Abstract
Autophagy is an important catabolic process that delivers cytoplasmic material to the lysosome for degradation. Autophagy promotes cell survival by elimination of damaged organelles and proteins aggregates, as well as by facilitating bioenergetic homeostasis. Although autophagy has been considered a cell survival mechanism, recent studies have shown that autophagy can promote cell death. The core mechanisms that control autophagy are conserved between yeast and humans, but animals also possess genes that regulate autophagy that are not present in yeast. These regulatory differences may be explained by the need to control autophagy in a cell context-specific manner in multicellular animals, such as during cell survival and cell death. Autophagy was thought to be a bulk cytoplasmic degradation mechanism, but recent studies have shown that specific cargo is recruited for degradation. This suggests the possibility that either cell survival or death may be regulated by selective autophagic clearance of cytoplasmic material. Here we summarize the mechanisms that regulate autophagy and how they may contribute to cell survival and death.
Animal-specific autophagy-regulatory genes may control cell survival and death in a context-dependent manner. The selective autophagic clearance of cytoplasmic material may also be involved.
Autophagy (self-eating) is an evolutionarily conserved catabolic process that is used to deliver cytoplasmic materials, including organelles and proteins, to the lysosome for degradation. Three types of autophagy have been described, including macroautophagy, microautophagy, and chaperone-mediated autophagy (Mizushima and Komatsu 2011). Although macroautophagy involves the fusion of the double membrane autophagosome and lysosomes, microautophagy is poorly understood and thought to involve direct uptake of material by the lysosome via a process that appears similar to pinocytosis. By contrast, chaperone-mediated autophagy is a biochemical mechanism to import proteins into the lysosome; it depends on a signature sequence and interaction with protein chaperones. Here we will focus on macroautophagy (hereafter called autophagy) because of our knowledge of this process in cell survival and cell death.
Autophagy was likely first observed when electron microscopy was used to observe “dense bodies” containing mitochondria in mouse kidneys (Clark 1957). Five years later, it was reported that rat hepatocytes exposed to glucagon possessed membrane-bound vesicles that were rich in mitochondria and endoplasmic reticulum (Ashford and Porter 1962). Almost simultaneously, it was shown that these membrane-bound vesicles contained lysosomal hydrolases (Novikoff and Essner 1962). In 1965 de Duve coined the term “autophagy” (Klionsky 2008).
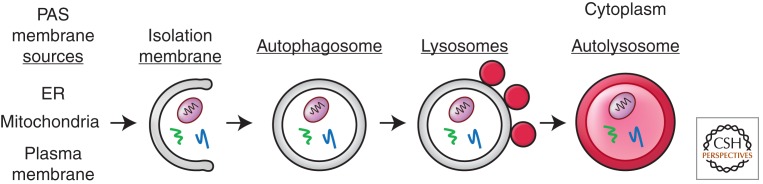
The delivery of cytoplasmic material to the lysosome by autophagy involves membrane formation and fusion events (Fig. 1). First an isolation membrane, also known as a phagophore, must be initiated from a membrane source known as the phagophore assembly site (PAS). de Duve suggested that the smooth endoplasmic reticulum could be the source of autophagosome membrane (de Duve and Wattiaux 1966), and subsequent studies have supported this possibility (Dunn 1990; Axe et al. 2008). Although controversial, mitochondria and plasma membrane could also supply membranes for the formation of the autophagosomes under different conditions (Hailey et al. 2010; Ravikumar et al. 2010). The elongating isolation membrane surrounds cargo that is ultimately enclosed in the double membrane autophagosome. Once the autophagosome is formed, it fuses with lysosomes (known as the vacuole in yeasts and plants) to form autolysosomes in which the cargo is degraded by lysosomal hydrolases. At this stage lysosomes must reform so that subsequent autophagy may occur (Yu et al. 2010).
Figure 1.
Macroautophagy (autophagy) delivers cytoplasmic cargo to lysosomes for degradation, and involves membrane formation and fusion. The isolation membrane is initiated from a membrane source known as the from the phagophore assembly site (PAS). The isolation membrane surrounds cargo, including organelles and proteins, to form a double membrane autophagosome. Autophagosomes fuse with lysosomes to form autolysosomes in which the cargo is degraded by lysosomal hydrolases.
AUTOPHAGY GENES
Autophagy is best characterized in the yeast Saccharomyces cerevisiae, in which genetic screens resulted in the identification of genes that are required for autophagy. Screens for yeast mutants with defects in either autophagic structures, degradation of cytoplasmic proteins, or possessing altered cytoplasm to vacuole targeting resulted in the identification of Apg, Aut, and Cvt mutants (Tsukada and Ohsumi 1993; Thumm et al. 1994; Harding et al. 1995). The recognition that some of these mutations were in common genes ultimately resulted in the renaming of these autophagy regulators as Atg genes (Harding et al. 1996; Klionsky et al. 2003). Over 30 autophagy genes have been identified in yeast, and many of these genes are conserved in animals (Weidberg et al. 2010).
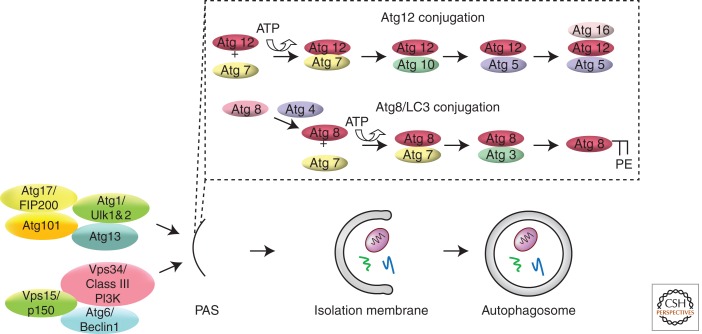
Autophagy is regulated by Atg1 and its interacting proteins, Vps34 and its interacting proteins, and two ubiquitin-like conjugation systems (Fig. 2). Atg1 (Ulk1 and 2 in mammals) is a serine–threonine protein kinase, and its kinase activity is required for autophagy (Matsuura et al. 1997; Kamada et al. 2000). Atg13 is the regulatory sub-unit of the Atg1 kinase complex that also includes FIP200 and Atg101 in animals (Weidberg et al. 2010). Atg1, Atg13 and FIP200 (Atg17) are present in yeast. However, the other Atg1 complex components, including Atg11, Atg20, Atg24, Atg29, and Atg31, do not appear to be encoded by animal genomes. Atg1 is necessary for the induction of autophagy in different cell types, and expression of Atg1 is also sufficient for the induction of autophagy in Drosophila (Scott et al. 2004, 2007; Berry and Baehrecke 2007; Chan et al. 2009).
Figure 2.
Core pathways that regulate autophagy. Atg1 and its interacting proteins, Vps34 and its interacting proteins, and two ubiquitin-like conjugation systems are required for the elongation of the isolation membrane and formation of an autophagosome.
The Vps34 regulatory complex is comprised of the lipid kinase Vps34 (also known as class III phosphatidylinositol 3 (PI3) kinase), Atg6 (known as Beclin1 in mammals), and the protein kinase Vps15 (p150 in mammals) (Simonsen and Tooze 2009). This core complex regulates the formation of PI3 phosphate (PI3P) lipids, and is required for multiple intracellular vesicle trafficking pathways, including endocytosis and autophagy. The Vps34 complex has different proteins associated with it that are thought to be specific to the vesicle process that is regulated, and in the context of autophagy these include Atg14 and Vps38 (UVRAG in mammals). The Vps34 complex components have been localized to the PAS, and are required for the formation of autophagosomes (Juhász et al. 2008). Although some studies have suggested that mTOR is in a common regulatory pathway with Vps34 (Byfield et al. 2005; Nobukuni et al. 2005), others have suggested that these complexes function in parallel genetic pathways (Juhász et al. 2008; Jaber et al. 2012).
Two ubiquitin-like conjugation pathways are required for autophagy, and involve the ubiquitin-like proteins Atg8 (LC3 in mammals) and Atg12 (reviewed in Ohsumi 2001). The carboxy-terminal glycine of Atg8 is covalently bound to phosphatidylethanolamine (PE) following processing by the cysteine protease Atg4 (Ichimura et al. 2000; Kabeya et al. 2000; Kirisako et al. 2000), whereas Atg12 is ultimately associated with Atg5 and Atg16 (Kuma et al. 2002). Both Atg8 and Atg12 conjugation systems use a common E1-like activating enzyme Atg7 (Tanida et al. 1999). Although the Atg8 conjugation system uses Atg3 as an E2-like conjugating enzyme, the Atg12 conjugation system uses Atg10 and associates with Atg5 and Atg16. Atg8-PE is associated with both the isolation membrane and autophagosome, whereas the Atg12, Atg5, and Atg16 complex is only associated with the isolation membrane and disassociates on formation of the autophagosome.
Although autophagy was long considered a bulk degradation process with limited specificity, recent studies have clearly shown that specific cargoes are recruited to autophagosomes for destruction (reviewed in Johansen and Lamark 2011). Several factors have been identified that are required for selection of proteins as cargo for autophagosomes, including p62/SQSTM1/Ref(2)P, Nbr1, and Alfy (Bjørkøy et al. 2005; Kirkin et al. 2009; Filimonenko et al. 2010). Autophagosomes can also consume large cargoes, including peroxisomes (pexophagy) (Manjithaya et al. 2010), mitochondria (mitophagy) (Elmore et al. 2001), ribosomes (ribophagy) (Kraft et al. 2008), and lipid droplets (lipophagy) (Singh et al. 2009). Although the elimination of these organelles may all influence cell survival and death, the removal of mitochondria is particularly interesting in this context given the role of this organelle in bioenergetics and the regulation of cell death. In yeast, Atg32 targets mitochondria to autophagosomes (Kanki et al. 2009; Okamoto et al. 2009), but this protein does not appear to be present in animals. In animals, Parkin and Nix mediate the selective recruitment of mitochondria to autophagosomes (Narendra et al. 2008; Novak et al. 2010). Recruitment of Parkin to damaged mitochondria requires PINK1 (Narendra et al. 2010), and the association of mutations in these genes with Parkinson disease families raises interesting possibilities about the role of autophagy in neurodegeneration.
The formation of autophagosomes is succeeded by docking and fusion with the lysosomes to form the autolysosome, and this process uses the Rab-SNARE system and other molecules that regulate membrane fusion (Nair et al. 2011). Unlike yeast, in which a single vacuole (lysosome) fuses with all autophagosomes, multiple lysosomes fuse with each autophagosome in animals (Yu et al. 2010). Subsequently, lysosomal hydrolases degrade the cargo, and the resulting macromolecules are released into the cytosol for further recycling. Therefore, the rate of autophagy (also known as autophagic flux) depends on both the number of autophagosomes that are formed and the degradative capacity of lysosomes and turnover of autophagic cargo within the cell. This is an important consideration during experimentation when increased numbers of autophagosomes may not necessarily indicate greater autophagic flux, as it may also reflect decreased degradation capacity. Once autolysosmes form, lysosome number is restored, and this process depends on mTOR function (Yu et al. 2010).
AUTOPHAGY GENES THAT ARE SPECIFIC TO MULTICELLULAR ANIMALS
Our knowledge of the core molecular mechanisms controlling autophagy is based on studies in yeast. However, several recent studies indicate that the regulation of autophagy may differ in multicellular animals. As mentioned above, the components and regulation of the Atg1 complex differs between yeast and animals (reviewed in Weidberg et al. 2010). In addition, novel regulators of the Vps34 complex are restricted to higher animals, with the best example being AMBRA1 that is present in mammals but absent in invertebrates (Fimia et al. 2007). It remains to be determined if some of the elegant emerging mechanisms for the regulation of autophagy in mammalian cells, including roles for Bcl-2 (Pattingre et al. 2005), lipid phosphatases (Vergne et al. 2009), and other factors, are conserved in diverse taxa.
The most comprehensive genetic screen for genes that are required for autophagy in animals was conducted by Zhang and colleagues (Tian et al. 2010). They screened for mutations that inhibited clearance of PGL granules in nematode embryos. In addition to the identification of many known core autophagy genes, they identified four ectopic PGL granule (epg) genes named epg-2, -3, -4, and -5 that are specific to multicellular animals. Although epg-3, -4, and -5 are required for starvation-induced autophagy, epg-2 mediates the recognition of cargo (e.g., aggregates of P granule proteins) for delivery to autophagosomes. epg-2 encodes a protein that appears to be specific to nematodes. By contrast, epg-3 encodes a protein that is conserved in Arabidopsis, Drosophila, and mammals, but no similar protein is present in S. cerevisiae. Like epg-3, epg-4 encodes a protein that is conserved in plants and animals, but no similar protein is present in yeast. epg-5 encodes a protein that is conserved in Drosophila and is known as VMP1 in mammals (Dusetti et al. 2002), but no similar proteins are present in either Dictyostelium, Arabidopsis, or S. cerevisiae. It is interesting to note that human homologs of the genes identified in this study have been implicated in cancer and other diseases (Gu et al. 2000; Dusetti et al. 2002; Sjöblom et al. 2006).
It is logical that multicellular animals may need specialized mechanisms for the regulation of autophagy in different situations. Although autophagy is a conserved catabolic process, this process may be adapted for use in specific cell contexts, such as cell survival and cell death. Although differences in autophagy may be specified at the level of recruitment of specific cargoes to autophagosomes, it is also possible that different types of autophagy are regulated by distinct activation and repression mechanisms. For example, the conserved immuno receptor Draper is required for autophagy in dying salivary glands in Drosophila, but not for autophagy in the fatbody where this process promotes nutrient utilization and cell survival (McPhee et al. 2010). A lack of experimental animal models to study autophagy in specific cell contexts is a limitation facing this research field.
AUTOPHAGY IN CELL SURVIVAL AND NUTRIENT UTILIZATION
Autophagy is involved in maintaining cellular homeostasis. Therefore, it is important to understand the regulation of basal autophagy under normal nutrient conditions. A recent genome-wide screen identified many genes that either suppress or enhance basal autophagy, including a mTOR-independent mechanism for the regulation of autophagy (Lipinski et al. 2010). Another high-throughput study identified numerous proteins that interact with the proteins known to regulate autophagy under basal conditions, thus providing a comprehensive parts list that will enable the dissection of the molecular mechanisms underlying basal autophagy (Behrends et al. 2010).
Studies in yeast pioneered our understanding of the genes that control autophagy, and much of this work has focused on stress-induced autophagy under nutrient-limiting conditions in which catabolism promotes cell survival. Like yeast, autophagy is induced by nutrient limitation in animals, and this influences the bioenergetics of the cell and possibly the organism (Lum et al. 2005). The importance of autophagy during animal starvation is exemplified by the important study showing that mice deficient for Atg5 appear almost normal at birth but die within 1 day of birth (Kuma et al. 2004).
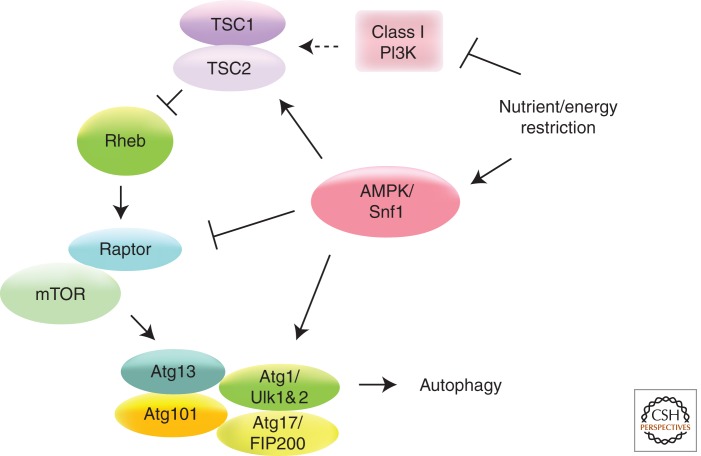
AMPK (SNF1 in yeast) and mTOR are conserved kinases that sense energy and nutrient stress. Both of these kinases influence the activity of Atg1 (ULKs in mammals) to regulate autophagy (Fig. 3) (Samari and Seglen 1998; Kamada et al. 2000; Wang et al. 2001; Scott et al. 2004; Meley et al. 2006). mTOR is inhibited upon withdrawal of growth factors, such as insulin or insulin-like growth factors, by a cascade of phosphorylation reactions involving Class I PI3K, Akt, TSC1/TSC2, and Rheb (Wullschleger et al. 2006). In mammalian cells, mTOR can also be regulated by a novel mechanism involving localization to the lysosome (Sancak et al. 2010). AMPK regulates autophagy by inhibition of mTOR by phosphorylation of TSC2 and Raptor (Inoki et al. 2003; Gwinn et al. 2008). Furthermore, three recent papers show that AMPK-dependent phosphorylation of Ulk1 can regulate autophagy in nutrient-limiting conditions (Lee et al. 2010; Egan et al. 2011; Kim et al. 2011), although the details of these studies vary. A direct interaction between AMPK and Ulk1 has also been shown (Behrends et al. 2010). The molecular players of starvation-induced autophagy have been studied in considerable detail when compared to autophagy that is induced by several other stresses, such as endoplasmic reticulum stress and hypoxia. Another important challenge that eukaryotic cells face is to combat microorganisms and hostile environments, and autophagy plays a major role in cellular defense and survival under these conditions (Deretic 2011).
Figure 3.
AMPK and mTOR are conserved kinases that sense energy and nutrient stress, and influence the activity of Atg1 to regulate autophagy. mTOR influences the activity of the Atg1 complex and autophagy. AMPK regulates autophagy by inhibition of mTOR by phosphorylation of TSC2 and Raptor. AMPK can also influence autophagy by phosphorylation of Atg1/Ulk1.
In addition to sensing stress, multicellular organisms appear to use developmental signals to regulate autophagy. It is possible that these signals, including hormones, are activated as part of a systemic stress response, but it is also possible that signals during development induce autophagy to regulate cell remodeling. In the context of some animals, this remodeling may be the most efficient method to recycle material for development while maintaining organism fitness and survival. Given the important role of autophagy in stress responses and maintenance of cellular homeostasis, more work is needed to understand if autophagy that is induced by developmental signals is part of a stress program.
AUTOPHAGY IN CELL DEATH
Schweichel and Merker identified three types of cell death based on the role and location of lysosomes inside the cell (Schweichel and Merker 1973). Type II, later called autophagic cell death, is distinguished from type I (apoptotic) cell death by the presence of abundant autophagic structures in the dying cell, a lack of phagocyte recruitment, and, in some instances, by caspase-independence (Schweichel and Merker 1973; Clarke 1990; Baehrecke 2005). The functional contribution of autophagy to cell death has been a subject of great controversy. The reason for controversy appears to be related to the historical focus on autophagy as a cell survival process that is described above. In addition, until relatively recently limited empirical studies had been done to test whether autophagy genes actually facilitate cell death.
Multiple experimental systems have contributed to our recent understanding of autophagy and cell death. Dictyostelium discoideum, for example, lacks apoptosis machinery that could participate in nonapoptotic cell death making this a simpler system for the interpretation of the role of autophagy in cell death. Dictyostelium exists as a unicellular organism when it is grown on rich media. Upon starvation, however, thousands of cells aggregate to form a multicellular fruiting body in which stalks support balls of spores. These stalk cells undergo developmental cell death via autophagy, as mutations in Atg genes prevent the death of stalk cells (Otto et al. 2003; Kosta et al. 2004). One limitation of this system is that Dictyostelium lacks apoptosis machinery, and an understanding of the relationship between autophagy and cell death in a system with intact apoptosis machinery is important to our understanding of how to modulate autophagy for therapeutic purposes in humans.
The contribution of autophagy to cell death has been studied most in Drosophila in which apoptosis machinery is involved in the death of multiple cell types (Ryoo and Baehrecke 2010). In Drosophila, an increase in a steroid hormone triggers the destruction of obsolete tissues at the end of larval development (Jiang et al. 1997). Dying larval midgut and salivary gland cells display markers of apoptosis, such as DNA fragmentation, acridine orange staining, and elevated levels of proapoptotic gene RNAs (Jiang et al. 1997; Lee and Baehrecke 2001; Lee et al. 2002, 2003). These cells also possess large numbers of autophagosomes and elevated levels of Atg RNAs (Lee and Baehrecke 2001; Lee et al. 2002, 2003; Li and White 2003; Denton et al. 2009). Surprisingly, midgut degradation is neither disrupted by expression of the pan-caspase inhibitor p35 nor by mutation of multiple caspases, indicating that apoptosis is dispensable for developmental midgut degradation (Denton et al. 2009). Interestingly, midgut destruction is blocked in animals with impaired Atg1, Atg2, or Atg18 function, directly implicating autophagy as a crucial process in steroid-induced degradation of midgut cells (Denton et al. 2009). Caspase deficiency does not enhance the Atg mutant midgut phenotypes, indicating that autophagic cell death in the midgut is caspase-independent (Denton et al. 2009).
In contrast to the Drosophila midgut, destruction of larval salivary glands requires both caspases and autophagy (Berry and Baehrecke 2007). Mutations in either Atg8 or Atg18 in addition to decreased function of a number of other Atg genes, all lead to the incomplete degradation of larval salivary glands. Similarly, Atg genes are required for cell death in the Drosophila amnioserosa and ovarian tissue (Hou et al. 2008; Mohseni et al. 2009; Nezis et al. 2009, 2010). It is important to note that the roles and relationship of autophagy and caspases in dying salivary gland, amnioserosa, and ovarian cells in flies is cell context-specific (discussed below). In addition, although larval salivary gland cell death requires both caspases and autophagy for completion of cell clearance, Atg1-induced autophagy in salivary glands is sufficient to induce premature cell death in a caspase-independent manner (Berry and Baehrecke 2007). Atg1 overexpression is also sufficient to cause cell death in the fat body and imaginal discs, but this death depends on caspase activity (Scott et al. 2007).
Studies in the nematode C. elegans also indicate that autophagy contributes to cell death (Kang et al. 2007). gbp-2 mutants show hyperactive muscarinic acetylcholine signaling in the pharyngeal muscle (You et al. 2006), are sensitive to starvation, and induce excess autophagy and cell death. This phenotype can be partially suppressed by either beclin-1 or Atg-7 RNAi indicating that autophagy contributes to cell death.
Autophagy is also observed in dying cells throughout mammalian development, including the regression of the corpus luteum, the involution of mammary and prostate gland and the regression of Mullerian duct structures during male genital development (reviewed in Clarke 1990). Studies of derived mammalian cell lines have shown that Atg genes are required for cell death that occurs in the absence of caspase activity (Shimizu et al. 2004; Yu et al. 2004), but no studies to date have shown that autophagy is required for the death of mammalian cells in vivo. However, studies of beclin1 mutant murine ES cells that form embroid bodies indicates that autophagy is required for lipid signaling that is required for clearance of dying cells (Qu et al. 2007).
AUTOPHAGY, CASPASES, AND CONTEXT SPECIFICITY FOR CELL DEATH
Autophagy promotes cell survival by catabolism of intracellular resources to maintain bioenergetics under nutrient limiting conditions. Furthermore, the elimination of damaged organelles and toxic protein aggregates by autophagy promotes cell survival. Therefore, loss of Atg gene function can promote cell death by apoptosis (Boya et al. 2005). In addition, autophagy can promote cell death, but this appears to occur in a cell type and context specific fashion (McPhee and Baehrecke 2009).
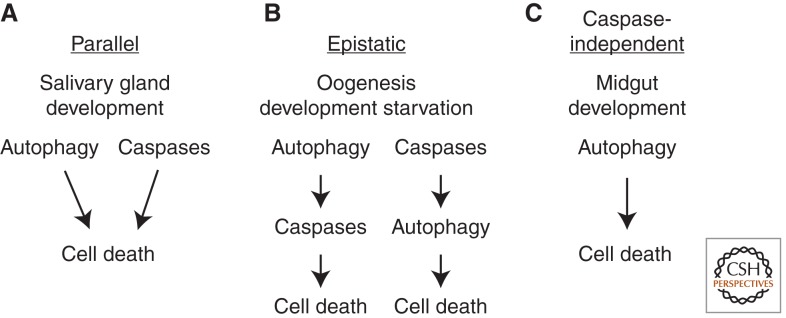
The cell context-specific function of autophagy in cell death has been best described in Drosophila in which autophagic cell death is known to occur in multiple cell types (Fig. 4). During larval salivary gland degradation, autophagy and caspases cooperate to efficiently clear dying cells. Therefore, impaired function of either autophagy or caspases results in partially degraded salivary gland cells, whereas decreased function of both of these processes results in intact salivary glands. These data indicate that autophagy and caspases function in parallel genetic pathways to degrade salivary glands. By contrast, caspases act upstream of autophagy to direct both starvation-induced ovarian cell death (Hou et al. 2008) and degradation of amnioserosa embryonic membrane (Mohseni et al. 2009). In addition, autophagy selectively degrades the caspase inhibitor dBruce to activate caspases and execute cell death in the Drosophila ovary (Nezis et al. 2009, 2010). As discussed above, autophagy plays a more prominent role in the death of fly midgut cells (Denton et al. 2009). Combined, these data indicate that multiple possible relationships exist between autophagy and caspases in dying fly cells, and it is important to determine if this is true in other organisms, including humans in which manipulation of autophagy could have therapeutic benefits.
Figure 4.
The relationship between autophagy and caspases is cell context specific during cell death. (A) During cell death of Drosophila larval salivary glands, autophagy and caspases function in parallel genetic pathways. (B) Autophagy degrades the inhibitor of apoptosis (IAP) protein Bruce enabling caspase activation during fly cell death in oogenesis. By contrast, starvation-induced autophagy leads to degeneration of egg chambers during oogenesis, and the caspase DCP-1 and IAP protein Bruce are required for autophagy to occur in this context. These studies indicate that autophagy and caspases function in an epistatic regulatory hierarchy. (C) Autophagy is essential for cell death during fly midgut cell death, while caspases do not appear to play a significant role in the elimination of this tissue.
Given the paucity of physiological in vivo models for autophagy and cell death in mammals, it is useful to consider what is known about this relationship in different types of cell lines that may reflect context-specificity. In mammalian cells, most reports of the involvement of autophagy in the execution of death are in cells that possess altered apoptotic pathways (Levine and Yuan 2005; Levine and Kroemer 2009). Treatment of MCF-7 mammary cancer cells that lack caspase-3 with 4-hydroxytamoxifen triggers cell death with autophagy, suggesting the possibility that autophagy can compensate for defects in apoptosis (Bursch et al. 1996). Lenardo and colleagues described a requirement for autophagy genes during cell death, and reported that U937 monocyte and L929 fibrosarcoma cells use Beclin1 and Atg7 for nonapoptotic cell death induced by caspase-8 inhibition (Yu et al. 2004). In another study, Tsujimoto and colleagues showed that Bax-/- and Bak-/- double knockout mouse embryonic fibroblasts undergo cell death accompanied by large scale autophagy, and this death was inhibited by knockdown of either Atg5 or Beclin1 (Shimizu et al. 2004). Ryan and colleagues showed that p53-induced cell death is mediated by a stress-induced regulator of autophagy termed DRAM (Crighton et al. 2006) further emphasizing the role of autophagy in cell death. Like many cell death regulators, DRAM levels are decreased in human cancers. In addition, a recent report shows that a human ovarian epithelial cell line that expresses oncogenic H-RasV12 undergoes caspase-independent autophagic cell death that relies on stress kinases, including MEK and ERK, Beclin-1 and Noxa (Elgendy et al. 2011). Interestingly, Debnath and colleagues identified a noncanonical Atg12- Atg3 complex that did not influence starvation-induced autophagy, but when disrupted resulted in increased mitochondrial mass and inhibition of cell death that is mediated by mitochondria (Radoshevich et al. 2010). Although the association of Beclin-1 with Bcl-2 provided one of the first molecular connections between autophagy and cell death (Liang et al. 1998), much remains to be learned about the relationship(s) between autophagy and cell death regulatory pathways. Clearly, a mechanistic understanding of the relationship between autophagy and cell death is critical to the design of rationale therapies.
CONCLUSIONS
Here we have described the regulation and function of autophagy in cell survival and cell death, two important processes involved in health and disease. Autophagy is often considered a cell survival process (Levine and Kroemer 2009), and it is clear that under nutrient restriction and cell stress, autophagy is augmented to protect the cell and maintain homeostasis. However, accumulating evidence indicates that autophagy can promote cell death, and how autophagy influences cell death appears to depend on the type and context of the cell.
The connection of autophagy to the control of metabolism, stress, survival, and death suggests that organism-specific utilization of this catabolic process is likely to occur. Autophagy is widely used to provide an internal source of nutrients under starvation conditions in organisms as diverse as yeast and humans, but this process may be augmented under organism-specific situations. In developing mice, for example, essential embryonic nutrients are supplied by the mother through the placental interface. At birth, when this supply is terminated, neonates face severe acute starvation, and autophagy is induced until mice are fed (Kuma et al. 2004). Similarly, C. elegans enter an alternative dauer larval form during unfavorable environmental conditions. Autophagy is elevated in dauer larvae, and decreased function of beclin-1, unc-51 (Atg1 in worms), Atg7, lgg-1(Atg8a in worms) and Atg18 inhibits the completion of dauer development (Melendez et al. 2003). Autophagy may also function to promote homeostasis by maintaining the health of stem cells. Indeed, a recent study showed that Atg7 plays a crucial role in adult mouse hematopoietic stem cell maintenance and survival by regulating mitochondrial quantity and quality (Mortensen et al. 2011). These studies show that autophagy is used in organism-specific biological programs to promote homeostasis. Although these programs appear to use conserved autophagy programs, it is possible that the stimuli that trigger autophagy are specialized. As mentioned above, steroids trigger autophagy in a stage and tissue-specific manner during Drosophila development. Furthermore, at least one factor Draper has been identified that is specifically required for autophagy in dying cells, but not during starvation-induced autophagy in Drosophila (McPhee et al. 2010). This study highlights the potential different roles and regulatory signaling mechanisms in different cell types. In addition, it is important to consider that differences in cell cargoes may also influence cell fates, including survival and death, as the depletion of survival factors is another way to kill a cell (Yu et al. 2006; Nezis et al. 2010).
Numerous reports suggest a role for autophagy in human diseases, including cancer, neurodegeneration and other disorders. These are age-associated disorders, and aging is associated with the accumulation of by-products of metabolism, cell damage and the inefficient function of the machinery that degrades damaged cell material. In this context, Beclin-1, Atg7, Atg8, and Atg12 have been shown to be involved in lifespan of worms and flies (Hars et al. 2007; Juhász et al. 2007; Simonsen et al. 2007), which is consistent with studies in mammals (reviewed in Cuervo 2008). Thus, either inhibitors or inducers of autophagy might play a prominent role as therapeutics in combating diseases associated with autophagy (Fleming et al. 2011). Promise exists to support autophagy as a therapeutic target, but caution is prudent when designing drugs that influence a fundamental catabolic process that appears to be involved in the health of all cells, particularly because of its cell context-specific functions in survival and death.
The multiple functions of autophagy are supported by complexity of disease phenotypes. For example, autophagy was first recognized as a potential tumor suppressor mechanism based on mono-allelic loss of Beclin1 in human tumors (Liang et al. 1999), and this is consistent in murine models (Qu et al. 2003; Yue et al. 2003). However, the mechanistic role of autophagy in tumor suppression is not completely clear. Although loss of autophagy can promote aneuploidy and the development of the transformed phenotype in cell lines (Mathew et al. 2009), it has also been implicated in tumor cell survival (Degenhardt et al. 2006). Significantly, loss of Atg5 leads to benign adenomas in livers, but this phenotype is not observed in other tissues (Takamura et al. 2011). In addition, the failure of these benign adenomas to cause cancer suggests that autophagy is required for tumor progression. These results are consistent with studies showing that both pancreatic and mammary tumors require autophagy for maintenance of tumorigenesis (Wei et al. 2011; Yang et al. 2011), but differ from models in which autophagy activation facilitates tumor cell killing by multiple agents (Martin et al. 2009; Hamed et al. 2010). Thus, the diametrically opposite roles of autophagy in tumor progression warrants further consideration for the development of rationale cancer therapies (Mah and Ryan 2012).
The complex roles of autophagy in survival and death should also be considered when designing therapies for other disorders. Autophagy promotes the clearance of protein aggregates (Hara et al. 2006; Komatsu et al. 2006) and has an important neuroprotective role in several neurodegenerative disease models, including Alzheimer's and Huntington's (Menzies et al. 2011). In addition, recent evidence suggests that mitochondrial autophagy plays an important role in the pathogenesis of Parkinson's disease (Nixon and Yang 2012). Although the promotion of autophagy in neurodegenerative disease models results in healthier individuals, it is also possible that too much autophagy could have deleterious effects, including problems with bioenergetics or even worse killing the cells while trying to protect them. Future work should not only consider how autophagy may promote cell survival or death, but what the impact of modulating autophagy may have on the health of the test subject and patient.
ACKNOWLEDGMENTS
We apologize to the many authors who were not cited because of space limitations. Research on this subject is supported by NIH grants GM079431 and CA159314 to EHB.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ashford TP, Porter KR 1962. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 12: 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke EH 2005. Autophagy: Dual roles in life and death? Nat Rev Mol Cell Biol 6: 505–510 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW 2010. Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. 2005. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS 1996. Active cell death induced by the anti-estrogens tamoxifen and ICI164384 in human mammary carcinoma cells (MCF-7) in culture: The role of autophagy. Carcinogenesis 17: 1595–1607 [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM 2005. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 280: 33076–33082 [DOI] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SLJ 1957. Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J Biophys Biochem Cytol 3: 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PGH 1990. Developmental cell death: Morphological diversity and multiple mechanisms. Anat Embryol 181: 195–213 [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM 2006. DRAM, a p53–induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134 [DOI] [PubMed] [Google Scholar]
- Cuervo AM 2008. Autophagy and aging: Keeping that old broom working. Trends Genet 24: 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Wattiaux R 1966. Functions of lysosomes. Annu Rev Physiol 28: 435–492 [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S 2009. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 19: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 240: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WAJ 1990. Studies on the mechanisms of autophagy: Formation of the autophagic vacuole. J Cell Biol 110: 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusetti NJ, Jiang Y, Vaccaro MI, Tomasini R, Azizi Samir A, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn JC, et al. 2002. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun 290: 641–649 [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ 2011. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell 42: 23–35 [DOI] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ 2001. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J 15: 2286–2287 [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. 2010. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 38: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. 2007. Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Fleming A, Noda T, Yoshimori T, Rubinsztein DC 2011. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Gu Z, Flemington C, Chittenden T, Zambetti GP 2000. ei24, a p53 response gene involved in growth suppression and apoptosis. Mol Cell Biol 20: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141: 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed HA, Yacoub A, Park MA, Eulitt P, Sarkar D, Dimitrie IP, Chen CS, Grant S, Curiel DT, Fisher PB, et al. 2010. OSU-03012 enhances Ad.7-induced GBM cell killing via ER stress and autophagy and by decreasing expression of mitochondrial protective proteins. Cancer Biol Ther 9: 526–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Bio 131: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ 1996. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein. J Biol Chem 271: 17621–17624 [DOI] [PubMed] [Google Scholar]
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF 2007. Autophagy regulates ageing in C. elegans. Autophagy 3: 93–95 [DOI] [PubMed] [Google Scholar]
- Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 182: 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al. 2012. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci 109: 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683 [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G, Erdi B, Sass M, Neufeld TP 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21: 3061–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 181: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, You YJ, Avery L 2007. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev 21: 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ 2009. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ 2008. Autophagy revisited: A conversation with Christian de Duve. Autophagy 4: 740–743 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WAJ, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. 2003. A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Kosta A, Roisin-Bouffay C, Luciani MF, Otto GP, Kessin RH, Golstein P 2004. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem 279: 48404–48409 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y 2002. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Lee C-Y, Baehrecke EH 2001. Steroid regulation of autophagic programmed cell death during development. Development 128: 1443–1455 [DOI] [PubMed] [Google Scholar]
- Lee C-Y, Cooksey BAK, Baehrecke EH 2002. Steroid regulation of midgut cell death during Drosophila development. Dev Biol 250: 101–111 [DOI] [PubMed] [Google Scholar]
- Lee C-Y, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol 13: 350–357 [DOI] [PubMed] [Google Scholar]
- Lee JW, Park S, Takahashi Y, Wang HG 2010. The association of AMPK with ULK1 regulates autophagy. PLoS ONE 5: e15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2009. Autophagy in aging, disease and death: The true identity of a cell death impostor. Cell Death Differ 16: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Yuan J 2005. Autophagy in cell death: An innocent convict? J Clin Invest 115: 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TR, White KP 2003. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell 5: 59–72 [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 72: 8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. 2010. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 18: 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB 2005. Autophagy in metazoans: Cell survival in the land of plenty. Nat Rev Mol Cell Biol 6: 439–448 [DOI] [PubMed] [Google Scholar]
- *.Mah LY, Ryan KM 2012. Autophagy and cancer. Cold Spring Harb Perspect Biol 4: a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farré JC, Subramani S 2010. Molecular mechanism and physiological role of pexophagy. FEBS Lett 584: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Mitchell C, Rahmani M, Nephew KP, Grant S, Dent P 2009. Inhibition of MCL-1 enhances lapatinib toxicity and overcomes lapatinib resistance via BAK-dependent autophagy. Cancer Biol Ther 8: 20842096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62 Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192: 245–250 [DOI] [PubMed] [Google Scholar]
- McPhee CK, Baehrecke EH 2009. Autophagy in Drosophila melanogaster. Biochim Biophys Acta 1793: 1452–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee CK, Logan MA, Freeman MR, Baehrecke EH 2010. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 465: 1093–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391 [DOI] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ 2006. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem 281: 34870–34879 [DOI] [PubMed] [Google Scholar]
- Menzies FM, Moreau K, Rubinsztein DC 2011. Protein misfolding disorders and macroautophagy. Curr Opin Cell Biol 23: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M 2011. Autophagy: Renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH 2009. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy 5: 329–338 [DOI] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. 2011. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med 208: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. 2011. SNARE proteins are required for macroautophagy. Cell 146: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T, Papassideri IS, Stravopodis DJ, Margaritis LH, Stenmark H, et al. 2009. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 5: 298–302 [DOI] [PubMed] [Google Scholar]
- Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H 2010. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 190: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Nixon RA, Yang D.-S 2012. Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a008839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci 102: 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff AB, Essner E 1962. Cytolysomes and mitochondrial degeneration. J Cell Biol 15: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y 2001. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 278: 17636–17645 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B 2007. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128: 931–946 [DOI] [PubMed] [Google Scholar]
- Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J 2010. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 12: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Baehrecke EH 2010. Distinct death mechanisms in Drosophila development. Curr Opin Cell Biol 22: 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samari HR, Seglen PO 1998. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J Biol Chem 273: 23758–23763 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweichel J-U, Merker H-J 1973. The morphology of various types of cell death in prenatal tissues. Teratology 7: 253–266 [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhász G, Neufeld TP 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD 2007. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4: 176–184 [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ 2009. Autophagy regulates lipid metabolism. Nature 458: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E 1999. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349: 275–280 [DOI] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141: 1042–1055 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 28: 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wilson WA, Fujino MA, Roach PJ 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol 21: 5742–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25: 1510–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z 2010. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125–156 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You YJ, Kim J, Cobb M, Avery L 2006. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab 3: 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304: 1500–1502 [DOI] [PubMed] [Google Scholar]
- Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo MJ 2006. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci 103: 4952–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. 2010. Autophagy termination and lysosome reformation regulated by mTOR. Nature 465: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]