mismatch repair protein MutLα (MLH1-PMS2 heterodimer) leads to genome instability and tumorigenesis, but the mechanisms controlling MutLα expression are unknown. Here we demonstrate in vitro and in vivo that MLH1 and miR-422a participate in a feedback loop that regulates the level of both molecules. Using a defined in-vitro miRNA processing system, we show that MutLα stimulates the conversion of pri-miR-422a to pre-miR-422a, as well as the processing of other miRNAs tested, implicating MutLα as a general stimulating factor for miRNA biogenesis. This newly identified MutLα function requires its ATPase and pri-miRNA binding activities. In contrast, miR-422a downregulates MutLα levels by suppressing MLH1 expression through base pairing with the MLH1 3′-untranslated region. A model depicting this feedback mechanism is discussed.
Keywords: mismatch repair, MLH1, MutLα, miRNA, Drosha, DGCR8, 3′-UTR
Introduction
MicroRNAs (miRNAs) are short (∼22s nucleotide) single-stranded RNAs that function as gene expression regulators by binding to and destabilizing or inhibiting translation of target mRNAs 1, 2, 3. In mammalian cells, primary miRNA transcripts (pri-miRNAs) are initially processed in the nucleus into the ∼70-nt hairpin RNAs termed precursor miRNAs (pre-miRNAs), which are further processed in the cytoplasm to yield mature miRNAs 1, 2, 3. The processing of pri-miRNAs to pre-miRNAs requires the RNase III Drosha and its partner DGCR8, with the former functioning as the catalytic subunit and the latter recognizing the RNA substrate 4, 5, 6, 7. Although recent studies have implicated other factors in miRNA biogenesis, which include p53 8, Smad 9, 10 and ATM 11, the molecular basis by which miRNA biogenesis is regulated is not fully understood.
miRNAs have been shown to regulate diverse cellular processes, including the DNA mismatch repair (MMR) pathway 12, 13, 14, a major genome maintenance system. The importance of MMR in genome maintenance is demonstrated by the fact that defects in the system cause genome instability and susceptibility to human cancers 15, 16, 17, 18, 19. The MMR functions that protect and maintain the integrity of the genome in mammalian cells include correcting DNA biosynthetic errors, suppressing homeologous recombination and mediating DNA damage signaling 20, 21, 22, 23. It is well established that MMR initiation factors, such as mammalian MutS and MutL homolog proteins, are essential in all MMR-mediated cellular processes. There are at least three functional MutL homolog proteins in human cells, which are MutLα (MLH1-PMS2), MutLβ (MLH1-PMS1) and MutLγ (MLH1-MLH3). Although the roles of MutLβ and MutLγ in MMR-associated genome maintenance remain to be characterized, the role of MutLα in these functions is well established 23. Because MLH1 is the obligating subunit in all these heterodimers, its cellular level is critical to the individual complexes and MMR-dependent genome-maintenance functions. Whereas no or low MLH1 results in genome instability and predisposition to cancer 16, 17, 19, overexpression of MLH1 induces apoptosis 24 and/or a mutator phenotype 25, 26. Therefore, precise regulation of the cellular level of MLH1 is critical for genome stability. Although miRNAs likely play an important role in regulating MLH1 expression 12, 13, 14, the mechanism of the regulation reaction is unclear. Interestingly, recent studies suggest that MLH1 may modulate miRNA processing, as MLH1 expression determines miRNA expression patterns in colorectal tumors 27, 28. These observations suggest a complex regulation relationship between MLH1 and miRNAs.
In this study, we demonstrate that miRNAs and MLH1 participate in a regulatory feedback loop, in which miRNA-422a negatively regulates the expression of MLH1 protein and the MLH1-PMS2 heterodimer (MutLα) positively regulates the processing of miR-422a and other miRNAs. In-vitro studies reveal that MutLα specifically binds to pri-miRNAs, interacts with the Microprocessor complex Drosha/DGCR8, and stimulates the Drosha/DGCR8-catalyzed processing of pri-miRNAs to pre-miRNAs in a manner dependent on MutLα ATPase and pri-miRNA-binding activities. These observations reveal a novel MutLα function in regulating miRNA biogenesis and a novel feedback mechanism that precisely regulates the cellular level of MutLα via miRNA functions.
Results
miR-422a regulates MLH1 expression by interacting with the MLH1 3′-UTR
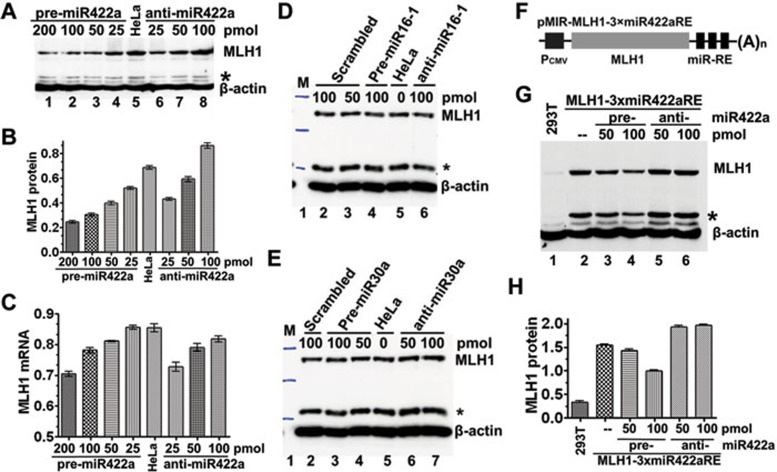
Our previous studies showed that a three-nucleotide deletion in the MLH13′-UTR is associated with leukemia relapse 12, 29. Computational analysis using MicroInspector 30 identified a putative binding site for miR-422a in the MLH1 3′-UTR that is disrupted by the 3-nucleotide deletion, suggesting a possible role for miR-422a in regulating expression of MLH1. To explore this possibility, HeLa cells were transfected with miR-422a precursor (pre-miR-422a) or anti-miR-422a, and the MLH1 protein level was quantified. The results showed that transfection of pre-miR-422a significantly lowered the level of MLH1 (P< 0.001), while transfection of anti-miR-422a increased the level of MLH1 (Figure 1A and 1B). The level of MLH1 mRNA was affected in a similar manner (Figure 1C). These results are consistent with the hypothesis that miR-422a negatively regulates MLH1 expression in HeLa cells.
Figure 1.
miR-422a regulates MLH1 expression via interaction with MLH1 3′-UTR. (A-C) Effect of miR-422a on endogenous MLH1 expression. Pre-miR-422a or anti-miR-422a was transfected into HeLa cells as indicated, and the endogenous MLH1 protein (A, B) and mRNA (C) were measured by western blotting or qRT-PCR, respectively. (D, E) Effect of miR-16-1 and miR-30a on exogenous MLH1 expression. The experiments were performed similarly as described in (A), but with pre-/anti-miR-16-1 and pre-/anti-miR-30a, as indicated. (F) Expression construct containing MLH1 and three tandem copies of the putative miR-422aRE (3×miR422aRE) in the 3′-UTR (G, H) Effect of miR-422a on exogenous MLH1 expression. pMIR-MLH1-3×miR-422aRE was co-transfected with pre-miR-422a or anti-miR-422a into MLH1-deficient 293T cells, and MLH1 protein was determined by western blotting and normalized against the amount of MLH1 protein in cells without miR-422a transfection. Data represent three independent experiments (mean ± SD). Statistical significance was determined by one-way ANOVA. *Degraded MLH1.
To determine if the repression of MLH1 expression is miR-422a-specific, the MLH1 protein levels were measured in HeLa cells transfected with pre-/anti-miR-16-1 or pre-/anti-miR-30a. The results show that the MLH1 level is essentially not interfered by either miR-16-1 or miR-30a (Figure 1D and 1E). These experiments ruled out the potential side effect of miR-422a transfection that might affect the MLH1 protein level. However, it is worth mentioning that HeLa cells transfected with 25 pmol of anti-miR-422a had an MLH1 protein level that is lower than that in untransfected cells (Figure 1A, compare lanes 5 and 6). This reduction in MLH1 expression is likely caused at the transcriptional level, but not at the translational level, as judged by the fact that (1) the lower dose anti-miRNA-transfected cells expressed almost the lowest level of MLH1 mRNA (Figure 1C), and (2) despite that, the cells transfected with 25 pmol of anti-miRNA and those transfected with 200 pmol of pre-miRNA expressed a comparable level of MLH1 mRNA (Figure 1C), the former cells exhibited an MLH1 protein level almost as twice as that in the latter cells (Figure 1B). However, how a low level of anti-miR-422a negatively influences the MLH1 transcription remains to be investigated.
To determine if miR-422a regulates MLH1 expression by binding to a putative miR-422a response element (RE) in the +18 to +50 region of the MLH1 3′-UTR, three tandem copies of the putative miR-422a RE (3×miR422aRE) were cloned into the pMIR-MLH1 vector to generate pMIR-MLH1-3×miR422aRE (Figure 1F). The resulting plasmid was co-transfected with pre- or anti-miR-422a into MLH1-defective 293T cells 31. Western blotting analysis showed that the level of MLH1 expression in 293T cells (Figure 1G and 1H) was down- or upregulated in a dose-dependent manner by pre- or anti-miR-422a, respectively. Comparable effects on ectopic luciferase expression were also observed when pMIR-Luc-3×miR422aRE was co-expressed with pre- or anti-miR-422a in 293T and HEK 293 cells (data not shown). These results are consistent with the notion that miR-422a represses MLH1 expression by binding to the region of +18 to +50 in the MLH1 3′-UTR.
Correlation between expression of MLH1 protein and pre-miR-422a
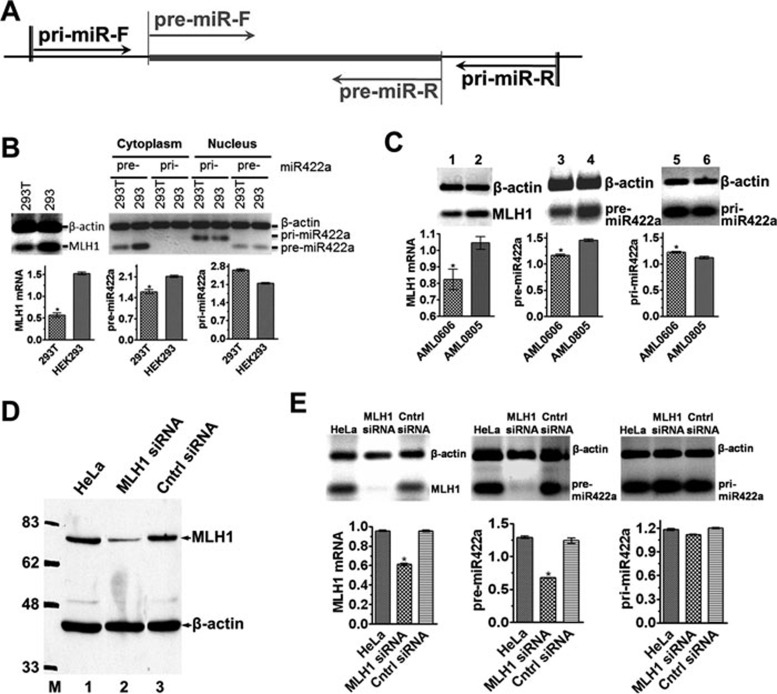
The suppression of MLH1 expression by miR-422a prompted us to think that a high level of endogenous miR-422a may have contributed to the MMR defect in 293T cells, which has previously been attributed to hypermethylation of the MLH1promoter 31. To test this possibility, we performed quantitative RT-PCR to determine the levels of pre- and pri-miR-422a in 293T cells using RNAs isolated from cytoplasm and nuclei, respectively. As shown in Figure 2B, pri-miRNAs appear to predominantly localize in nuclei, while pre-miRNAs are mainly present in cytoplasm, indicating translocation of pre-miRNAs from the nucleus to cytoplasm upon their production 32. Surprisingly, despite that the level of pri-miR-422 in both 293T and 293 cells is comparable, the level of pre-miR-422a in MLH1-deficient 293T cells is significantly lower than in MLH1-proficient 293 cells (Figure 2B). It was noted that the amount of pre-miR-422a amplified from nuclear RNAs of 293T and 293 cells was essentially the same (Figure 2B). We believe that these products were derived mostly from the pri-miR-422a template in the nucleus, as the primers for pre-miR-422a could also amplify the pre-miRNA portion of the pri-miRNA (see Figure 2A). Similar results were also obtained with MLH1-deficient and MLH1-proficient leukemia patient cells, i.e., MLH1-deficient AML0606 cells express significantly less pre-miR-422a than their isogenic MLH1-proficient AML0805 cells (P < 0.01; Figure 2C, lanes 3 and 4). These results strongly suggest that MLH1 positively regulates miR-422a processing from pri-miR-422a to pre-miR422a.
Figure 2.
Correlation between levels of MLH1 protein and pre-miR-422a. (A) Schematic diagram of primers used to quantify pre-miR-422a and pri-miR-422a. Primers for pri-miR422a amplify a 198-bp fragment, but primers for pre-miR422a-amplify an 83-bp fragment. (B) Endogenous expression of MLH1 mRNA, pre- and pri-miR422a in 293T and 293 cells. (C) Expression of MLH1mRNA, and pre- and pri-miR422a in an AML patient before and after relapse. (D) MLH1 knockdown in HeLa cells by anti-MLH1-siRNA. (E) Effect of MLH1 knockdown on expression of pre- and pri-miR-422a in HeLa cells. The levels of pre- and pri-miR-422a were determined by qRT-PCR using RNAs isolated from cytoplasm and nuclei as templates, respectively. PCR products from β-actin mRNA were used as internal controls. Data represent three independent experiments (mean ± SD). Statistical significance was determined by either Student's t-test (C) or one-way ANOVA (E). MLH1 protein was determined by western blotting.
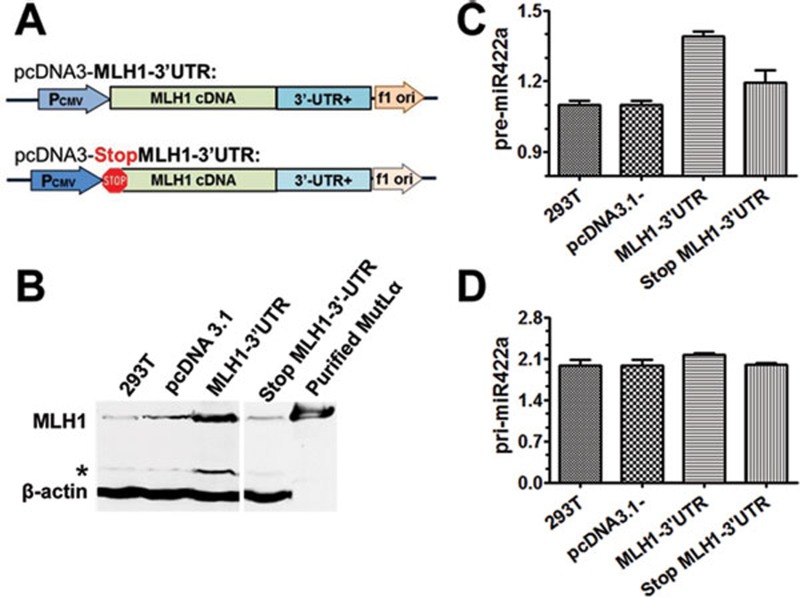
To test this hypothesis, we first knocked down MLH1 expression by siRNA in HeLa cells. As expected, the knockdown led to significant MLH1 reduction at both protein (Figure 2D) and mRNA levels (Figure 2E, left panel). We then quantified the pre- and pri-miR422a levels by qRT-PCR in the MLH1-knockdown cells. The results show that the level of pre-miR-422a (Figure 2E, middle), but not that of pri-miR-422a (Figure 2E, right), decreased significantly in the knockdown cells (P < 0.01). We further analyzed the levels of pri- and pre-miR-422a in MLH1-deficient 293T cells transfected with a plasmid (pcDNA3.1) carrying either a wild-type (WT) MLH1 gene or an MLH1 gene whose translational start codon (ATG) was changed to a stop codon (Figure 3A). As expected, the ectopic MLH1 expression was only observed in 293T cells transfected with WT MLH1 but not the mutant MLH1 (Figure 3B). Conversely, level of increased pre-miR-422a (Figure 3C, ANOVA, P < 0.001), but not pri-miR-422a (Figure 3D), was detected in cells transfected with WT MLH1. Similar results were also obtained in MLH1-deficient HCT116 cells transfected with these plasmids (data not shown). These observations indicate that higher ectopic MLH1 expression correlates with higher endogenous pre-miR-422a level, supporting the notion that MLH1 expression promotes the production of pre-miR-422a.
Figure 3.
Expression of ectopic MLH1 protein increases the level of pre-miR-422a in 293T cells. (A) Plasmid pcDNA3.1 constructs carrying WT MLH1 cDNA and a mutant MLH1 gene whose ATG start codon was changed to a TAA stop codon by site-directed mutagenesis. (B) MLH1 expression in MLH1-deficient 293T cells transfected with pcDNA3.1 alone, pcDNA3.1 carrying WT or mutant MLH1, as indicated. Ectopic MLH1 protein was detected by western blotting, with β-actin serving as an internal loading control. (C, D) qPCR quantification of pre-miR-422a (C) and pri-miR-422a (D) in 293T cells transfected with the indicated plasmids. Data shown are from three independent experiments (mean ± SD). Statistical significance was determined by one-way ANOVA. *Degraded MLH1.
MutLα interacts with pri-miRNA and Microprocessor
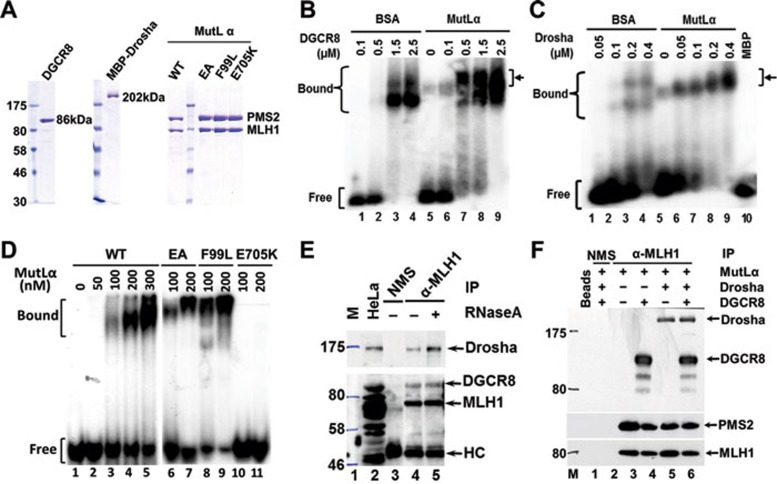
The conversion of pri-miRNA to pre-miRNA is catalyzed by the Drosha/DGCR8 complex 1, 2, 3, also known as Microprocessor. Because MutLα (MLH1-PMS2 heterodimer) is the most important and abundant protein among MLH1-formed MutL heterodimers 23, MutLα was analyzed for its potential role to stimulate pri-miR-422a processing by Microprocessor. We first tested if purified MutLα enhances the interaction between purified Microprocessor (Figure 4A) and pri-miR-422a, using electrophoretic mobility shift assays (EMSAs). In the absence of MutLα, efficient shift of 32P-labeled pri-miR-422a required a DGCR8 concentration of 1.5 μM (Figure 4B, lane 3). However, addition of 0.15 μM MutLα to the reaction reduced the required DGCR8 concentration for pri-miRNA binding to 0.5 μM (Figure 4B, lane 7), suggesting that MutLα enhances DGCR8 interaction with pri-miR-422a. Interestingly, a band that is different from the DGCR8-pri-miR-422a complexes appeared in the reaction containing MutLα, but no DGCR8 (Figure 4B, lane 5), indicating binding of the pri-miRNA by MutLα. Additionally, when DGCR8 concentration increased, more slowly migrated complexes were evident (see arrows in Figure 4B, lanes 7-9), suggesting that MutLα induces supershifts, which could be due to MutLα interactions with the pri-miRNA and/or DGCR8-pri-miRNA complex. Similar results were also observed when MutLα was added to the Drosha-pri-miR-422a complex (Figure 4C). Taken together, these observations suggest that MutLα interacts with both pri-miR-422a and the Microprocessor proteins.
Figure 4.
MutLα interacts with Microprocessor and pri-miR-422a. (A) Purified recombinant proteins used in this study. (B, C) Effect of MutLα on pri-miR-422a interactions with DGCR8 (B) and Drosha (C). (D) Binding of pri-miR-422a by wild-type (WT) and mutant MutLα proteins. (E, F) Co-immunoprecipitation (Co-IP) of MLH1 and Microprocessor in HeLa whole-cell extracts (E) and purified proteins (F). Co-IP reactions were performed using an MLH1 antibody and individual proteins were detected using their corresponding antibodies, as indicated. NMS, non-immune mouse serum; HC, IgG heavy chain.
EMSAs were performed to determine direct binding of MutLα to pri-miR-422a. As shown in Figure 4D, MutLα indeed specifically shifted 32P-labeled pri-miR-422a even in the presence of excess, unlabeled competitor tRNA. Pri-miR-422a-binding reactions were also carried out with several MMR-deficient MutLα mutants, MutLαEA (MLH1E34A-PMS2E41A) 33, 34, MutLαF99L (MLH1F99L-PMS2) 29 and MutLαE705K (MLH1-PMS2E705K) 35. While MutLαEA (Figure 4D, lanes 6-7) and MutLαF99L (Figure 4D, lanes 8-9) bound pri-miR-422a as efficiently as the WT MutLα, MutLαE705K failed to interact with pri-miR-422a (Figure 4D, lanes 10-11). These results demonstrate that MutLα has significant in-vitro affinity for pri-miR-422a, which may play an important role in regulating miRNA processing.
Co-immunoprecipitation experiments were performed to determine MutLα interactions with Drosha and DGCR8 in HeLa extracts. The result revealed that an MLH1 antibody could pull down not only MLH1, but also Drosha and DGCR8 (Figure 4E, lane 4). This pull-down was not due to the binding of the individual proteins to endogenous pri-miRNAs, as RNase A-treated co-immunoprecipitation gave the same outcome (compare lanes 4 and 5). Similarly, the MLH1 antibody could pull down Drosha and DGCR8, as well as PMS2, when all purified proteins were used in the co-immunoprecipitation experiment (Figure 4F). These results suggest that MutLα may form a complex with Microprocessor.
MutLα stimulates pri-miRNA processing in vitro and in vivo
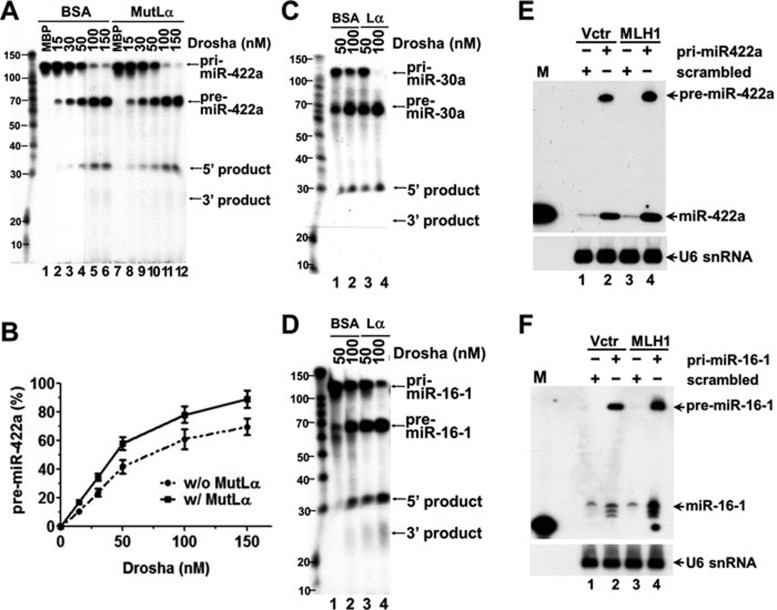
Based on the fact that MutLα interacts with both the substrate and enzymes of miRNA processing, we hypothesize that the ability for MLH1 to facilitate miR-422a production in cells is likely due to direct participation of MutLα in miRNA biogenesis. This hypothesis was first tested using an in-vitro assay. For this purpose, we established conditions that support miRNA processing in vitro, and demonstrated that Drosha and DGCR8 efficiently convert 32P-labeled pri-miRNAs to pre-miRNAs (Figure 5). To our knowledge, a successful in-vitro reconstitution of miRNA processing using purified proteins has not been reported. In this reconstituted miRNA-processing assay, we incubated 32P-labeled pri-miRNA with recombinant Drosha and DGCR8 proteins in the presence or absence of MutLα. As shown in Figure 5A, addition of purified MutLα into the reconstituted reactions stimulated conversion of pri-miR-422a to pre-miR-422a by Drosha and DGCR8 (compare lanes 1-6 with lanes 8-12). Quantification of these data reveals an increase of approximately 20%-25% in pre-miR-422a in reactions containing MutLα (Figure 5B). Similar results were obtained in the processing reactions with pri-miR-30a (Figure 5C) and pri-miR-16-1 (Figure 5D) in the presence of MutLα.
Figure 5.
MutLα promotes pri-miR-422a processing. (A, B) In-vitro reconstitution of miRNA processing. Reconstitution of miRNA processing was performed in reactions containing pri-miR-422a, Drosha and DGCR8 in the presence or absence of MutLα, as indicated. Three independent experiments were performed. Data were quantified, presented as mean ± SD, and plotted in (B). (C, D) Stimulation of Drosha/DGCR8-mediated processing of pre-miR-30a and pre-miR-16-1 by MutLα, respectively. (E, F) MutLα stimulates the processing of miR-422a and miR-16-1 in vivo, respectively. HCT116 cells with or without MLH1 expression were transfected with plasmids carrying the indicated pri-miRNAs, and RNAs were isolated 48 h after transfection and analyzed for conversion of the exogenous pri-miRNAs to their corresponding pre- and mature miRNAs using northern blotting analysis as described 49. U6 snRNA was used as a loading control. M, a 23-oligonucleotide size marker; Lα, MutLα Vctr, vector alone.
To determine the MutLα effect on miRNA processing in vivo, we transfected a plasmid carrying either a pri-miRNA gene (pri-miR-422a or pri-miR-16-1) or a scrambled sequence into MLH1-deficient HCT116 cells with or without MLH1 expression, and the conversion to pre-miRNA and mature miRNA from the exogenous pri-miRNA was detected by northern blotting analysis using a DNA oligomer complementary to the mature miRNA as a probe. It was found that enhanced production of pre-miRNA and mature miRNA was observed in HCT116 cells transfected with MLH1 for both miR-422a (Figure 5E) and miR-16-1 (Figure 5F). Taken together, our results shown here suggest that MutLα stimulates Drosha/DGCR8-mediated processing of pri-miRNA to pre-miRNA in vitro and in vivo.
MutLα ATPase and pri-miRNA binding activities are required for stimulation of miRNA processing
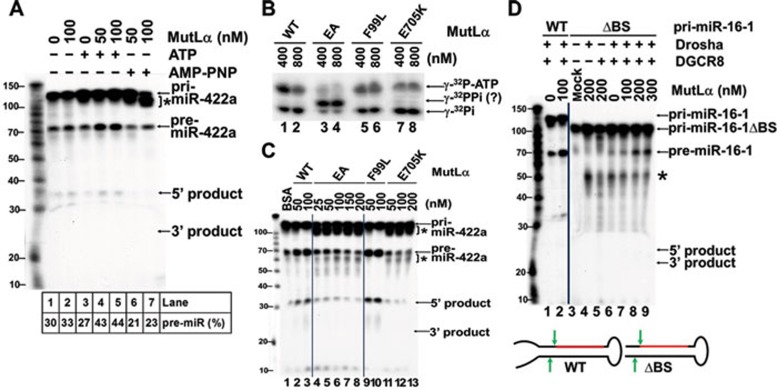
MutLα has an intrinsic ATPase activity that is required for its function in MMR 33, 34. We therefore examined whether ATP hydrolysis by MutLα is essential for its ability to promote pri-miRNA processing. As shown in Figure 6A, MutLα stimulates pri-miRNA processing in the presence of ATP (compare lanes 4 and 5 with lane 2). However, the stimulation was not observed when ATP was replaced with a non-hydrolyzable ATP analog, AMP-PNP, in the reaction (Figure 6A, lanes 6 and 7). Instead, an uncharacterized slow-migrating RNA species accumulated in the presence of AMP-PNP (Figure 6A, asterisk). These observations indicate that ATP hydrolysis by MutLα is essential for its ability to stimulate miRNA processing.
Figure 6.
Processing enhancement by MutLα requires integrity of the MutLα ATPase and pri-miRNA binding activity. (A) Effect of ATP on miRNA processing stimulation by MutLα. In-vitro processing of pri-miR-422a was carried out in the presence of 2 mM ATP or AMP-PNP, as indicated. pri-miR-422a and the resulting pre-miR-422a and 3′-, 5′-products are indicated with arrows. (B) ATPase assay of wild-type (WT) and mutant MutLα proteins. The indicated proteins were incubated with 330 mM (γ-32P)-ATP in reactions containing 50 mM HepesċKOH, pH7.6 and 10 mM MgCl2, and the hydrolysis products were analyzed in a 20% polyacrylamide gel. (C) In-vitro processing of pri-miR-422a using WT MutLα, MutLαEA (EA), MutLαF99L (F99L) or MutLαE705K (E705K). (D) In-vitro processing of WT pri-miR-16-1 and flanking basal segment deleted pri-miR-16-1ΔBS (ΔBS). *Non-specific reaction products.
This idea was further examined using MMR-deficient MutLα proteins MutLαEA, MutLαF99L and MutLαE705K 29, 34, 35. Our results show that MutLαEA and MutLαF99L are proficient, but MutLαE705K is deficient in pri-miRNA binding (Figure 4D). In addition, MutLαEA hydrolyzes ATP, but generates different products than the WT MutLα (Figure 6B, compare lanes 1 and 2 with lanes 3 and 4), while MutLαF99L (Figure 6B, lanes 5, 6) and MutLαE705K (Figure 6B, lanes 7, 8) exhibit nearly normal ATPase activity. Figure 6C shows reconstituted miRNA processing reactions in the presence or absence of these MutLα mutants. MutLαF99L stimulates the reaction as efficiently as WT MutLα (Figure 6C, compare lanes 2-3 with 9-10). However, MutLαEA and MutLαE705K have lost the ability to stimulate this reaction; instead, two novel species accumulate, one migrating immediately below the substrate and the other below the normal reaction product (Figure 6C, asterisks). These species, similar to those generated in reactions containing AMP-PNP (Figure 6A, lanes 6 and 7), may be nonspecific reaction products produced by aborted processing reactions 36. These observations suggest that MutLα must retain both pri-miRNA binding and normal ATPase activities in order to stimulate miRNA processing by Microprocessor.
MutLα compensates for the loss of basal segments of pri-miRNA
Previous studies indicate that the single-stranded RNA segments at the 3′- and 5′-termini of pri-miRNA, also known as basal segments, are essential for pri-miRNA processing by Drosha and DGCR8 36, 37. Our studies confirm that the Drosha-DGCR8 Microprocessor converted pri-miR-16-1 to pre-miR-16-1 with high efficiency (Figure 6D, lanes 1-2), but the same substrate lacking basal segments (i.e., pri-miR-16-1ΔBS) was poorly processed by the Microprocessor complex, as judged by the fact that a very small amount pre-miR-16-1 and several other fast-migrating RNA species (likely nonspecific reaction products) were produced (Figure 6D, lane 6). However, MutLα caused a dose-dependent increase in the specific product and decrease in the non-specific reaction products (Figure 6D, lanes 7-9). These findings suggest that pri-miRNA basal segments are essential for miRNA processing by Drosha/DGCR8 in the absence of MutLα, but are not required when MutLα is present. In other words, MutLα restores normal reaction efficiency and specificity when the pri-miRNA substrate lacks basal segments. EMSA assays confirmed that MutLα binds efficiently to pri-miR-16-1ΔBS (data not shown). We also examined the effect of MutLα on the processing of pri-miRNAs with a nicked terminal loop, as whether or not the terminal loop is required for efficient processing of pri-miRNAs is rather controversial 36, 37, 38, 39. However, our results reveal that in the purified system, Microprocessor by itself can efficiently process pri-miR-16-1 containing a nicked terminal loop, although MutLα can stimulate the reaction (data not shown). Collectively, we demonstrate that MutLα promotes a Microprocessor-mediated processing of miR-422a, as well as several other miRNAs tested, including those lacking basal segments.
Discussion
In this study, we made the following interesting observations: (1) discovery of a novel MutLα function in stimulating miRNA processing; (2) identification of a regulatory feedback between miR-422a and MLH1; (3) reconstitution of the miRNA processing reaction in a defined system.
We initially observed a positive correlation between miR-422a processing and MLH1 expression (Figures 2 and 3), and later showed that the correlation is due to direct stimulation of the miRNA processing by MutLα (Figure 5). Since MutLα interacts with pri-miRNAs and the Microprocessor complex (Figure 4), and also promotes the processing of several other miRNAs tested (Figure 5), we believe that the newly identified MutLα activity in miRNA biogenesis likely applies to many other miRNAs, if not all. This idea is consistent with the fact that MLH1 dictates many miRNA expression patterns in colorectal tumors 27, 28.
The mechanism by which MutLα stimulates miRNA processing is not clear. Nonetheless, the data shown in this study indicate that the stimulation reaction requires MutLα ATPase activity and two novel activities identified in this study (Figure 6), i.e., pri-miRNA binding and interactions with Drosha and DGCR8. Because ATP hydrolysis by MutL leads to conformational changes of the protein, which modulate interactions between MutL and other MMR components 40, 41, it is possible that ATP hydrolysis by MutLα during the pri-miRNA processing reaction promotes a required conformational change of MutLα and/or its binding partners in the context of this reaction. This possibility is consistent with the observation that MutLαEA, which is defective in normal ATPase activity (Figure 6B), does not stimulate, but inhibits, miRNA processing (Figure 6C). Similarly, the MutLα mutant that lacks pri-miRNA-binding activity also fails to stimulate miRNA processing (see MutLαE705K in Figure 6C). The most important aspect of the MutLα-mediated stimulation of miRNA processing may be the interaction between MutLα and Microprocessor, which may allow MutLα to independently enhance the affinity of Drosha and/or DGCR8 for its pri-miRNA substrate (Figure 4B and 4C). However, the mechanism underlying these interactions remains to be investigated.
It is worth mentioning that MutLα has recently been shown to possess a latent endonuclease activity, but the E705K substitution in the PMS2 subunit of MutLα abolishes the endonuclease activity 35. Therefore, it is possible that the observed stimulation of miRNA processing by MutLα may be attributed to the protein endonuclease activity. However, based on the fact that miRNA processing requires site-specific cleavages and that MutLα is a non-specific nuclease, whose activation requires replication factor C and proliferating cellular nuclear antigen 35, which were not present in our reactions, we believe that the MutLα endonuclease activity is unlikely involved in stimulating miRNA processing.
A second way by which MutLα enhances pre-miRNA production is to ensure the conversion of pri-miRNAs that lack basal segments to pre-miRNAs. Previous studies have shown that miRNA basal segments are required for Drosha/DGCR8-mediated processing of pri-miRNA to pre-miRNA 36. We demonstrate here that the presence of MutLα in the miRNA processing reaction is sufficient to compensate for the loss of miRNA basal segments. However, further studies are required to understand the molecular basis of this reaction.
While MutLα (MLH1-PMS2) facilitates miRNA processing, we also show that expression of MLH1 is regulated by at least miR-422a. MutLα is known to be involved in all known genome-maintenance functions of the MMR system, including correction of DNA biosynthetic errors, suppression of homeologous recombination and DNA damage signaling 20, 21, 22, 23, 42. In addition, MLH1 plays a role in DNA interstand crosslink repair 43 and spermatogenesis 44, 45. Interestingly, overexpression of MLH1 does not enhance genome stability, but promotes hypermutability 25, 26. Because MLH1 is a critical component of multiple protein complexes and has multiple important cellular functions, the cellular level of MLH1 is likely to be subject to tight regulation. The reciprocal feedback regulation between MLH1 and miR-422a may represent one of the mechanisms by which cells regulate MLH1 expression. A model describing this feedback regulation is depicted in Figure 7.
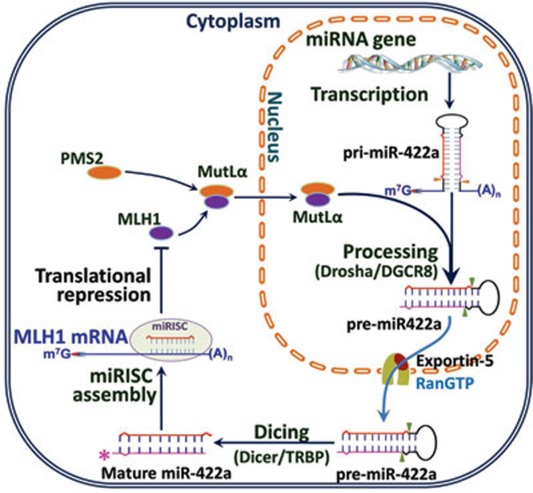
Figure 7.
Model for reciprocal feedback regulation between MLH1 and miR-422a. MLH1 dimerizes with PMS2 to form MutLα in the cytoplasm, and the MutLα heterodimer then translocates into the nucleus to participate in genome maintenance via the MMR system. When MutLα reaches to a level that is toxic to cells, it begins to stimulate the conversion of pri-miR-422a into pre-miR-422a, leading to increasing production of pre-miR-422a and subsequently mature miR-422a. The resulting high level of mature miR-422a interacts with MLH1mRNA by pairing with a putative miR-422a interaction sequence in the MLH13′-UTR, which inhibits MLH1 translation.
In this model, MLH1 and miR-422a reciprocally regulate each other's cellular level. This relationship has several implications: First, a high level of MLH1 stimulates miR-422a processing. MLH1 dimerizes with PMS2 in the cytoplasm to form MutLα, and the MutLα heterodimer then translocates into the nucleus 46, where it participates in genome maintenance via the MMR system. However, as the concentration of MutLα increases, it stimulates conversion of pri-miR-422a into pre-miR-422a, leading to increased level of pre-miR-422a. Pre-miR-422a is transported into the cytoplasm and further processed into mature miR-422a. Second, as the concentration of mature miR-422a increases, it inhibits MLH1 expression. This inhibition is through pairing of the mature miRNA with the putative miR-422a-binding sequence in the MLH1 3′-UTR, suppressing MLH1 translation (this study). We believe that this mechanism is critical for precise regulation of the cellular level of MLH1. An important feature of this model is that high or low thresholds of MLH1 concentration trigger a cellular response. When MLH1 protein falls below a critical level, all MLH1 molecules are recruited into genome-maintenance activities, and MLH1 is not available to stimulate Drosha/DGCR8-mediated miRNA processing reactions. Thus, the rate of pri-miR-422a processing decreases, leading to a reduced level of mature miR-422a. In contrast, when the MLH1 concentration increases above the level required for genome-maintenance, it becomes toxic and cell viability declines 24, 25, 26. In this situation, excess MutLα stimulates processing of pri-miR-422a, and the resulting mature miR-422a suppresses MLH1 translation, thus preventing toxic effects of MLH1 overexpression. However, whether or not other miRNAs participate in a similar feedback regulation with MLH1 awaits further investigations.
Materials and Methods
DNA constructs
A synthetic DNA oligonucleotide was prepared using the DNA sequence of the putative miR-422aRE located between +18 and +50 of the MLH1 3′-UTR. Three tandem copies of this olignocleotide (3×miR422aRE) were subcloned into the HindIII and SpeI sites of pMIR-REPORT Luciferase (Ambion, Inc), to produce pMIR-Luc-3×miR422aRE. pMIR-MLH1-3×miR422aRE was constructed from pMIR-Luc-3×miR422aRE by replacing the Luciferase gene with the MLH1 cDNA.
The MLH1 cDNA, with or without the MLH1 3′-UTR or 3×miR422aRE, was subcloned into pcDNA3.1/His ABC (Invitrogen Corp.). The stop codon TAA was substituted for the ATG start codon, as indicated in Figure 3B, using QuikChange Site-Directed Mutagenesis Kit (Stratagene). Cloning strategies for all reporter and MLH1 expression constructs used in this study are shown in Figures 1D, 3A and 3B. Primers used to detect pri- and pre-miR-422a are shown in Figure 2A, and their specific sequences are provided in Supplementary information, Tables S1,S2,S3,S4. The sequences of all plasmids were confirmed by DNA sequencing.
Cell culture and transfection
HeLa, HEK293, 293T and HCT116 cells were cultured in media recommended by ATCC supplemented with 10% fetal bovine serum. Transfections were carried out using 25-200 pmol of the indicated pre-miR-422a or anti-miR-422a, with or without 2 μg plasmid DNA, as indicated, and FuGENE® HD Transfection Reagent (Roche Diagnostics GmbH, Germany). The medium was switched 48 h after transfection. Total RNA and cellular protein was isolated from transfected cells. Alternatively, luciferase activity was measured in transfected cells.
Total RNA extraction, RT-PCR and qRT-PCR
Unless mentioned otherwise, total RNAs were used to determine mRNA levels of MLH1 and β-actin (internal control), and RNAs isolated from cytoplasm and nuclei were employed to determine the levels of pre- and pri-miRNAs, respectively, using Qiagen RNeasy Mini Kit (Qiagen). RNAs (1 μg) were treated with RQ1 DNase (Promega Corp.) and used for first-strand cDNA synthesis in a 100-μl reaction containing TaqMan Reverse Transcription Reagents (Applied Biosystems). cDNA from each sample was diluted 1:10 in qRT-PCR buffer. qRT-PCR reactions were performed using a 7300 Real-Time PCR System (Applied Biosystems), with SYBR® Green PCR Master Mix (2×) Kit (Applied Biosystems). All reactions were carried out in quadruplicate with reference dye normalization, and the median Ct value was used for statistical analysis. qRT-PCR was carried out in triplicate. All values were normalized to β-actin. Primer sequences used for quantitative PCR studies are listed in Supplementary information, Tables S1, S2, S3, S4.
RNA interference
Human MLH1 siRNA duplex was from Santa Cruz Biotechnology, Inc. HeLa cells were transfected with 100 pmol MLH1 siRNA duplex in six-well plates with FuGENE® HD Transfection Reagent (Roche Diagnostics GmbH, Germany). After 48 h, cells were harvested and processed for RNA quantification by RT- and/or qRT-PCR or protein quantification by western blotting.
Protein expression and purification
Drosha cDNA was subcloned into pFastBac-1 (Invitrogen) with an MBP-tag at N-terminus, and a tandem Flag- and 6× His-tag at C-terminus. The resulting expression construct, pFB-MBP-Drosha-FH, was used to overexpress MBP-Drosha in High Five™ insect cells. Drosha was purified from the insect cells through affinity column chromatography, initially with a nickel HisTrap™ HP column (GE Healthcare) and later with an Amylose Resin column (New England Biolabs, Inc.), followed by dialysis against the PBE100 Buffer (25 mM HEPES-NaOH, pH 7.8, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA and 10% glycerol) for 4 h at 4 °C. Aliquots of MBP-Drosha were added 1 mg/ml BSA, and frozen in liquid nitrogen and stored at −80 °C for use.
Similarly, DGCR8 cDNA was cloned into the EcoRI and HindIII sites of pFastBac-HTc (Invitrogen), and the recombinant protein containing an N-terminal tandem 6× His- and a Flag-tag C-terminus was expressed in High Five™ insect cells and purified using a HisTrap™ HP nickel column (GE Healthcare). The peak fractions containing DGCR8 were dialyzed against the PBE100 buffer for 4 h at 4 °C, and further purified using an ANTI-FLAG M2 affinity gel (Sigma-Aldrich). Briefly, the dialyzed DGCR8 was incubated with the affinity gel at 4 °C overnight, washed with five-resin volumes of the PBE100 buffer, and eluted with 3× FLAG peptide (0.3 mg/ml in PBE100). Aliquots of purified DGCR8 were adjusted to 1 mg/ml BSA, frozen in liquid nitrogen and stored at −80 °C for future use.
Constructs of expression of MutLα and MutLαEA were gifts from Michael Liskay (Oregon Health Sciences University), and constructs for MutLαF99L and MutLαE705K were generated using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). All MutL heterodimers were expressed in pFastBac Dual vector and purified as described 34, 47. The purity of all recombinant proteins was assessed by SDS-PAGE and protein concentrations were determined by Bradford assay.
Preparation of miRNA processing substrates
pri-miR-422a, pri-miR-145, pri-miR-30a, pri-miR-16-1, pri-miR16-sTL1 and pri-miR-16-1ΔBS were designed, generated and purified as described previously 36. PCR products contained the T7-promoter sequence and two tandem G's right before pri-miRNA sequences, and were amplified from a normal human whole-blood genomic DNA sample. The T7-primer sequences are available upon request. The T7-G2-pri-miRNA PCR products were used directly as templates for in-vitro transcription of pri-miRNA substrates with MEGAshortscript™ Kit (Ambion, Inc.). The pri-miRNA transcripts were radiolabeled internally by incubating with [α-32P]-ATP in the transcription reactions according to the manufacturer's instruction.
Electrophoretic mobility shift assay
EMSA was performed in 20-μl reactions containing buffer PBE100, Drosha, DGCR8, 1.0 U SUPERase•In™ RNase Inhibitor (Ambion, Inc.), and 200 pmol 32P-labeled pri-miR-422a in the presence or absence of MutLα or MutLα mutants, as indicated. Unlabeled tRNAs (10× excess) were included as non-specific competitor. Reactions were incubated for 20 min on ice, followed by 5 min at room temperature. Reaction products were loaded onto 4% non-denaturing polyacrylamide gels in 0.5× TBE. Gels were run at 150 V for 3 h at 4 °C, and then quantified by phosphorimaging using a KODAK Molecular Imager System.
Immunoprecipitation analysis
Co-immunoprecipitation of Drosha, DGCR8 and MutLα was conducted by incubating either HeLa whole-cell extracts (500 μg) or purified proteins (100 nM Drosha, 200 nM DGCR8 and 100 nM MutLα) with an MLH1 antibody (BD Pharmingen, 554073) under the conditions for in-vitro pri-miRNA processing (see below). MBP or BSA was used as negative controls for the purified system. Immunoprecipitates were analyzed by 8% SDS-PAGE and simultaneously immunoblotted with rabbit antibodies against DGCR8 (ProteinTech Group, Inc), Drosha (Bethyl Laboratories, Inc) and MBP (ProteinTech Group, Inc).
Reconstitution of pri-miRNA processing
The conditions for in-vitro pri-miRNA processing were essentially the same as described 6, 48, except that purified proteins were used in this study. Unless indicated otherwise, pri-miRNA processing assays were performed in 30-μl reactions, containing 100 nM Drosha, 200 nM DGCR8, 100 nM MutLα (or mutant variants indicated), 6.4 mM MgCl2, 2 mM ATP, 1.0 U SUPERase•In RNase Inhibitor and 200 pmol 32P-labeled pri-miRNA substrate in buffer PBE100. BSA was included in reactions lacking MutLα, and MBP was included in reactions lacking Drosha. The reaction mixture was incubated at 37 °C for 90 min (or as indicated) and terminated by phenol extraction. The RNA products were analyzed by 12.5% denaturing PAGE in 0.5× TBE. Gels were run at 350 V until bromophenol blue reached the bottom of the gel (about 2 h). Radiolabeled species were visualized by autoradiography and/or PhosphorImager, and quantified using KODAK Molecular Imager Systems (KODAK MI) version 5.0 (EASTMAN KODAK).
Statistical analysis
The data shown in this study presented an average of at least three independent experiments, each performed in triplicate. All statistical assays, Student's t-test and one-way ANOVA with Tukey's multiple comparison, were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.). Data were considered statistically significant if P-values were less than 0.05 or 0.001, as indicated.
Acknowledgments
We are grateful to VN Kim (Seoul National University, Korea) for providing human Flag-Drosha and Flag-DGCR8 cDNAs, to Ramin Shiekhattar (The Wistar Institute, USA) for pCMV-DGCR8 clone, to Feng Guo (University of California, Los Angeles, USA) for pFastBac-6×His-Drosha vector, to Michael Liskay (Oregon Health & Science University, USA) for baculovirus and plasmid constructs expressing WT and mutant MutLα proteins, and to H-S Yang (University of Kentucky, USA) for technical assistance on the luciferase analysis. This work was supported by National Institutes of Health Grants CA104333 (to LG), GM072756 and CA115942 (to GML). GML is the James-Gardner Chair in Cancer Research.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
RT-PCR Primers for Detecting MLH1 and β-actin mRNA, premiR-422a, and pri-miR-422a (related to Figures 1, 2, 3).
PCR Primers Used for Template Preparation of the In vitro Transcription of pri-miRNAs (related to Figures 4, 5, 6, 7).
Primers for Cloning Drosha and DGCR8 (related to Figures 4, 5, 6, 7).
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Pan X, Gu L. Evidence that a mutation in the MLH1 3′-untranslated region confers a mutator phenotype and mismatch repair deficiency in patients with relapsed leukemia. J Biol Chem. 2008;283:3211–3216. doi: 10.1074/jbc.M709276200. [DOI] [PubMed] [Google Scholar]
- Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Kolodner RD. Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Alani E. Mismatch repair and cancer susceptibility. Curr Opin Biotechnol. 1994;5:585–594. doi: 10.1016/0958-1669(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Zhang H, Richards B, Wilson T, et al. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- Shcherbakova PV, Hall MC, Lewis MS, et al. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg AL, French AJ, Sarver AL, et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6:e 20465. doi: 10.1371/journal.pone.0020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver AL, French AJ, Borralho PM, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Yuan F, Absher K, et al. Preferential loss of mismatch repair function in refractory and relapsed acute myeloid leukemia: potential contribution to AML progression. Cell Res. 2008;18:281–289. doi: 10.1038/cr.2008.14. [DOI] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Stojic L, Mojas N, et al. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem. 2002;277:21810–21820. doi: 10.1074/jbc.M108787200. [DOI] [PubMed] [Google Scholar]
- Tomer G, Buermeyer AB, Nguyen MM, Liskay RM. Contribution of human mlh1 and pms2 ATPase activities to DNA mismatch repair. J Biol Chem. 2002;277:21801–21809. doi: 10.1074/jbc.M111342200. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Wu Q, Vasquez KM. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4:e 1000189. doi: 10.1371/journal.pgen.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Wu X, Platt JL, Cascalho M. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLalpha. Mol Cell Biol. 2003;23:3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Koscianska E, Starega-Roslan J, Czubala K, Krzyzosiak WJ. High-resolution northern blot for a reliable analysis of microRNAs and their precursors. Sci World J. 2011;11:102–117. doi: 10.1100/tsw.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR Primers for Detecting MLH1 and β-actin mRNA, premiR-422a, and pri-miR-422a (related to Figures 1, 2, 3).
PCR Primers Used for Template Preparation of the In vitro Transcription of pri-miRNAs (related to Figures 4, 5, 6, 7).
Primers for Cloning Drosha and DGCR8 (related to Figures 4, 5, 6, 7).