Abstract
Complex social communication is expected to evolve whenever animals engage in many and varied social interactions; that is, sociality should promote communicative complexity. Yet, informal comparisons among phylogenetically independent taxonomic groups seem to cast doubt on the putative role of social factors in the evolution of complex communication. Here, we provide a formal test of the sociality hypothesis alongside alternative explanations for the evolution of communicative complexity. We compiled data documenting variations in signal complexity among closely related species for several case study groups—ants, frogs, lizards and birds—and used new phylogenetic methods to investigate the factors underlying communication evolution. Social factors were only implicated in the evolution of complex visual signals in lizards. Ecology, and to some degree allometry, were most likely explanations for complexity in the vocal signals of frogs (ecology) and birds (ecology and allometry). There was some evidence for adaptive evolution in the pheromone complexity of ants, although no compelling selection pressure was identified. For most taxa, phylogenetic null models were consistently ranked above adaptive models and, for some taxa, signal complexity seems to have accumulated in species via incremental or random changes over long periods of evolutionary time. Becoming social presumably leads to the origin of social communication in animals, but its subsequent influence on the trajectory of signal evolution has been neither clear-cut nor general among taxonomic groups.
Keywords: animal communication, phylogenetic comparative methods, adaptation, sexual selection, natural selection
1. Introduction
Complex communication is classically linked to the evolution of complex animal societies [1]: as the number and context of social interactions increase, communication mediating those interactions tends to become increasingly elaborate. Indeed, it is difficult to think of any highly social animal that does not possess a complex system of communication. Humans are an obvious example, as are many other primates and long-lived mammals. Chimpanzees form complex hierarchies and alliances among troop members and rely on an elaborate array of vocal and visual signals to mediate those relationships [2–5]. Elephants have similarly complex social interactions and use an extensive repertoire of social signals as an apparent consequence [6–9]. But is it correct to say that sociality is a necessary prerequisite for the evolution of communicative complexity? Are less social species really predisposed to basic forms of communication more than socially complex species?
These questions lie at the very foundation of how we believe communication evolved in animals. If communicative complexity and sociality are tightly coupled, then both the origin and direction of communication evolution has essentially been a by-product of factors driving the evolution of sociality more generally. Alternatively, communication might have originated to mediate social interactions among conspecifics, but its ultimate elaboration has been driven by other factors independent of changes in sociality. Freeberg et al. [1] have discussed in detail the possible non-social pressures that might produce communicative complexity. These include environmental factors that influence signal fidelity, the need for reliable species recognition and neutral or non-adaptive evolutionary processes. Some of these factors have empirical support (species recognition; [10,11]), whereas others have very little (neutral processes; reviewed by Freeberg et al. [1]). The challenge is to test the sociality hypothesis alongside its alternatives and in a way that allows the weight of evidence for each hypothesis to be directly compared.
One can try to make broad taxonomic comparisons to argue that group-living primates, for example, have significantly more elaborate systems of communication relative to claw-waving territorial crabs because primates form complex societies, whereas crabs do not (by comparison). Yet there are many other non-primate groups that do exhibit extraordinary complexity in their communication, and some researchers have even gone as far as to say have levels of signal complexity comparable to primates [12,13]. Consider the exquisite song and courtship dances of many birds [14], the elaborate headbobbing and colourful dewlap displays of anole lizards [11,15], the rippling colour and movement displays of cuttlefish [16,17] or the elaborate foot thumping signals of ornate wolf spiders [18,19]. These are all examples of complex communication—complex in repertoire, the number and type of components making up a signal and, in some cases, the number of sensory modalities used for communication (e.g. signals that are both auditory and visual). Yet, these animals do not exhibit the same level of sociality seen in primate societies.
Such examples seem to weaken the putative link between communicative complexity and sociality. However, broad comparisons among disparate groups like these (primates versus crabs or birds) actually provide little insight into the evolution of animal communication. The vast number of attributes that vary among species at these broad phylogenetic scales make it virtually impossible to determine what factors might or might not account for taxonomic differences in communication. More informative are comparisons among closely related species within broad taxonomic groups. In almost any group that depends on social communication, closely related species differ, to a lesser or greater degree, in their social behaviour and complexity of communication. Furthermore, at this finer phylogenetic scale, the specific selection pressures that direct the evolution of communicative complexity become more apparent (reviewed by Ord [20]). Rather than conducting a literature review of the evidence for and against the sociality hypothesis, we chose to test the hypothesis directly alongside other potential causal factors, using empirical data collected for closely related species.
To this end, we developed six datasets or ‘case studies’ to represent a variety of taxonomic groups (frogs, birds, lizards and ants) and signal classes (acoustic, visual and chemical). The data for each case study documented variations among closely related species in signal complexity, social behaviour and other factors predicted to influence the evolution of communicative complexity (see §1a–d). We then used new phylogenetic comparative approaches and assembled new phylogenies to evaluate alternative evolutionary models of how communicative complexity might have evolved in each case study. The models corresponded to one of the four hypotheses and were not mutually exclusive. We assembled findings across the case studies to determine the generality of each hypothesis in describing the evolution of communicative complexity over diverse taxa and signal classes. A secondary goal of our study was to provide communication biologists, who might not be familiar with recent advances in phylogenetic techniques, with a heuristic example of how multiple hypotheses can be considered jointly, using methods that also inform on the probable mode of evolution.
(a). Social drivers of signal complexity
When the frequency or importance of social interactions increases, signals used to mediate those interactions should become more complex for a range of reasons: to convey information more reliably, to manipulate social partners more effectively or to provide relevant social cues in different contexts [1]. For instance, as sexual selection intensifies, either because mates become choosy in what they find attractive or as competition for mates and territories increases, signal repertoires are expanded or the design of signals are elaborated to help the signaller outcompete courtship and territorial rivals [11,21–25]. For species living in groups or otherwise interacting with a range of different social partners (e.g. mates, rivals, juveniles, adults, dominates and subordinates), different signals or cues are often required for different contexts, leading to an increase in repertoire size or the complexity of existing signals to convey multiple messages [26]. The sociality hypothesis predicts variation among species in the design or repertoire of signals whenever species differ in the frequency or context of social interactions.
(b). Ecological influences on signal complexity
The range of factors that make up the ecology of species is diverse and there are a number of possible ways in which ecology can direct the evolution of communicative complexity. The influence of the environment on the transmission of animal signals is well documented [27–30]. Background acoustic or visual noise masks calls [31,32] and displays [33,34] and obstructions in the environment deflect and scatter sound waves [28] and obscure visual signals [35,36], as does the reduced visibility imposed by poor light [37–39]. The strategies animals adopt to enhance signal fidelity in noisy, cluttered or dim habitats are varied. Background noise can limit the range of frequencies heard by birds [40] or the types of movements that can be seen by lizards [34], and this restricts the range of signal designs that can be readily detected by receivers. Namely, difficult environmental conditions limit signals to those that are simple in design. There are, however, some instances where noisy environments can facilitate signal complexity by promoting the evolution of alert components. These are new components added to signals to attract the attention of receivers, before the more information rich portion of the signal is delivered [39,41]. Here, poor signal environments promote the evolution of more complex signals (i.e. increases in component number through the addition of alerts and other components to enhance signal detection).
Microhabitat or the site within the environment from which animals communicate with one another can increase or decrease the severity of environmental conditions that affect signal efficiency [42]. Acoustically communicating species living near streams are susceptible to noise generated by flowing water in what otherwise might be a quiet macrohabitat (e.g. a sheltered temperate forest). In the case of some frogs, species near streams facilitate call detection by producing calls at ultrasonic frequencies above the broadband frequency of running water [43]. Calls given near the ground are also more susceptible to degradation through muffling and deflection [28]. Likewise, the visibility of displays is enhanced from perches above undergrowth and other low-lying visual obstructions [44]. On the one hand, the constraints acting on signal efficiency in microhabitats near noise sources or physical obstructions limit the range of signal designs that can be detected by receivers. On the other hand, animals can minimize masking by adding alert or amplifier components to signals and, in the process, increase the complexity of their signals [39,41]. Whether environmental variables constrain or promote signal complexity is unclear (but see [35]). Yet it is reasonable to expect environmental conditions to play some part in directing the evolution of communicative complexity.
Finally, when animals frequently encounter heterospecific congeners in the environment and are not in direct competition with those congeners for resources (e.g. mates), the need for accurate species recognition becomes important (reviewed by Ord et al. [45]). The design of social signals often provides the best cues for ascertaining species identity. The number of sympatric species an animal encounters should prompt increases in signal complexity to facilitate recognition [10,11]. That is, signal elaboration results in a unique, species-typical signal that can be easily distinguished from the signals of sympatric congeners.
(c). The allometry of signal complexity
Body size influences numerous features of an animal. Of special relevance to communication is the allometry of physical structures and the physiological mechanisms that govern signal production. For example, the length of the vocal tract is heavily dependent on body size (larger animals have longer vocal tracts [46]). The vocal tract in turn determines the types of sounds that can be produced by an animal [47,48]. The allometry of the vocal tract should therefore lead to disparity in vocal signals among species that differ in size. In addition, size-specific metabolic rate seems to explain a large portion of the variance among species in the rate, frequency and duration of acoustic signals in groups as diverse as insects and mammals: large species typically produce low-frequency calls of long duration and at low rates [49]. A similar physiological mechanism has been implicated in the evolution of movement-based displays in lizards [50]. Larger lizards have higher energetic costs associated with movement compared with smaller lizards, and this has been suggested to constrain the number, duration or type of movements that large lizards can include in displays [50]. However, in the case of static visual signals, possessing a large body might instead provide more surface area for the expression of big or elaborate ornaments.
Taken together, the allometry of communicative complexity will depend on the modality and type of signal characteristic examined. Larger bodied acoustically communicating species will have vocalizations of longer duration than smaller bodied species [49]. Whether size-specific metabolic rate, or body size more generally, influences other indices of vocal complexity such as repertoire size or note number is unknown. In visually communicating lizards, size-specific energetic costs should limit the evolution of movement-based display complexity [50], but larger body sizes should allow the evolution of large or more numerous ornaments because of increased surface area.
(d). Non-adaptive signal complexity
Divergence in song complexity can occur ‘passively’ among populations in wide-ranging species through cultural or genetic drift (e.g. birds: [51,52]; mammals: [53]). This leads to the unusual hypothesis that stochastic processes can generate signal complexity in the absence of selection (see also [54]). Genetic drift and neutral mutation (mutations that are neither deleterious nor beneficial) may incidentally increase signal complexity over evolutionary time or following bouts of rapid evolution (e.g. during speciation). If true, variations in communicative complexity will tend to track phylogeny (closely related species will tend to share similar levels of signal complexity, whereas distantly related species will not) and, in particular, match a pattern of stochastic evolution. Recent developments in phylogenetic comparative analyses allow the joint estimation of both phylogenetic inertia and stochasticity in evolutionary diversification [55]. This neutrality hypothesis will be novel to most communication biologists, but it is an important null model missing from most studies of signal adaptation.
2. Material and methods
We identified recent studies in which the design or repertoire of communication had been surveyed for a large number of closely related species. Our criterion for selecting case studies was dependent on several factors. First, we needed to be fairly certain that we could obtain adequate social, ecological or morphological data for the species in question. Some of this information was reported in the original sources, or the authors of those sources were willing to share unpublished data with us when contacted directly (see Acknowledgements). In other cases, we used electronic databases and other published sources to supplement datasets. Second, to construct phylogenies for each case study, there had to be adequate and comprehensive genetic markers for species in GenBank. Third, we wished to cover several diverse taxonomic groups and obtain representatives of both the most commonly studied and least-studied forms of communication (in decreasing order of research attention: acoustic signals, static visual ornaments, movement-based displays and chemical signals). We excluded primates and other mammals because these systems were either well represented in earlier tests of the sociality hypothesis (e.g. [56–58]; reviewed in Freeberg et al. [1]) or were the focus of other studies included in this theme issue [59–61]. Finally, in some cases, we selected two case studies for the same taxonomic group (specifically birds and lizards), one that represented phylogenetically diverse species from multiple genera and families, while the second consisted of closely related species from the same genus or phylogenetically adjacent genera. We refer to these case studies as ‘distantly related’ and ‘closely related’ examples, respectively. Our motivation here was to assess the sensitivity of our estimates of phylogenetic inertia and stochasticity to taxon sampling.
Details on the data and analyses used are described in the following sections. All data and GenBank accession numbers for DNA sequences used to assemble phylogenies have been deposited in files assigned to this article in the public electronic database Dryad Digital Repository (see Acknowledgements).
(a). Communication data and social indicators
We compiled data on signal and social characteristics that we believed were consistent with current definitions of complexity [1]. Indices of signal complexity were: call amplitude modulation (frogs); call, song or display duration (frogs, birds and lizards); song or syllable repertoire size (birds); number of ornaments (lizards); number of separate components making up signals (lizards and ants; this includes colour dichromatism in lizards, measured as the number of colourful body patches exhibited by males but not seen in females [62]). Indices of sociality were: sexual size dimorphism (frogs and lizards; in these taxa, size dimorphism is believed to reflect the intensity of competition among males for mates and territories [24]); levels of extra-pair paternity (birds); mating system (in birds this was coded as ‘monogamy’, ‘irregular polygyny’ and ‘regular polygyny’; in ants, ‘no polygyny or polyandry’ versus ‘polygyny or polyandry present’); and colony size (ants). All data on signal complexity were compiled directly from published studies [25,62–67] except for frogs (see below). Indices of sociality were obtained from the same sources used for communication data (frogs [68]; lizards, distantly related [25]; birds [66]), unpublished data from the authors of these studies (ants—E. van Wilgenburg, M. R. E. Symonds & M. A. Elgar 2011, unpublished data) or compiled separately from other literature (lizards, closely related [69]).
To obtain data on the signals of frogs, we used oscillograms of species-typical frog calls to estimate call duration and amplitude modulation from a comprehensive field guide on the anuran fauna of the Kaiteur National Park in Guyana [68]. Oscillograms were digitally scanned and ImageJ v. 1.42q (W. Rasband 1997–2009, NIH) used to measure the duration of calls in seconds from the start of the first note to the end of the last note. Call amplitude modulation was estimated as the coefficient of variation (CV) of the peak sound pressure computed across pulses making up a call. That is, a call with many pulses varying in peak sound pressure had a higher estimated CV than a call with pulses that peaked at consistent sound pressure levels.
(b). Ecological and morphological data
Most ecology data were obtained from sources reporting communication data and included: macrohabitat for the ‘distantly related’ lizards [25]; microhabitat for both lizard case studies [63,64] as well as the frogs [68]; environmental noise for the frogs [68]; whether species were migratory for the ‘closely related’ bird case study [65]; species geographical range for the ‘distantly related’ lizard case study [62]; the number of sympatric species for frogs [68], and two climatic variables for ants [67]. Macrohabitat was generally coded as ‘open’ (e.g. grasslands, deserts and plains) or ‘closed’ habitats (e.g. forests and woodlands). This is a biologically relevant categorization of habitat for communication because closed environments generally reduce the range of detection for acoustic and visual signals compared with open environments (see §1). Microhabitat in frogs was the average height above ground of call sites for each species, while in lizards it was whether species were arboreal or terrestrial. In frogs, species that were reported preferring environments near streams were assumed to experience environmental noise from running water and categorized as occupying ‘high noise’ environments, whereas all other species were categorized as occupying ‘low noise’ environments. Whether species were migratory or occupying large geographic ranges was used as an index of the likely habitat heterogeneity experienced by species, as well as the likelihood of interacting with heterospecifics. Habitat heterogeneity was expected to influence the evolution of communicative complexity because heterogeneity will either truncate the set of signals that are detectable across different habitats or because heterogeneity facilitates the evolution of new signal components or larger repertoire sizes (see §1). Widely distributed species can also be expected to interact with more sympatric species compared to localized species, and sympatry is in turn predicted to promote the evolution of signal complexity (see §1). In frogs, the number of sympatric species was estimated as the number of co-occurring frog species reported at sites where a species was stated to occur. Climatic variables expected to influence the composition of cuticular hydrocarbons in ants are rainfall and temperature [67], which may limit the range of chemical components included in an olfactory signal.
We used a GIS approach to obtain information on the ecology of species for two case studies for which data were not reported in original sources. For one lizard case study (communication data from Martins [63]), we used GIS distribution data to compute the geographic range of species, the percentage range overlap with congeneric heterospecifics (species of the same genus; this index is hereafter referred to as ‘sympatry: overlap’), the number of sympatric species (hereafter ‘sympatry: number’) and the level of habitat heterogeneity likely experienced by species. Specifically, geographical distribution data for 79 species of Sceloporus (88% of described species within the genus) were obtained from the IUCN red list database (http://www.iucnredlist.org/technical-documents/spatial-data) and mapped using a cylindrical equal area projection. The range for the species of interest was measured in square kilometres and estimated as the area enclosed by the polygon corresponding to each species distribution. We then calculated the percentage area of a species range that was overlapped by at least one Sceloporus congener. We also estimated the number of sympatric species by tallying the number of overlapping species. Habitat heterogeneity was computed by intersecting the ecoregion GIS layer described in Olson et al. ([70]; available electronically at http://www.worldwildlife.org/science/data/item1875.html) with the projected distribution of species. Initially, the percentage overlap of ecoregions for a given species was grouped into four broad categories—forest, shrubland, grassland and desert—but these were later merged into ‘forest’ and ‘non-forest’ environments (forest versus all others) because this dichotomous categorization was more biologically appropriate for testing our hypotheses (see opening paragraph in this section).
The same methods were used to obtain ecological data for one bird case study (communication data from Soma & Garamszegi [66]). In this case, species distributions were obtained from the BirdLife International and NatureServe database ([71]; http://www.birdlife.org/datazone/info/spcdownload3, accessed on August 19, 2011). Our projected distributions were based on the known ‘breeding range’ if species were migratory (given that song charactersitics were used in mate choice and territory defence [66], activities specific to the breeding season) or the ‘resident range’ if species were non-migratory. Ecoregion data and analyses were the same as those of the lizard example described earlier. However, we were unable to obtain data on sympatric species for this bird case study, so sympatry was not evaluated. All GIS analyses were performed using ArcGIS v. 9.3 (ESRI, Redlands CA).
Data on body size were largely obtained from studies reporting communication data (the only exception being the Soma & Garamszegi [66] case study; see above). Measures of size included snout-to-vent length for frogs and lizards, and tarsus length in birds (tarsus length is a common metric of size in birds—e.g. [72,73]—and is a useful measure because it reflects both the overall size and mass of the bird).
(c). Phylogenies
We developed phylogenies for each case study using the two most comprehensive mitochondrial DNA markers deposited in GenBank [74]. The longest sequences available for a given species were selected and aligned using the program Mafft v. 6 ([75]; http://www.ebi.ac.uk/Tools/msa/mafft/) with the following parameter settings: gap penalty = 1.53; gap extension = 0.123; tree rebuilding number = 100; maximum number of iterations = 100; and fast Fourier transforms (FFTS) = local pair. When sequence data for a given species were not available in GenBank, we used surrogate sequences from two species of the same genus (when possible; in rare instances only one species of the same genus was represented in GenBank). Surrogate sequences were required only for five of the 199 species included in our study.
For every case study, we generated two ultrametric phylogenies using BEAST v. 1.6.1 [76]. We used two trees rather than a single phylogeny for each case study to incorporate alternative phylogenetic hypotheses into our analyses (results from comparative analyses are partly dependent on the phylogeny used). The first tree was based on species relationships informed exclusively by the two mitochondrial DNA genes downloaded from GenBank. The second tree used additional information from the most recently published phylogenies for a given group to ‘constrain’ the topology of the tree such that only branch lengths were estimated in our phylogeny using the mitochondrial DNA genes from GenBank. This lead to an overall topology in the second tree that was congruent with existing phylogenies (frogs: [77]; birds: [78–83]; lizards: [84–88]; ants: [67]). We refer to these trees as ‘unconstrained’ and ‘constrained’ phylogenies, respectively.
Topology, node supports (in the unconstrained trees) and branch lengths (in both the unconstrained and constrained trees) were inferred using the Bayesian algorithms implemented in BEAST. A Yule branching process with a uniform prior was used and an uncorrelated branch rate variation was modelled using a lognormal distribution. For both genes, the model of evolution was set to GTRGAMMAI. The mean global substitution rate was set to unity and produced ultrametric trees with branch lengths expressed in units of substitutions per site. The analysis consisted of two to four independent Markov Chain Monte Carlo (MCMC) chains that ran for 20 000 000 generations with parameters and trees sampled every 3000 generations. Independent runs in all cases converged on very similar posterior estimates and were combined using LogCombiner v. 1.5.4 (included in the package BEAST). In all runs, the first 10 per cent of generations were considered to belong to the burn-in phase of the analysis and were excluded. The program Tracer v. 1.2 [89] was then used to confirm convergence and good mixing of the combined MCMC chains. Finally, summary trees were obtained with mean node heights computed using TreeAnnotator v. 1.5.4 (in the package BEAST), with a posterior probability limit set to 0.5.
(d). Analysis
We used model fitting within a phylogenetic framework to assess the extent to which different predictor variables explained variation among species in communicative complexity. We used the second-order Akaike's Information Criterion (AICc) to determine the level of support for each model. AICc is a modification of AIC that corrects for sample size; as sample size increases, AICc values converge on those of AIC [90]. As applied here, an AICc value reflects the likelihood that a given model fits the observed variation in signal complexity among species, given the phylogenetic relationships among those species. The model with the lowest AICc value is the model that best fits the data, although any model within two AICc units of this lowest value is by convention considered to fit the data just as well [90]. We also computed model weights, AICw, to provide a more intuitive indication of the support of a given model relative to all models considered. See Burnham & Anderson [90] for details on the calculation of AICc and AICw.
Model fitting and subsequent selection using information and likelihood theory is becoming increasingly common in evolutionary ecology studies (see [91] for review). This is because it offers an enticing alternative approach to traditional p-value-driven statistical analyses, with several statistical advantages that are particularly amenable for biological study. First, there is often a range of biologically plausible variables that might account for the observed patterns we see in the natural world. These variables might be alternative indices associated with a specific hypothesis or correspond to different hypotheses in their own right. Model fitting incorporates this notion of multiple potential causal factors as an explicit part of its analytic philosophy by allowing simultaneous evaluation of a number of alternative models. Second, such multiple comparisons are a menace for the interpretation of p-values because of the increased chance of falsely concluding a ‘significant’ relationship when many statistical comparisons are performed (i.e. type II error rates increase with the number of statistical analyses performed on a dataset). While there are various corrections that can be applied to p-value thresholds for judging significance (Bonferroni, false discovery rate), the problem is circumvented entirely using model fitting methods such as AIC.
This ability to consider a range of different models of how evolution might have occurred in a group is especially useful for phylogenetic comparative analyses in which there are typically several potential causal factors. In our study, we had a number of predictor variables associated with several hypotheses. These hypotheses were not mutually exclusive. While the model fitting framework allows all possible multivariate combinations of variables to be considered, we choose to focus on a relatively simple set of models to facilitate interpretation and avoid generating large unwieldy AICc tables. We therefore relied on model weights to identify which hypothesis, or combination of hypotheses, most likely accounted for the evolution of signal complexity in a given case study.
Models were fitted individually to the communication data using the phylogenetic comparative software SLOUCH v. 1.1 [55] run in R v. 2.8.1 (R Development Core Team). Thomas Hansen provides an introduction to his program and its analyses in accessible non-technical language in the accompanying user manual. A more detailed description of his approach is given in Hansen et al. [55] (see also [92]). We elaborate here on some of the key aspects of the program that are relevant for the interpretation our results.
SLOUCH has several features that make it especially attractive for comparative study among the daunting array of methods currently available. In particular, rather than assume a particular mode of evolution at the outset (a critical flaw of the popular independent contrasts method ([93]; see discussion in Ord & Martins [94]), SLOUCH uses likelihood to estimate the level of phylogenetic inertia and stochasticity in the evolution process based on the distribution of species data across the tips of the phylogeny and the nature of the phylogeny itself (e.g. its topology, length of branches). In the case of phylogenetic inertia, SLOUCH computes the phylogenetic half-life, t1/2, of the phenotype (here signal complexity), which is the time the phenotype would likely take to evolve halfway towards an adaptive optimum. The value of t1/2 is a direct function of phylogenetic inertia: strong phylogenetic inertia is reflected in large values of t1/2 and is consistent with incremental phenotypic change over long periods of evolutionary time, whereas weak phylogenetic inertia is reflected in small values of t1/2 and bursts of phenotypic change over short periods of evolutionary time. Physiological or morphological constraints, genetic correlations or low mutation rate are some of the factors that might contribute to phylogenetic inertia and large values of t1/2.
SLOUCH parametrizes t1/2 from zero to infinity, a range that includes a rapid and directed phenotypic change in response to selection, to gradual phenotypic change occurring via a process of Brownian motion (which may or may not be directed by selection). In the instance of adaptive evolution, the phenotype might track a continuously shifting selection regime or be maintained at an optimum phenotype through stabilizing selection. This provides considerable versatility because it allows for the possibility of non-adaptive evolutionary change, adaptive evolutionary change through directional selection and adaptive evolutionary change through stabilizing selection.
In the context of signal evolution, values of t1/2 approaching infinity imply that signals have evolved via an incremental change culminated over long periods of evolutionary time. In this scenario, species tend to share similar levels of signal complexity as a function of phylogenetic relatedness. Adaptive evolution may have occurred, but the process has been slowed by strong phylogenetic inertia. Intermediate values of t1/2 imply that signal evolution has tended to proceed towards an optimal phenotype via Brownian motion and, if reached, stabilizing selection has kept signal designs at or near this optimum. Values of t1/2 approaching zero suggest that signal designs among species are effectively independent and have little relationship to phylogeny. In this situation, animal signals have been free to vary adaptively and retain no signature of evolutionary history.
There are other phylogenetic comparative methods that use likelihood to estimate parameters reflecting the extent phenotypic evolution has been influenced by factors associated with phylogeny. For example, Martin & Hansen's [95] phylogenetic generalized least squares model (PGLS) estimates α, or the rate of adaptation, which can be used to compute t1/2 [55]. Other methods compute parameters such as K [96] or λ [97] that estimate the extent phenotypic evolution has followed a Brownian motion process, commonly interpreted as the level of phylogenetic signal in phenotypic traits. The utility of SLOUCH lies in its foundation on the Ornstein–Uhlenbeck process, which includes both Brownian motion and the possibility of adaptive evolution towards an optimum. Furthermore, unlike other methods, SLOUCH computes a separate estimate of stochasticity. Stochasticity can generate non-adaptive phenotypic variations among living species despite the presence of strong directional or stabilizing selection and contributes to a random phenotypic change under Brownian motion.
SLOUCH estimates stochasticity, vy, over a range from zero to infinity. Values approaching infinity imply that stochastic processes have been highly influential in the course of phenotypic evolution. Stochasticity can be generated by factors such as genetic drift or secondary selection pressures acting on other traits that are genetically correlated or during the process of evolutionary change more generally. Values approaching zero imply that there have been few perturbations from an adaptive optimum or during the process of evolutionary change generally.
There is always the danger of over interpreting t1/2 and vy parameters and others like them in phylogenetic comparative methods (K or λ). It must be kept in mind that these parameters are not direct measures of the evolutionary process. Rather they are statistical parameters measuring patterns in the data that are consistent with phylogenetic inertia and stochasticity in phenotypic evolution. But these parameter values can reflect other non-biological factors, such as the level of measurement error in data or patchiness in taxon sampling. Practitioners of phylogenetic comparative methods should consider carefully the warnings of Revell et al. [98] and Freckleton [99]. With a reasonable degree of caution, parameters potentially reflecting phylogenetic inertia and stochasticity can still provide some insight into the evolutionary processes that might have lead to species variation in signal complexity. To facilitate interpretation, we provide a measure of confidence in t1/2 and vy estimates by reporting the range of values within two support units of the best estimate [55]. As with AICc, values within this two-unit range are those that are essentially equally well supported.
Finally, SLOUCH offers two methods for examining phenotypic evolution. The first assumes that the phenotype has evolved towards discrete stationary optima (either a common optimum or several alternative optima; e.g. a specific phenotype selected for and maintained in a given environment via stabilizing selection). The method relies on ancestor state reconstructions of a categorical predictor variable onto the phylogeny (e.g. habitat type), which is then used to assess whether different phenotypes have been favoured in different regions of the phylogeny specified by that predictor variable. We used this approach to fit models with the following variables: open versus closed macrohabitats, arboreal versus terrestrial microhabitats, high-noise versus low-noise environments, migration and mating system. These categories were reconstructed onto the phylogeny using parsimony in the program Mesquite v. 2.74 [100]. Ideally, likelihood reconstructions should be used here, but likelihood assigns a probability that a categorical variable is present in a given ancestor. To implement the optimality analysis in SLOUCH, these probabilities would have to be manually assigned using some arbitrary cut-off (e.g. an ancestor with a probability greater than 50 per cent coded as having lived in an open rather than closed habitat). To avoid this, we followed current convention and assigned ancestor states as present or absent using parsimony [101,102]. The method of reconstruction does influence ancestor assignments. However, our preliminary analyses showed that changes to the phylogeny had a greater effect on ancestor reconstructions than the method of assignment (parsimony versus likelihood). All of our analyses were repeated on alternative phylogenies for each case study.
Reconstructions were imported into SLOUCH and used to fit models that assumed that reconstructed variables (e.g. open versus closed habitats) have selected for different adaptive optima in signal complexity (e.g. complex signals in open habitats, simple signals in closed habitats).
The second method in SLOUCH is similar in the respect that it also assumes that the phenotype has evolved towards an adaptive optimum, but differs in the sense that this optimum is not stationary and varies as a function of fluctuations in a continuous predictor variable. The model fitted is essentially a regression of signal complexity on the predictor variable and was used for all continuous predictors. SLOUCH also provides output for an optimal and evolutionary regression. The optimal regression depicts the relationship between the predictor variable and signal complexity assuming phylogenetic inertia was absent, whereas the evolutionary regression provides the ‘observed’ relationship between the predictor and signal complexity as a function of both adaptation and the constraining force of phylogenetic inertia. We report the results from evolutionary regressions only.
For all best supported models, we report phylogenetic effect sizes (r-values) to provide an indication of the direction and magnitude of relationships in the data.
3. Results
Support for models applied to each case study are reported in tables 1–3. We provide a brief summary of key findings below and in figures 1 and 2, and elaborate on the combined findings of the analyses in the discussion (§4).
Table 1.
Factors influencing the evolution of complexity in vocal communication. SSD, sexual size dimorphism.
| case-study, signal variable | model, rank | constrained tree |
unconstrained tree |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AICc | AICw | r | t1/2 (support region) | vy (support region) | AICc | AICw | r | t1/2 (support region) | vy (support region) | ||
| frogs, n = 32 species | |||||||||||
| call amplitude modulation | 1. phylogeny: null | 292.1 | 0.37 | n.a. | 0 (0–10) | 440 (260–760) | 290.5 | 0.37 | n.a. | 20 (0–140) | 440 (300–∞) |
| 2. ecology: sympatry | 294.4 | 0.12 | ∼0.16a | 1 (0–10) | 420 (260–720) | 292.9 | 0.11 | ||||
| 3. ecology: microhabitatb | 294.2 | 0.13 | 292.5 | 0.14 | −0.14 | 20 (0–160) | 430 (280–∞) | ||||
| 4. ecology: noisec | 294.2 | 0.13 | 292.8 | 0.12 | |||||||
| 5. social: SSD | 294.5 | 0.11 | 293.1 | 0.10 | |||||||
| 6. allometry: body size | 294.7 | 0.10 | 292.6 | 0.13 | |||||||
| 7. ecology: macrohabitat | 296.7 | 0.04 | 294.8 | 0.04 | |||||||
| call duration | 1. ecology: microhabitatb | −11.82 | 0.81 | 0.45 | 900 (170–∞) | 30 (0–70) | 57.1 | 0.12 | |||
| 2. phylogeny: null | −7.20 | 0.08 | 54.6 | 0.42 | n.a. | 460 (870–∞) | 10 (0–20) | ||||
| 3. ecology: sympatry | −5.16 | 0.03 | 57.3 | 0.11 | |||||||
| 4. allometry: body size | −5.20 | 0.03 | 57.2 | 0.11 | |||||||
| 5. ecology: noisec | −4.63 | 0.02 | 57.3 | 0.11 | |||||||
| 6. social: SSD | −4.59 | 0.02 | 57.3 | 0.11 | |||||||
| 7. ecology: macrohabitat | −3.53 | 0.01 | 60.0 | 0.03 | |||||||
| birds (distantly related), n = 23 species | |||||||||||
| syllable repertoire size | 1. allometry: body size | 46.2 | 0.35 | 0.35 | 75 (25–∞) | 25 (0–700) | 47.4 | 0.34 | 0.34 | 500 (25–∞) | 175 (0–700) |
| 2. phylogeny: null | 46.3 | 0.33 | n.a. | 300 (25–∞) | 125 (0–825) | 47.4 | 0.34 | n.a. | 175 (25–∞) | 75 (0–825) | |
| 3. ecology: breeding range | 48.6 | 0.11 | 49.7 | 0.11 | |||||||
| 4. ecology: macrohabitat | 48.7 | 0.10 | 50.0 | 0.09 | |||||||
| 5. social: extra-pair paternity | 48.9 | 0.09 | 49.9 | 0.10 | |||||||
| 6. social: mating system | 52.1 | 0.02 | 53.0 | 0.02 | |||||||
| song repertoire size | 1. phylogeny: null | 55.0 | 0.46 | n.a. | 975 (0–∞) | 600 (0–∞) | 56.3 | 0.46 | n.a. | 975 (0–∞) | 575 (0–∞) |
| 2. ecology: breeding range | 57.0 | 0.17 | 0.21 | 950 (0–∞) | 550 (0–∞) | 58.3 | 0.17 | 0.20 | 975 (0–∞) | 575 (0–∞) | |
| 3. allometry: body size | 57.9 | 0.11 | 59.2 | 0.11 | |||||||
| 4. social: extra-pair paternity | 58.0 | 0.10 | 59.2 | 0.11 | |||||||
| 5. ecology: macrohabitat | 58.0 | 0.10 | 59.1 | 0.11 | |||||||
| 6. social: mating system | 59.2 | 0.06 | 61.3 | 0.04 | |||||||
| birds (closely related), n = 23 species | |||||||||||
| syllable repertoire | 1. ecology: migration | −41.4 | 0.42 | ∼0.52d | ∞ (630–∞) | 10 (0–20) | −37.5 | 0.13 | |||
| 2. phylogeny: null | −41.1 | 0.36 | n.a. | 940 (500–∞) | 10 (0–30) | −40.5 | 0.57 | n.a. | 910 (480–∞) | 10 (0–30) | |
| 3. ecology: macrohabitat | −38.7 | 0.11 | −37.8 | 0.15 | |||||||
| 4. allometry: body size | −38.7 | 0.11 | −38.0 | 0.16 | |||||||
| syllable duration | 1. phylogeny: null | −64.1 | 0.66 | n.a. | ∞ (830–∞) | 10 (0–20) | −64.2 | 0.66 | n.a. | ∞ (830–∞) | 10 (0–20) |
| 2. ecology: macrohabitat | −61.2 | 0.15 | −61.3 | 0.16 | |||||||
| 3. allometry: body size | −61.2 | 0.15 | −61.2 | 0.15 | |||||||
| 4. ecology: migration | −57.9 | 0.03 | −58.1 | 0.03 | |||||||
| song duration | 1. phylogeny: null | −57.8 | 0.64 | n.a. | ∞ (780–∞) | 10 (0–20) | −52.2 | 0.11 | |||
| 2. allometry: body size | −55.2 | 0.27 | −55.0 | 0.43 | 0.28 | ∞ (780–∞) | 10 (0–20) | ||||
| 3. ecology: macrohabitat | −54.9 | 0.15 | −54.6 | 0.35 | 0.05 | ∞ (780–∞) | 10 (0–20) | ||||
| 4. ecology: migration | −52.1 | 0.06 | −52.2 | 0.11 | |||||||
aThere are several prominent outliers making direct interpretation of this coefficient misleading (figure 1a).
bMicrohabitat was defined as the height (m) of call sites above ground.
cThe level of background noise potentially experienced by a species was determined by whether species were reported to call near streams.
dThe standard errors of parameter values for migration were very large making biological interpretation difficult.
Table 3.
Factors influencing the evolution of complexity in chemical communication. CHCs, cuticular hydrocarbons.
| case-study, signal variable | model, rank | constrained tree |
unconstrained tree |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AICc | AICw | r | t1/2 (support region) | vy (support region) | AICc | AICw | r | t1/2 (support region) | vy (support region) | ||
| ants, n = 40 species | |||||||||||
| no. different CHCs | 1. phylogeny: null | 298.9 | 0.46 | n.a. | 0 (0–10) | 90 (50–150) | 298.9 | 0.46 | n.a. | 0 (0–10) | 90 (50–150) |
| 2. social: colony size | 301.2 | 0.14 | 301.2 | 0.14 | |||||||
| 3. ecology: rainfall | 301.3 | 0.14 | 301.3 | 0.14 | |||||||
| 4. social: mating system | 301.4 | 0.13 | 301.4 | 0.13 | |||||||
| 5. ecology: temperature | 301.4 | 0.13 | 301.4 | 0.13 | |||||||
Figure 1.
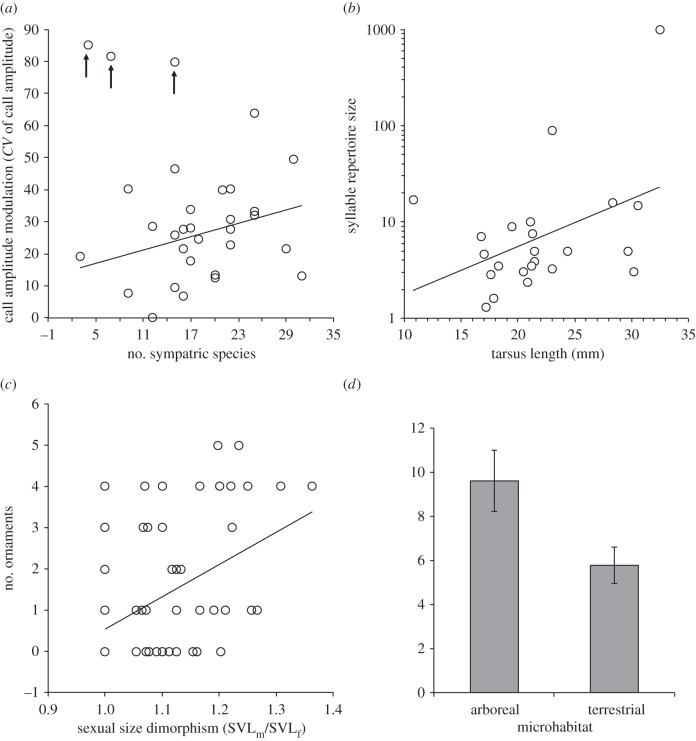
Factors predicting variations in signal complexity in (a) frogs, (b) distantly related birds, (c) distantly related agamid lizards and (d) closely related Sceloporus lizards. The arrows in (a) highlight outliers not included in the computation of the trend line depicted in the plot.
Figure 2.
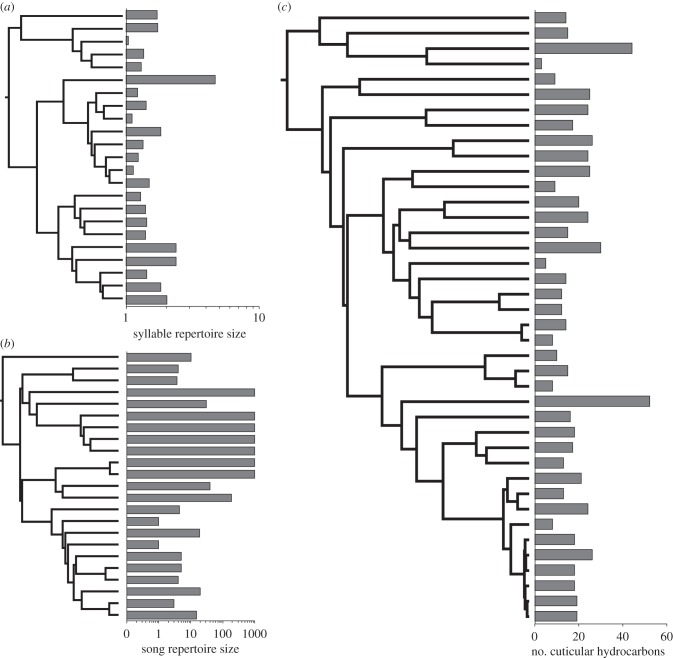
The phylogeny of signal complexity in (a) closely related birds, (b) distantly related birds and (c) ants. Strong phylogenetic inertia was estimated for the repertoires of both bird groups, while virtually no phylogenetic inertia was found for the pheromones of ants.
(a). Complexity in vocal communication
There was little support for the role of sociality in the evolution of complexity in vocal signals (mean ± s.e. AICw = 0.07 ± 0.01). Instead, phylogenetic null models and a range of different ecological models—the probability of encountering heterospecifics, migration/geographic range and habitat preference—were among the best-supported models (table 1). For example, the phylogeny of frogs, their probable encounter rate with heterospecifics and call site position above ground fit variations in call complexity well. Plots of sympatry and call modulation revealed several prominent outliers (figure 1a), but the overall trend was consistent with the species recognition hypothesis. Call site was positively correlated to call duration and, to some extent, levels of call modulation, implying that calling high above the ground has facilitated the evolution of more complex calls. There was some support for the allometry hypothesis in birds (e.g. body size correlated positively with syllable repertoire size; figure 1b).
There were high levels of possible phylogenetic inertia in most vocal signals. It was particularly striking that the estimates of phylogenetic inertia (implying evolution via Brownian motion) were consistent across measures of signal complexity and for data-sets in which species were sampled quite differently (distantly related versus closely related). Figure 2a,b illustrates repertoire size in birds as it relates to phylogeny. Both examples recorded high phylogenetic inertia, but differed in estimates of stochasticity: stochasticity in repertoire evolution was low for the closely related species (figure 2a), but high among distantly related species (figure 2b). This difference in stochasticity probably reflects differences in taxon sampling between the two case studies.
(b). Complexity in visual communication
Visual communication in lizards provided the only compelling support for models of sociality (mean ± s.e. AICw = 0.44 ± 0.13). Effect sizes showed that sexual size dimorphism, a proxy for the intensity of male–male competition within species, was positively correlated with the number of ornaments (e.g. horns, spines, tail crests; figure 1c), colour dichromatism and headbob display duration (table 2). In the case of colour dichromatism, our analyses were focused on colour signals occurring on two separate body regions because previous studies have shown that the selection pressures acting on dorsal coloration (‘exposed’; these signals are visible to aerial predators) have been different from those acting on ventral coloration (‘concealed’; these signals are only exposed during headbob displays to territorial rivals [25]). Phylogeny and whether species were arboreal or terrestrial were other models that received good support. On the latter, consistent with call evolution in frogs, headbob display evolution in lizards seems to have been facilitated by an arboreal lifestyle (figure 1d).
Table 2.
Factors influencing the evolution of complexity in visual communication.
| case-study, signal variable | model, rank | constrained tree |
unconstrained tree |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AICc | AICw | r | t1/2 (support region) | vy (support region) | AICc | AICw | r | t1/2 (support region) | vy (support region) | ||
| lizards (distantly related), n = 55–59 species | |||||||||||
| no. ornaments | 1. social: SSD | 210.7 | 1.00 | 0.58 | 40 (0–450) | 100 (20–∞) | 208.8 | 1.00 | 0.58 | 60 (0–400) | 140 (20–∞) |
| 2. allometry: body size | 230.7 | 0 | 228.6 | 0 | |||||||
| 3. phylogeny: null | 234.4 | 0 | 231.9 | 0 | |||||||
| 4. ecology: macrohabitat | 236.5 | 0 | 233.8 | 0 | |||||||
| 5. ecology: range | 236.6 | 0 | 221.1 | 0 | |||||||
| colour dichromatism, exposed | 1. phylogeny: null | 161.8 | 0.37 | n.a. | 125 (0–800) | 225 (25–∞) | 150.6 | 0.39 | n.a. | 50 (0–∞) | 75 (25–∞) |
| 2. social: SSD | 162.6 | 0.24 | 0.15 | 100 (0–825) | 175 (25–∞) | 152.2 | 0.17 | 0.11 | 175 (0–∞) | 250 (25–∞) | |
| 3. allometry: body size | 163.8 | 0.13 | 0.07 | 277 (0–800) | 225 (25–∞) | 152.6 | 0.14 | 0.07 | 50 (0–∞) | 75 (25–∞) | |
| 4. ecology: macrohabitat | 163.9 | 0.13 | 152.7 | 0.14 | |||||||
| 5. ecology: range | 163.9 | 0.13 | 152.4 | 0.16 | 50 (0–∞) | 75 (25–∞) | |||||
| colour dichromatism, concealed | 1. social: SSD | 172.2 | 0.79 | 0.31 | 25 (0–800) | 50 (25–∞) | 169.7 | 0.76 | 0.31 | 25 (0–800) | 50 (25–∞) |
| 2. phylogeny: null | 176.7 | 0.08 | 174.0 | 0.09 | |||||||
| 3. allometry: body size | 177.1 | 0.07 | 174.1 | 0.08 | |||||||
| 4. ecology: range | 178.8 | 0.03 | 175.8 | 0.04 | |||||||
| 5. ecology: macrohabitat | 178.9 | 0.03 | 176.2 | 0.03 | |||||||
| lizards (closely related), n = 22 species | |||||||||||
| bob no. | 1. ecology: microhabitata | 125.3 | 0.67 | 0.49 | 0 (0–10) | 10 (0–30) | 125.3 | 0.69 | 0.49 | 0 (0–10) | 10 (0–30) |
| 2. ecology: sympatry, overlap | 128.5 | 0.14 | 128.5 | 0.14 | |||||||
| 3. phylogeny: null | 129.3 | 0.09 | 129.3 | 0.09 | |||||||
| 4. social: SSD | 131.2 | 0.04 | 132.3 | 0.02 | |||||||
| 5. ecology: sympatry, no. | 131.2 | 0.04 | 132.4 | 0.02 | |||||||
| 6. allometry: body size | 132.4 | 0.02 | 132.4 | 0.02 | |||||||
| 7. ecology: macrohabitat | 134.1 | 0.01 | 132.3 | 0.02 | |||||||
| display duration | 1. phylogeny: null | 132.7 | 0.35 | n.a. | 0 (0–10) | 20 (5–35) | 132.7 | 0.35 | n.a. | 0 (0–50) | 20 (0–∞) |
| 2. social: SSD | 134.1 | 0.18 | 0.29 | 1 (0–50) | 20 (0–∞) | 134.1 | 0.17 | 0.29 | 0 (0–70) | 20 (0–∞) | |
| 3. allometry: body size | 134.8 | 0.12 | 134.8 | 0.12 | |||||||
| 4. ecology: sympatry, overlap | 135.0 | 0.11 | 135.0 | 0.11 | |||||||
| 5. ecology: microhabitata | 135.2 | 0.10 | 135.1 | 0.10 | |||||||
| 6. ecology: sympatry, no. | 135.6 | 0.08 | 135.4 | 0.09 | |||||||
| 7. ecology: macrohabitat | 136.7 | 0.05 | 136.4 | 0.05 | |||||||
aMicrohabitat was defined as arboreal or terrestrial (that is, typically displaying from trees or from the ground).
Visual signal evolution in lizards was associated with estimates of phylogenetic inertia close to zero. Our analyses also suggested very little stochasticity in evolutionary diversification. Taken together, visual signals have potentially been free to respond quite rapidly to selection.
(c). Complexity in chemical communication
We found little support for the role of sociality in the evolution of signal complexity in ants (mean ± s.e. AICw = 0.14 ± 0.003). The ecological models faired no better (table 3). Phylogeny appeared to be the only model that fit the data at all and even then the level of support was only marginally better than the other models considered (table 3). Almost no phylogenetic inertia or stochasticity was detected in the data. That is, evolutionary changes in the number of cuticular hydrocarbons appear to have occurred extremely rapidly and not stochastically. This implies that these signals could have been targets for selection. What selection pressure(s) this might have been is unknown. Variations in signal complexity across the ant phylogeny is depicted in figure 2c.
4. Discussion
The goal of our study was twofold: (i) to evaluate the relative support of several hypotheses for the evolution of communicative complexity, and (ii) to illustrate how new phylogenetic approaches can be incorporated into the study of animal communication more generally. With this second goal in mind, we briefly discuss some practical points first to provide readers who are not familiar with the techniques used with a better perspective on how to interpret our findings, before elaborating on the broader implications of our study.
An important point to remember for any statistical analysis is that the validity of results is contingent on the extent to which the data accurately reflect the biological variables being investigated. For example, the particular index of signal complexity we used for a given taxonomic group may or may not be functionally relevant for the species in question. Communication systems are also often complex in ways that are not easily quantified by a single metric. For example, males might be the predominant signallers in one species, whereas both sexes might rely heavily on social communication in another species. In this instance, it is not immediately obvious whether repertoire size (for example) should be summed across the sexes to provide a common metric for both species, or whether each should be sex-evaluated separately. Similar difficulties can exist for metrics of sociality. As an example, the evolution of sexual size dimorphism can reflect other factors aside from sexual selection. The sexes might appear monomorphic despite male size being the target of sexual selection because natural selection on females has lead to the evolution of large female body size for enhanced fecundity [103]. Alternatively, the presence of size dimorphism might reflect sex-specific divergence in the ecological resources exploited (e.g. differences in the size of prey eaten by the sexes [104]). In comparative analyses, errors in the phylogeny can influence findings as well, as can the assumed mode by which evolution has proceeded. To circumvent these latter issues, we relied on alternative phylogenies and a comparative method that assessed the fit of a range of parameter values designed to assay varying levels of phylogenetic inertia and stochasticity in the evolution process.
The consequence of error in either the phenotypic data or the phylogeny is the generation of noise in analyses. Data noise makes it difficult to detect relationships of small effect that might otherwise exist in nature (i.e. enhanced type I statistical error). For this reason, it is hard to conclusively reject a hypothesis if it does not gain compelling support in a comparative analysis. This is exacerbated when data are complied from a range of different sources (noise in the data can increase because sources define variables differently or use different methods for measuring variables). By the same token, broad trends that are in fact revealed in comparative analyses will probably reflect major evolutionary phenomena because only factors leading to strong biological effects will tend to be detected. In this respect, phylogenetic comparative analyses can offer conservative support for a hypothesis. This support can then be used to justify a more refined comparative analysis in which direct measures or more accurate data are collected by the comparative biologists themselves (e.g. see exploratory literature-based analysis of Ord & Martins [11] and subsequent follow-up field study by Ord et al. [105]) or focused experimental research that confirms causal links between a putative selective force and its adaptive outcome [39,106].
Our study is therefore an exploratory analysis that identifies potentially productive avenues for future research. Our findings are not meant to provide definitive conclusions on the specific evolutionary causes of signal complexity in the groups studied (although several strong candidates are highlighted (figure 1a–d). Rather the purpose of our study was to use these diverse groups as case studies to offer broader insight into the evolution of communicative complexity generally. On this front, we found that sociality – based on the metrics we were able to compile from the literature and electronic databases – was not as influential in the evolution of signal complexity as we had anticipated. Indeed, it appears to have only been an important factor in the evolution of signal complexity in lizards (table 2 and figure 1c; this is consistent with earlier comparative analyses [24,25]).
Table 4 provides an overview of the median model rank associated with each of the four hypotheses. It should be noted that the table weighs evidence from each of the case studies equally, and the communication systems and ecology of these groups do differ in a number of potentially important ways (e.g. dueting songbirds versus chorusing male frogs; colonially living ants versus territorial lizards). However, if a hypothesis is a general explanation for the evolution of communicative complexity, then it should be largely independent of the social or ecological peculiarities of a given species or taxonomic group. That is, support for a given hypothesis should be apparent across broad taxonomic groups rather than exclusive to select species. Table 4 shows that the phylogenetic null model, not social factors, was most often the best-supported model, followed closely by models reflecting ecology. In some instances, phylogenetic inertia and stochastic processes can explain variations in signal complexity among species. Signal complexity therefore has the potential to accumulate in lineages via neutral or non-adaptive processes (e.g. syllable duration in birds; table 1). When evidence of adaptation was found, it was more often associated with variations in ecological factors and (to a lesser extent) allometry than it was with social pressures. For example, signal complexity increased in frogs as a possible function of the number of sympatric species encountered (table 1 and figure 1a) or the height of call sites above the ground (table 1). In lizards, the switch from a terrestrial lifestyle to being arboreal appears to have facilitated the evolution of more elaborate territorial displays (table 2 and figure 1d; see also [63]). The difficulty in interpreting the role of ecology here is whether variables such as call or display site drive or constrain signal complexity. On the one hand, the range over which signals must remain effective increases with perch elevation, leading to potential directional selection on signals for longer duration or more components to facilitate detection by distant receivers. On the other hand, the environmental constraint leading to simple signals when communication is conducted close to the ground is no longer present or reduced for species signalling from elevated perches, opening the door for more complex signals to evolve via other factors. Semantically the distinction is subtle, but biologically it is important for inferring causality. But again, causality can only really be confirmed by means of empirical study and experimental manipulation [106].
Table 4.
A summary of the relative support for factors expected to influence the evolution of communicative complexity. Shown are median ranks of the highest ranked model for a given hypothesis for each case study based on the results from the constrained trees only or both the constrained and unconstrained trees. Calculations used the actual ranks of models when those models were ranked within two AICc units of the best-supported model or an assigned rank of 100 if models fell outside this region. Subsequent median ranks of 100 were then classified as ‘unranked’.
| hypothesis | signal modality |
|||
|---|---|---|---|---|
| acoustic | visual | chemical | all modalities | |
| constrained trees only | ||||
| phylogeny | 1 | 1 | 1 | 1 |
| ecology | 1 | 1-unranked | unranked | 1–2 |
| social | unranked | 1–2 | unranked | unranked |
| allometry | unranked | 3-unranked | not tested | unranked |
| constrained or unconstrained trees | ||||
| phylogeny | 1 | 1 | 1 | 1 |
| ecology | 1 | 2 | unranked | 1–2 |
| allometry | 1 | 3-unranked | not tested | 3 |
| social | unranked | 1–2 | unranked | unranked |
That said, empirical and experimental studies within species alone do not adequately identify the selection pressures that have directed signal evolution. Experimental studies can demonstrate current utility, but they cannot reveal the evolutionary history of communication, which in itself can have important consequences on how species adapt to contemporary selection pressures [107]. Consider the phylogeny of signal complexity in the case studies we examined. Table 5 illustrates remarkable consistency within signal classes in possible levels of phylogenetic inertia. If these estimates are accurate, they imply that the evolution of complexity in vocal signals exhibits far greater phylogenetic inertia, and subsequently lower rates of adaptation, than other signal modalities. This could reflect major physiological or metabolic constraints on the production of auditory signals [49,108,109]. We also found evidence for possible selection on the pheromone signals of ants, even though none of our selection models obtained any compelling support. The evolution of pheromone complexity appears to have been non-random and not the product of the gradual accumulation of cuticular hydrocarbons over long periods of evolutionary time (indicated by low values of stochasticity in the evolution process and low values of phylogenetic half-life, respectively). We reiterate that the interpretation of statistical parameters reflecting phylogenetic patterns need to be made with caution. With this in mind, our results are consistent with the notion that an unknown selection pressure has promoted rapid, predictable changes in pheromone complexity in ants. However, these results may also reflect that the hydrocarbons assayed have little functional relevance for communication or fitness generally.
Table 5.
A summary of phylogenetic patterns associated with estimates of communicative complexity. CHCs, cuticular hydrocarbons.
| case-study, signal variable | Nspecies | phylogenetic inertia (t1/2) | stochasticity (vy) |
|---|---|---|---|
| vocal signals | |||
| frogs | |||
| call amplitude modulation | 32 | low | high |
| call duration | 32 | high | low |
| birds (distantly related) | |||
| syllable repertoire size | 23 | moderate | moderate |
| song repertoire size | 23 | high | high |
| birds (closely related) | |||
| syllable repertoire | 23 | high | low |
| syllable duration | 23 | high | low |
| song duration | 23 | high | low |
| visual signals | |||
| lizards (distantly related) | |||
| number of ornaments | 59 | low | low–moderate |
| colour dichromatism, exposed | 55 | low–moderate | low–moderate |
| colour dichromatism, concealed | 55 | low | low |
| lizards (closely related) | |||
| bob number | 22 | virtually zero | low |
| display duration | 22 | virtually zero | low |
| chemical signals | |||
| ants | |||
| no. different CHCs | 40 | virtually zero | low |
Future research will be needed to clarify the significance of our findings in relation to the specific case studies examined. But the general outcome of our investigation is that sociality is not always required for the evolution of communicative complexity. Or at least, communicative complexity is the product of an intricate evolutionary process that cannot be distilled to a single factor. The complexity of form in the way animals communicate with one another fascinates biologists and amateur naturalists alike. On an informal level, communicative complexity implies richness in the social lives of animals. This sophistication in social behaviour probably enticed many of us into a career of studying animal communication in the first place. Indeed, in some regards, the study of communication in non-avian and non-primate species might be under-represented in the literature because of the perceived notion that such systems lack a degree of sociality and are therefore less attractive systems for the study of communication. Yet the generality of any hypothesis of why and how animal communication evolved is reliant on testing hypotheses on diverse taxa. We conducted such a study here and found compelling evidence—ironically, it seems—only for social factors in the evolution of visual signals in lizards. Whether sociality is a prerequisite for the evolution of communicative complexity in other systems awaits further investigation. Our results suggest that it may not be (see [56–58] for positive tests in mammals).
Acknowledgements
We thank Matthew Symonds, Carlos Botero and László Garamszegi for generously providing us access to their original data, Victor Soria-Carrasco for his helpful advice in GIS matters, and Todd Freeberg and two anonymous reviewers for comments on a previous version of this paper. J.G.P. was supported by a pre-doctoral research fellowship (JAE) from the CSIC. All data used in this study have been archived in the Dryad Digital Repository (doi:10.5061/dryad.nf7697j6).
References
- 1.Freeberg T. M., Dunbar R. I. M., Ord T. J. 2012. Social complexity as a proximate and ultimate factor in communicative complexity. Phil. Trans. R. Soc. B 367, 1785–1801 10.1098/rstb.2011.0213 (doi:10.1098/rstb.2011.0213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollick A. S., de Waal F. B. M. 2007. Ape gestures and language evolution. Proc. Natl Acad. Sci. USA 104, 8184–8189 10.1073/pnas.0702624104 (doi:10.1073/pnas.0702624104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vick S.-J, Waller B. M., Parr L. A., Smith Pasqualini M. C., Bard K. A. 2007. A cross-species comparison of facial morphology and movement in humans and chimpanzees using the facial action coding system (FACS). J. Nonverb. Behav. 31, 1–10 10.1007/s10919-006-0017-z (doi:10.1007/s10919-006-0017-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbib M. A., Liebal K. &, Pika S. 2008. Primate vocalization, gesture, and the evolution of human language. Curr. Anthropol. 49, 1053–1076 10.1086/593015 (doi:10.1086/593015) [DOI] [PubMed] [Google Scholar]
- 5.Dunbar R. I. M. 2012. Bridging the bonding gap: the transition from primates to humans. Phil. Trans. R. Soc. B 367, 1837–1846 10.1098/rstb.2011.0217 (doi:10.1098/rstb.2011.0217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McComb K., Reby D., Baker L., Moss C. J., Sayialel S. 2003. Long-distance communication of acoustic cues to social identity in African elephants. Anim. Behav. 65, 317–329 10.1006/anbe.2003.2047 (doi:10.1006/anbe.2003.2047) [DOI] [Google Scholar]
- 7.Soltis J., Leong K., Savage A. 2005. African elephant vocal communication. I. Antiphonal calling behaviour among affiliated females. Anim. Behav. 70, 579–587 10.1016/j.anbehav.2004.11.015 (doi:10.1016/j.anbehav.2004.11.015) [DOI] [Google Scholar]
- 8.Soltis J., Leong K., Savage A. 2005. African elephant vocal communication. II. Rumble variation reflects the individual identity and emotional state of callers. Anim. Behav. 70, 589–599 10.1016/j.anbehav.2004.11.016 (doi:10.1016/j.anbehav.2004.11.016) [DOI] [Google Scholar]
- 9.Meyer J. M., Goodwin T. E., Schulte B. A. 2008. Intrasexual chemical communication and social responses of captive female African elephants, Loxodonta africana. Anim. Behav. 76, 163–174 10.1016/j.anbehav.2007.12.019 (doi:10.1016/j.anbehav.2007.12.019) [DOI] [Google Scholar]
- 10.Figuerola J., Green A. J. 2000. The evolution of sexual dimorphism in relation to mating patterns, cavity nesting, insularity and sympatry in the Anseriformes. Funct. Ecol. 14, 701–710 10.1046/j.1365-2435.2000.00474.x (doi:10.1046/j.1365-2435.2000.00474.x) [DOI] [Google Scholar]
- 11.Ord T. J., Martins E. P. 2006. Tracing the origins of signal diversity in anole lizards: phylogenetic approaches to inferring the evolution of complex behaviour. Anim. Behav. 71, 1411–1429 10.1016/j.anbehav.2005.12.003 (doi:10.1016/j.anbehav.2005.12.003) [DOI] [Google Scholar]
- 12.Hailman J., Ficken M. 1986. Combinatorial animal communication with computable syntax: chick-a-dee calling qualifies as ‘language’ by structural linguistics. Anim. Behav. 34, 1899–1901 10.1016/S0003-3472(86)80279-2 (doi:10.1016/S0003-3472(86)80279-2) [DOI] [Google Scholar]
- 13.Martins E. P. 1994. Structural complexity in a lizard communication system: the Sceloporus graciosus ‘push-up’ display. Copeia 194, 944–955 10.2307/1446717 (doi:10.2307/1446717) [DOI] [Google Scholar]
- 14.Catchpole C. K., Slater P. J. B. 2008. Bird song: biological themes and variations, 2nd edn. Cambridge, UK: Cambridge University Press [Google Scholar]
- 15.Nicholson K. E., Harmon L. J., Losos J. B. 2007. Evolution of Anolis lizard dewlap diversity. PLoS ONE 2, e274. 10.1371/journal.pone.0000274 (doi:10.1371/journal.pone.0000274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamo S. A., Hanlon R. T. 1996. Do cuttlefish (Cephalopoda) signal their intentions to conspecific during agonistic encounters? Anim. Behav. 52, 73–81 10.1006/anbe.1996.0153 (doi:10.1006/anbe.1996.0153) [DOI] [Google Scholar]
- 17.Mäthger L. M., Shashar N., Hanlon R. T. 2009. Do cephalopods communicate using polarized light reflections from their skin? J. Exp. Biol. 212, 2133–2140 10.1242/jeb.020800 (doi:10.1242/jeb.020800) [DOI] [PubMed] [Google Scholar]
- 18.Hebets E. A., Uetz G. W. 2000. Leg ornamentation and the efficacy of courtship display in four species of wolf spider (Araneae: Lycosidae). Behav. Ecol. Sociobiol. 47, 280–286 10.1007/s002650050667 (doi:10.1007/s002650050667) [DOI] [Google Scholar]
- 19.Gordon S. D., Uetz G. W. 2011. Multimodal communication of wolf spiders on different substrates: evidence for behavioural plasticity. Anim. Behav. 81, 367–375 10.1016/j.anbehav.2010.11.003 (doi:10.1016/j.anbehav.2010.11.003) [DOI] [Google Scholar]
- 20.Ord T. J. 2010. Phylogeny and the evolution of communication. In Encyclopedia of animal behavior (eds Breed M. D., Moore J.), pp. 652–660 Oxford, UK: Academic Press [Google Scholar]
- 21.Krebs J. R., Ashcroft R., Webber M. 1978. Song repertoires and territory defence in the great tit. Nature 271, 539–543 10.1038/271539a0 (doi:10.1038/271539a0) [DOI] [Google Scholar]
- 22.Read A. F., Weary D. M. 1992. The evolution of bird song: comparative analyses. Phil. Trans. R. Soc. Lond. B 338, 165–187 10.1098/rstb.1992.0137 (doi:10.1098/rstb.1992.0137) [DOI] [Google Scholar]
- 23.MacDougall-Shackleton S. A. 1997. Sexual selection and the evolution of song repertoires. Curr. Ornithol. 14, 81–124 [Google Scholar]
- 24.Ord T. J., Blumstein D. T., Evans C. S. 2001. Intrasexual selection predicts the evolution of signal complexity in lizards. Proc. R. Soc. Lond. B 268, 737–744 10.1098/rspb.2000.1417 (doi:10.1098/rspb.2000.1417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart-Fox D., Ord T. J. 2004. Sexual selection, natural selection and the evolution of dimorphic coloration and ornamentation in agamid lizards. Proc. R. Soc. Lond. B 271, 2249–2255 10.1098/rspb.2004.2802 (doi:10.1098/rspb.2004.2802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins E. P., Ord T. J., Davenport S. W. 2005. Combining motions into complex displays: playbacks with a robotic lizard. Behav. Ecol. Sociobiol. 58, 351–360 10.1007/s00265-005-0954-2 (doi:10.1007/s00265-005-0954-2) [DOI] [Google Scholar]
- 27.Wiley R. H., Richards D. G. 1982. Adaptations for acoustic communication in birds: sound transmission and signal detection. In Acoustic communication in birds, vol. 1 (eds Kroodsma D. E., Miller E. H., Ouellet H.), pp. 131–181 New York, NY: Academic Press [Google Scholar]
- 28.Brenowitz E. A. 1986. Environmental influences on acoustic and electric animal communication. Brain Behav. Evol. 28, 32–42 10.1159/000118690 (doi:10.1159/000118690) [DOI] [PubMed] [Google Scholar]
- 29.Romer H. 1993. Environmental and biological constraints for the evolution of long-range signalling and hearing in acoustic insects. Phil. Tran. R. Soc. Lond. B 340, 179–185 10.1098/rstb.1993.0056 (doi:10.1098/rstb.1993.0056) [DOI] [Google Scholar]
- 30.Forrest T. G. 1994. From sender to receiver: propagation and environmental effects on acoustic signals. Am. Zool. 34, 644–654 [Google Scholar]
- 31.Potvin D. A., Parris K. M., Mulder R. A. 2011. Geographically pervasive effects of urban noise on frequency and syllable rate of songs and calls in silvereyes (Zosterops lateralis). Proc. R. Soc. B 278, 2464–2469 10.1098/rspb.2010.2296 (doi:10.1098/rspb.2010.2296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis C. D., Ortega C. P., Cruz A. 2011. Different behavioural responses to anthropogenic noise by two closely related passerine birds. Biol. Lett. 7, 850–852 10.1098/rsbl.2011.0359 (doi:10.1098/rsbl.2011.0359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ord T. J., Peters R. A., Clucas B., Stamps J. A. 2007. Lizards speed up visual displays in noisy motion habitats. Proc. R. Soc. B 274, 1057–1062 10.1098/rspb.2006.0263 (doi:10.1098/rspb.2006.0263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters R. A. 2008. Environmental motion delays the detection of movement-based signals. Biol. Lett. 4, 2–5 10.1098/rsbl.2007.0422 (doi:10.1098/rsbl.2007.0422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ord T. J., Blumstein D. T., Evans C. S. 2002. Ecology and signal evolution in lizards. Biol. J. Linn. Soc. 77, 127–148 10.1046/j.1095-8312.2002.00100.x (doi:10.1046/j.1095-8312.2002.00100.x) [DOI] [Google Scholar]
- 36.Eifler D. A., Eilfer M. A., Karnes T. 2008. Effect of habitat visibility on movements and displaying by side-blotched lizards (Uta stansburiana). Ethol. Ecol. Evol. 20, 283–289 10.1080/08927014.2008.9522527 (doi:10.1080/08927014.2008.9522527) [DOI] [Google Scholar]
- 37.Marchetti K. 1993. Dark habitats and bright birds illustrate the role of the environment in species divergence. Nature 362, 149–152 10.1038/362149a0 (doi:10.1038/362149a0) [DOI] [Google Scholar]
- 38.Gomez D., Thery M. 2004. Influence of ambient light on the evolution of colour signals: comparative analysis of a neotropical rainforest bird community. Ecol. Lett. 7, 279–284 10.1111/j.1461-0248.2004.00584.x (doi:10.1111/j.1461-0248.2004.00584.x) [DOI] [Google Scholar]
- 39.Ord T. J., Stamps J. A. 2008. Alert signals enhance animal communication in ‘noisy’ environments. Proc. Natl Acad. Sci. USA 105, 18 830–18 835 10.1073/pnas.0807657105 (doi:10.1073/pnas.0807657105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halfwerk W., Bot S., Buikx J., van der Velde M., Komdeur J., ten Cate C., Slabbekoorn H. 2011. Low-frequency songs lose their potency in noisy urban conditions. Proc. Natl Acad. Sci. USA 108, 14 549–14 554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards D. G. 1981. Alerting and message components in songs of rufous-sided towhees. Behaviour 76, 223–249 10.1163/156853981X00095 (doi:10.1163/156853981X00095) [DOI] [Google Scholar]
- 42.Endler J. A. 1992. Signals, signal conditions, and the direction of evolution. Am. Nat. 139, S125–S153 10.1086/285308 (doi:10.1086/285308) [DOI] [Google Scholar]
- 43.Feng A. S., Narins P. M., Xu C.-H., Lin W.-Y., Yu Z.-L., Qiu Q., Xu Z.-M., Shen J.-X. 2006. Ultrasonic communication in frogs. Nature 440, 333–336 10.1038/nature04416 (doi:10.1038/nature04416) [DOI] [PubMed] [Google Scholar]
- 44.Ramírez-Bautista A., Benabib M. 2001. Perch height of the arboreal lizard Anolis nebulosus (Sauria: Polychrotidae) from a tropical dry forest of Mexico: effect of the reproductive season. Copeia 2001, 187–193 10.1643/0045-8511(2001)001[0187:PHOTAL]2.0.CO;2 (doi:10.1643/0045-8511(2001)001[0187:PHOTAL]2.0.CO;2) [DOI] [Google Scholar]
- 45.Ord T. J., King L., Young A. R. 2011. Constrasting theory with the empirical data of species recognition. Evolution 65, 2572–2591 10.1111/j.1558-5646.2011.01319.x (doi:10.1111/j.1558-5646.2011.01319.x) [DOI] [PubMed] [Google Scholar]
- 46.Fitch W. T. 2000. Skull dimensions in relation to body size in nonhuman mammals: the causal bases for acoustic allometry. Zoology 103, 40–58 [Google Scholar]
- 47.Charlton B. D., Ellis W. A. H., McKinnon A. J., Cowin G. J., Brumm J., Nilsson K., Fitch W. T. 2011. Cues to body size in the formant spacing of male koala (Phascolarctos cinereus) bellows: honesty in an exaggerated trait. J. Exp. Biol. 214, 3414–3422 10.1242/jeb.061358 (doi:10.1242/jeb.061358) [DOI] [PubMed] [Google Scholar]
- 48.Frey R., Volodin I., Volodina E., Soldatova N. V., Juldaschev E. T. 2011. Descended and mobile larynx, vocal tract elongation and rutting roars in male goitred gazelles (Gazella subgutturosa Güldenstaedt, 1780). J. Anat. 218, 566–585 10.1111/j.1469-7580.2011.01361.x (doi:10.1111/j.1469-7580.2011.01361.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillooly J. F., Ophir A. G. 2010. The energetic basis of acoustic communication. Proc. R. Soc. B 277, 1325–1331 10.1098/rspb.2009.2134 (doi:10.1098/rspb.2009.2134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ord T. J., Blumstein D. T. 2002. Size constraints and the evolution of display complexity: why do large lizards have simple displays? Biol. J. Linn. Soc. 76, 145–161 10.1111/j.1095-8312.2002.tb01721.x (doi:10.1111/j.1095-8312.2002.tb01721.x) [DOI] [Google Scholar]
- 51.Irwin D. E., Thimgan M. P., Irwin J. H. 2008. Call divergence is correlated with geographic and genetic distance in greenish warblers (Phylloscopus trochiloides): a strong role for stochasticity in signal evolution? J. Evol. Biol. 21, 435–448 10.1111/j.1420-9101.2007.01499.x (doi:10.1111/j.1420-9101.2007.01499.x) [DOI] [PubMed] [Google Scholar]
- 52.Benedict L., Bowie R. C. K. 2009. Macrogeographical variation in the song of a widely distributed African warbler. Biol. Lett. 5, 484–487 10.1098/rsbl.2009.0244 (doi:10.1098/rsbl.2009.0244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell P., Pasch1 B., Pino1 J. L., Crino1 O. L., Phillips1 M., Phelps S. M. 2010. Geographic variation in the songs of neotropical singing mice: testing the relative importance of drift and local adaptation. Evolution 64, 1955–1972 10.1111/j.1558-5646.2010.00962.x (doi:10.1111/j.1558-5646.2010.00962.x) [DOI] [PubMed] [Google Scholar]
- 54.Grant P. R., Grant B. R. 2009. The secondary contact phase of allopatric speciation in Darwin's finches. Proc. Natl Acad. Sci. USA 106, 20 141–20 148 10.1073/pnas.0911761106 (doi:10.1073/pnas.0911761106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen T. F., Pienaar J., Orzack S. H. 2008. A comparative method for studying adaptation to a randomly evolving environment. Evolution 62, 1965–1977 10.1111/j.1558-5646.2008.00412.x (doi:10.1111/j.1558-5646.2008.00412.x) [DOI] [PubMed] [Google Scholar]
- 56.Blumstein D. T., Armitage K. B. 1997. Does sociality drive the evolution of communicative complexity? A comparative test with ground-dwelling sciurid alarm calls. Am. Nat. 150, 179–200 10.1086/286062 (doi:10.1086/286062) [DOI] [PubMed] [Google Scholar]
- 57.McComb K., Semple S. 2005. Coevolution of vocal communication and sociality in primates. Biol. Lett. 1, 381–385 10.1098/rsbl.2005.0366 (doi:10.1098/rsbl.2005.0366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobson S. D. 2009. Socioecological correlates of facial mobility in nonhuman anthropoids. Am. J. Physic. Anthropol. 139, 413–420 10.1002/ajpa.21007 (doi:10.1002/ajpa.21007) [DOI] [PubMed] [Google Scholar]
- 59.delBarco-Trillo J., Sacha C. R., Dubay G. R., Drea C. M. 2012. Eulemur, me lemur: the evolution of scent-signal complexity in a primate clade. Phil. Trans. R. Soc. B 367, 1909–1922 10.1098/rstb.2011.0225 (doi:10.1098/rstb.2011.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gustison M. L., le Roux A., Bergman T. J. 2012. Derived vocalisations of geladas (Theropithecus gelada) and the evolution of vocal complexity in primates. Phil. Trans. R. Soc. B 367, 1847–1859 10.1098/rstb.2011.0218 (doi:10.1098/rstb.2011.0218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramsier M. A., Cunningham A. J., Finneran J. J., Dominy N. J. 2012. Social drive and the evolution of primate hearing. Phil. Trans. R. Soc. B 367, 1860–1868 10.1098/rstb.2011.0219 (doi:10.1098/rstb.2011.0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stuart-Fox D., Owens I. P. F. 2003. Species richness in agamid lizards: chance, body size, sexual selection or ecology? J. Evol. Biol. 16, 659–669 10.1046/j.1420-9101.2003.00573.x (doi:10.1046/j.1420-9101.2003.00573.x) [DOI] [PubMed] [Google Scholar]
- 63.Martins E. P. 1993. A comparative study of the evolution of Sceloporus push-up displays. Am. Nat. 142, 994–1018 10.1086/285585 (doi:10.1086/285585) [DOI] [PubMed] [Google Scholar]
- 64.Ord T. J., Stuart-Fox D. 2006. Ornament evolution in dragon lizards: multiple gains and widespread losses reveal a complex history of evolutionary change. J. Evol. Biol. 19, 797–808 10.1111/j.1420-9101.2005.01050.x (doi:10.1111/j.1420-9101.2005.01050.x) [DOI] [PubMed] [Google Scholar]
- 65.Botero C. A., Boogert N. J., Vehrencamp S. L., Lovette I. J. 2009. Climatic patterns predict the elaboration of song displays in mockingbirds. Curr. Biol. 19, 1–8 10.1016/j.cub.2008.11.058 (doi:10.1016/j.cub.2008.11.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soma M., Garamszegi L. Z. 2011. Rethinking birdsong evolution: meta-analysis of the relationship between song complexity and reproductive success. Behav. Ecol. 22, 363–371 10.1093/beheco/arq219 (doi:10.1093/beheco/arq219) [DOI] [Google Scholar]
- 67.Van Wilgenburd E., Symonds M. R. E., Elgar M. A. 2011. Evolution of cuticular hydrocarbon diversity in ants. J. Evol. Biol. 24, 1188–1198 10.1111/j.1420-9101.2011.02248.x (doi:10.1111/j.1420-9101.2011.02248.x) [DOI] [PubMed] [Google Scholar]
- 68.Kok P. J. R., Kalamandeen R. 2008. Introduction to the taxonomy of the amphibians of the Kaieteur National Park, Guyana. Brussels: ABC Taxa [Google Scholar]
- 69.Fitch H. S. 1978. Sexual size differences in the genus Sceloporus. Univ. Kansas Sci. Bull. 51, 441–461 [Google Scholar]
- 70.Olson D. M., et al. 2001. Terrestrial ecoregions of the world: a new map of life on earth. BioScience 51, 933–938 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2 (doi:10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2) [DOI] [Google Scholar]
- 71.BirdLife International & NatureServe 2011. Bird species distribution maps of the world. Cambridge, UK: BirdLife International; Arlington, VA: NatureServe [Google Scholar]
- 72.Husby A., Hille S. M., Visser M. E. 2011. Testing mechanisms of Bergmann's rule: phenotypic decline but no genetic change in body size in three passerine bird populations. Am. Nat. 178, 202–213 10.1086/660834 (doi:10.1086/660834) [DOI] [PubMed] [Google Scholar]
- 73.Ferree E. D., Dickinson J. L. 2011. Natural extrapair paternity matches receptivity patterns in unguarded females: evidence for importance of female choice. Anim. Behav. 82, 1167–1173 10.1016/j.anbehav.2011.08.016 (doi:10.1016/j.anbehav.2011.08.016) [DOI] [Google Scholar]
- 74.Benson D.A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. 2010. GenBank. Nucleic Acids Res. 38, D46–D51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K., Misawa K., Kuma K., Miyata T. 2002. Mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 30, 3059–3066 10.1093/nar/gkf436 (doi:10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pyron R. A., Wiens J. J. 2011. A large-scale phylogeny of Amphibia including over 2,800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phyl. Evol. 61, 543–583 10.1016/j.ympev.2011.06.012 (doi:10.1016/j.ympev.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 78.Lovette I. J., et al. 2011. Phylogenetic relationships of the mockingbirds and thrashers (Aves: Mimidae). Mol. Phyl. Evol. 63, 219–229 10.1016/j.ympev.2011.07.009 (doi:10.1016/j.ympev.2011.07.009) [DOI] [PubMed] [Google Scholar]
- 79.Ericson P. G. P., Johansson U. S. 2003. Phylogeny of Passerida (Aves: Passeriformes) based on nuclear and mitochondrial data. Mol. Phyl. Evol. 29, 126–138 10.1016/S1055-7903(03)00067-8 (doi:10.1016/S1055-7903(03)00067-8) [DOI] [PubMed] [Google Scholar]
- 80.Immler S., Pitnick S., Parker G. A., Durrant K. L., Lüpold S., Calhim S., Birkhead T. R. 2011. Resolving variation in the reproductive tradeoff between sperm size and number. Proc. Natl Acad. Sci. USA 108, 5325–5330 10.1073/pnas.1009059108 (doi:10.1073/pnas.1009059108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klicka J., Burns K., Spellman G. M. 2007. Defining a monophyletic Cardinalini: a molecular perspective. Mol. Phyl. Evol. 45, 1014–1032 10.1016/j.ympev.2007.07.006 (doi:10.1016/j.ympev.2007.07.006) [DOI] [PubMed] [Google Scholar]
- 82.Fregina S., Haasea M., Olssonb U., Alströmc P. 2009. Multi-locus phylogeny of the family Acrocephalidae (Aves: Passeriformes): the traditional taxonomy overthrown. Mol. Phyl. Evol. 52, 866–878 10.1016/j.ympev.2009.04.006 (doi:10.1016/j.ympev.2009.04.006) [DOI] [PubMed] [Google Scholar]
- 83.Jønsson K. A., Irestedt M., Fuchs J., Ericson P. G. P., Christidis L., Bowie R. C. K., Norman J. A., Pasquet E., Fjeldså J. 2008. Explosive avian radiations and multi-directional dispersal across Wallacea: evidence from the Campephagidae and other Crown Corvida (Aves). Mol. Phyl. Evol. 47, 221–236 10.1016/j.ympev.2008.01.017 (doi:10.1016/j.ympev.2008.01.017) [DOI] [PubMed] [Google Scholar]
- 84.Collar D. C., Schulte J. A., II, O'Meara B. C., Losos J. B. 2010. Habitat use affects morphological diversification in dragon lizards. J. Evol. Biol. 23, 1033–1049 10.1111/j.1420-9101.2010.01971.x (doi:10.1111/j.1420-9101.2010.01971.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ota H., Kobayashi M., Nabhitabhata J., Yong H.-S., Sengoku S., Hikida T. 2000. Phylogenetic relationships of the family Agamidae (Reptilia: Iguania) inferred from mitochondrial DNA sequences. Zool. Sci. 17, 527–537 [Google Scholar]
- 86.Macey J. R., Schulte J. A., II, Larson A., Ananjeva N. B., Wang Y., Pethiyagoda R., Rastegar-Pouyani N., Papenfuss T. J. 2000. Evaluating trans-tethys migration: an example using acrodont lizard phylogenetics. Syst. Biol. 49, 233–256 10.1093/sysbio/49.2.233 (doi:10.1093/sysbio/49.2.233) [DOI] [PubMed] [Google Scholar]
- 87.Leaché A. D., Koo M. S., Spencer C. L., Papenfuss T. J., Fisher R. N., McGuire J. A. 2009. Quantifying ecological, morphological, and genetic variation to delimit species in the coast horned lizard species complex (Phrynosoma). Proc. Natl Acad. Sci. USA 106, 12 418–12 423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hugall A. F., Foster R., Hutchinson M., Lee M. S. Y. 2008. Phylogeny of Australasian agamid lizards based on nuclear and mitochondrial genes: implications for morphological evolution and biogeography. Biol. J. Linn. Soc. 93, 343–358 10.1111/j.1095-8312.2007.00911.x (doi:10.1111/j.1095-8312.2007.00911.x) [DOI] [Google Scholar]
- 89.Rambaut A., Drummond A. J.2007. Tracer v. 1.4. See http://beast.bio.ed.ac.uk/Tracer .
- 90.Burnham K. P., Anderson D. R. 2002. Model selection and multimodal inference: a practial information-theoretic approach, 2nd edn. New York, NY: Springer [Google Scholar]
- 91.Johnson J. B., Omland K. S. 2004. Model selection in ecology and evolution. Trends Ecol. Evol. 19, 101–108 10.1016/j.tree.2003.10.013 (doi:10.1016/j.tree.2003.10.013) [DOI] [PubMed] [Google Scholar]
- 92.Hansen T. F. 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51, 1341–1351 10.2307/2411186 (doi:10.2307/2411186) [DOI] [PubMed] [Google Scholar]
- 93.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15 10.1086/284325 (doi:10.1086/284325) [DOI] [Google Scholar]
- 94.Ord T. J., Martins E. P. 2010. The evolution of behavior: phylogeny and the origin of present-day diversity. In Evolutionary behavioral ecology (eds Westneat D. F., Fox C. W.), pp. 108–128 New York, NY: Oxford University Press [Google Scholar]
- 95.Martins E. P., Hansen T. F. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667 10.1086/286013 (doi:10.1086/286013) [DOI] [Google Scholar]
- 96.Blomberg S. P., Garland T., Jr, Ives A. R. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 [DOI] [PubMed] [Google Scholar]
- 97.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884 10.1038/44766 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 98.Revell L. J., Harmon L. J., Collar D. C. 2008. Phylogenetic signal, evolutionary process, and rate. Syst. Biol. 57, 591–601 10.1080/10635150802302427 (doi:10.1080/10635150802302427) [DOI] [PubMed] [Google Scholar]
- 99.Freckleton R. P. 2009. The seven deadly sins of comparative analysis. J. Evol. Biol. 22, 1367–1375 10.1111/j.1420-9101.2009.01757.x (doi:10.1111/j.1420-9101.2009.01757.x) [DOI] [PubMed] [Google Scholar]
- 100.Maddison W. P., Maddison D. R. 2010. Mesquite: a modular system for evolutionary analysis, v. 2.74. See http://mesquiteproject.org
- 101.Butler M. A., King A. A. 2004. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am. Nat. 164, 683–695 10.1086/426002 (doi:10.1086/426002) [DOI] [PubMed] [Google Scholar]
- 102.Labra A., Piernaar J., Hansen T. F. 2009. Evolution of thermal physiology in Liolaemus lizards: adaptation, phylogenetic inertia, and niche tracking. Am. Nat. 174, 204–220 10.1086/600088 (doi:10.1086/600088) [DOI] [PubMed] [Google Scholar]
- 103.Prenter J., Elwood R. W., Montgomery W. I. 1999. Sexual size dimorphism and reproductive investment by female spiders: a comparative analysis. Evolution 53, 1987–1994 10.2307/2640458 (doi:10.2307/2640458) [DOI] [PubMed] [Google Scholar]
- 104.Schoener T. W. 1967. The ecological significance of sexual dimorphism in size in the lizard Anolis conspersus. Science 155, 474–477 10.1126/science.155.3761.474 (doi:10.1126/science.155.3761.474) [DOI] [PubMed] [Google Scholar]
- 105.Ord T. J., Stamps J. A., Losos J. B. 2010. Adaptation and plasticity of animal communication in fluctuating environments. Evolution 64, 3134–3148 10.1111/j.1558-5646.2010.01056.x (doi:10.1111/j.1558-5646.2010.01056.x) [DOI] [PubMed] [Google Scholar]
- 106.Clucas B., Ord T. J., Owings D. H. 2010. Fossils and phylogeny uncover the evolutionary history of a unique antipredator behaviour. J. Evol. Biol. 23, 2197–2211 10.1111/j.1420-9101.2010.02083.x (doi:10.1111/j.1420-9101.2010.02083.x) [DOI] [PubMed] [Google Scholar]
- 107.Ord T. J., Charles G. K., Hoffer R. K. 2011. The evolution of alternative adaptive strategies for effective communication in noisy environments. Am. Nat. 177, 54–64 10.1086/657439 (doi:10.1086/657439) [DOI] [PubMed] [Google Scholar]
- 108.Ophir A. G., Schrader S. B., Gillooly J. F. 2010. Energetic cost of calling: general constraints and species-specific differences. J. Evol. Biol. 23, 1564–1569 10.1111/j.1420-9101.2010.02005.x (doi:10.1111/j.1420-9101.2010.02005.x) [DOI] [PubMed] [Google Scholar]
- 109.Stoddard P. K., Salazar V. 2010. Energetic cost of communication. J. Exp. Biol. 214, 200–205 10.1242/jeb.047910 (doi:10.1242/jeb.047910) [DOI] [PMC free article] [PubMed] [Google Scholar]