Abstract
The eukaryotic porin superfamily consists of two families, voltage-dependent anion channel (VDAC) and Tom40, which are both located in the mitochondrial outer membrane. In Trypanosoma brucei, only a single member of the VDAC family has been described. We report the detection of two additional eukaryotic porin-like sequences in T. brucei. By bioinformatic means, we classify both as putative VDAC isoforms.
Keywords: porin, mitochondria, outer membrane, Euglenozoa
1. Introduction
With a few known exceptions [1,2] the protein families of voltage-dependent anion channel (VDAC), Tom40 and Sam50 are common to all eukaryotic organisms. These proteins reside in the mitochondrial outer membrane (MOM) and facilitate metabolite exchange, protein translocation or membrane insertion. Sam50 is homologous to the bacterial Omp85 [3] while VDAC and Tom40 belong to the eukaryotic porin superfamily [4,5].
In general, the transmembrane β-barrel domain of porins is characterized by a so-called in-plug, which is responsible for function and stability of the pore [6]. In prokaryotic porins with 16 or 18 β-strands, in-plugs of α-helical or β-sheet character have been described, which are formed by an inward-folded loop of a β-hairpin within the β-barrel domain [7]. In eukaryotic porins, however, the in-plug is located N-terminally before the 19-stranded β-barrel domain and so far only α-helical structure has been reported [5,8].
Eukaryotic porins are responsible for transport of solutes across the MOM [8] and in plants VDAC has been implicated in tRNA import [9]. In Trypanosoma brucei, an ancient parasitic eukaryote, belonging to the phylum Euglenozoa (class: Kinetoplastida), only a single eukaryotic porin has been identified [4]. As the deletion mutant of this porin is still capable of solute transport [4], we searched for unidentified members of the eukaryotic porin family.
2. Material and methods
Sequences were collected from NCBI (nr database, 6 September 2011) and Bodo saltans genome project (http://www.sanger.ac.uk/resources/downloads/protozoa/bodo-saltans.html; [10]) by HMMER (http://hmmer.org/) and BLAST [11]. Redundant sequences were removed with cd-hit [12]. As input for the search with HMMER, we used the porin_3 HMM (PF01459) from PFAM (v. 24.0/v. 25.0 from 2009/2010; [13]) and an HMM generated from a multiple sequence alignment (MSA) of VDAC and Tom40 [5]. Our final database consists of 370 Tom40, 590 VDAC and 41 Euglenozoa sequences. Clustering was done with CLANS [14] in two dimensions at a p-value cutoff of 0.1. MSAs were calculated with MAFFT v. 6.847b (E-INS-I; [15]) from our database (maximal sequence identity 90%). As an extension of our previous method [5], we collected the alignment information from screening the parameter space described by a range of gap extension/insertion penalties (ep:0-0.9/op:1-3) in steps of 0.1 and amino acid substitution matrices (JTT100-300 [16]) in steps of 50.
3. Results
(a). Multiple porin-like sequences in Kinetoplastida
The high divergence of eukaryotic porin sequences among sequenced Protozoa and the low similarity to eukaryotic porins from Opisthokonta and Viridiplantae complicate their detection. In contrast to a recent conservative approach [17], we aimed for an enhanced detection capability and generated an HMM of the eukaryotic porin superfamily from an MSA containing both VDAC and Tom40 sequences [5]. In short, this MSA was constructed as follows. Based on the fact that the topology of the guide tree used for refinement in iterative, progressive MSA depends on the succession of sequences in the input file, we calculated multiple MSAs [5]. Regions most frequently aligned without insertions or deletions to β-strand core regions of the mVDAC-1 structure [8] were defined as transmembrane β-strands. Subsequently, we selected a single MSA, which contained all those assigned regions aligned to transmembrane β-strands of mVDAC-1. Thus, this MSA is—owing to the method applied—more accurate than a single MSA by chance [5]. Consequently, the resulting HMM is more sensitive than the porin_3 HMM from PFAM (electronic supplementary material, table S1). The HMM was used to search against NCBI's nr database. We found 32 different sequences in Euglenozoa, 23 being significant (electronic supplementary material, table S1). Therein, we confirmed the known VDAC of the euglenid Euglena gracilis and of the kinetoplastid T. brucei (TbVDAC; [4,18]), and identified two further porin-like sequences in the genome of T. brucei. We performed BLAST searches with all identified sequences and detected nine additional Kinetoplastida sequences with significant E-values (less than 1e–35).
To classify the identified sequences in Kinetoplastida, we created a CLANS clustering with all Tom40 and VDAC sequences obtained from our searches (figure 1). In the clustering 18 of our sequences, representing significant hits, are located near the main VDAC cluster with moderate connections (greater than 1e–9), including the experimentally described TbVDAC. The Euglenida sequence is also located near the main VDAC cluster, but connections to Kinetoplastida VDAC sequences (greater than 1e–7) are not as strong as to the main VDAC cluster (greater than 1e–10).
Figure 1.
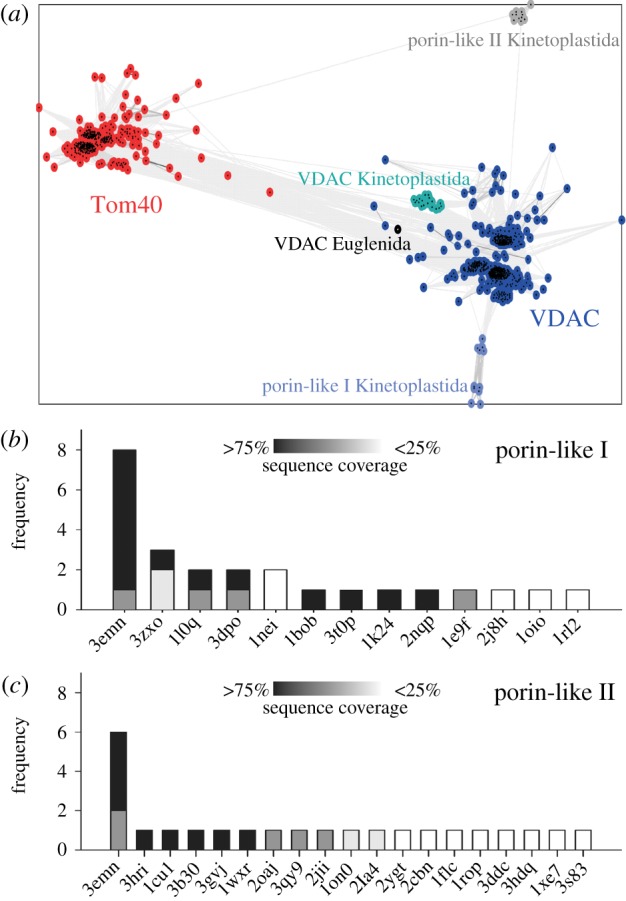
Classification of eukaryotic porin-like sequences. (a) Sequences of our database were clustered with CLANS by their pairwise similarities (BLAST p-values). Colour indicates cluster membership of sequences. Connections between sequences reflect p-values < 1e−5. (b and c) We have summed up how many times the same fold was detected by nine different fold recognition servers (electronic supplementary material, tables S2 and S3) for (b) Tb11.01.255 and (c) Tb927.4.1610. Sequence coverage of query sequence by the template is indicated by different shades of grey (from black (greater than 75%) to white (less than 25%)).
Two additional clusters of 11 Kinetoplastida sequences each are observed (figure 1). The ‘porin-like I’ (p1) cluster shows weak connections to other VDAC sequences (greater than 1e–5), whereas the ‘porin-like II’ (p2) cluster shows weak connections to both the Tom40 and VDAC cluster (p-values > 1e–5).
(b). The porin-like sequences from Kinetoplastida form a voltage-dependent anion channel-like fold
To substantiate the classification of the two newly identified porin-like sequences from the Kinetoplastida clusters as VDAC-like proteins, we submitted the two representative sequences from T. brucei TREU927 each to nine fold recognition servers (electronic supplementary material, table S2). For the p1 sequence eight out of nine servers detected a VDAC-like fold as best hit with good sequence coverage (figure 1b). No other transmembrane β-barrel was among the top three hits and the other reported folds were identified at most three times with often low sequence coverage. These additional folds have high β-sheet content and the models exhibit clear defects. For p2 five out of nine servers reported a VDAC-like fold as best hit again with good sequence coverage (figure 1c). All other folds were identified by at most a single server and, thus, most likely represent false positives.
From our database of eukaryotic porins, we calculated a composite MSA comprising the N-terminal helical region and transmembrane β-strands by the method described (figure 2; electronic supplementary material, figure S1). The N-terminal helical region shows the high similarity among the three kinetoplastid sequence clusters, exhibiting conserved properties of VDAC. All 19 β-strands can be assigned for the TbVDAC, p1 and p2 clusters. For a few of them, however, the transmembrane positioning might deviate by a few residues. As the N-terminal region of transmembrane β-barrels is less conserved than their C-terminal region [3], and as eukaryotic porins from available Protozoa genomes show a high degree of sequence diversity, future sequencing projects will enhance sequence space coverage and thereby improve an MSA of eukaryotic porins.
Figure 2.
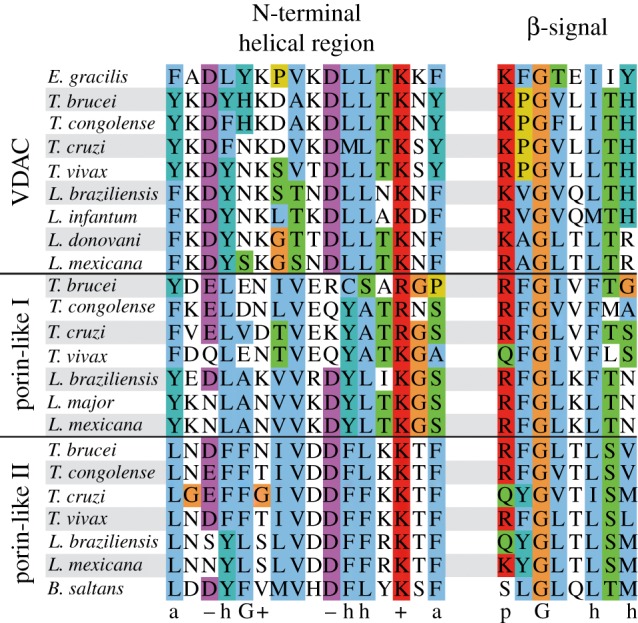
Conserved elements in eukaryotic porins. Euglenozoa sequences from the composite MSA of eukaryotic porins are shown. For each sequence the region is shown, which aligns most frequently to the helical region (left) and to the last β-strand (right) of the crystal structure of mVDAC-1. Amino acids are coloured with Jalview (Clustalx colour code), reflecting the conservation of the respective alignment column. On the bottom conserved properties derived from the MSA of VDAC [5] and the β-signal definition [19,20] are displayed: polar (p), aromatic (a), glycine (G), hydrophobic (h), positive (plus symbol), negative charge (minus symbol).
In all known mitochondrial β-barrels the last β-strand encodes a so-called β-signal serving as a targeting signal [19,20]. In our sequences, the last β-strand contains a motif, which can be interpreted as the β-signal—only its last position is polar instead of hydrophobic in VDAC and p1 sequences (figure 2). This supports the possibility that the identified sequences could be targeted and integrated into the MOM.
4. Discussion
In summary, we suggest that all three eukaryotic porin-like sequences have a VDAC-like fold, because they contain the N-terminal helical region, 19 β-strands and a putative β-signal in the last β-strand. Orthologues of the two newly identified eukaryotic porin-like sequences in T. brucei are also present in other Kinetoplastida (electronic supplementary material, table S1). Further support is provided by the fact that multiple isoforms developed several times independently within the VDAC family [21]. Also, the two identified genes are transcribed in both life stages of T. brucei at a similar albeit much lower level than TbVDAC [22,23]; both proteins were detected via mass spectrometry in the mitochondrial fraction [24,25].
In T. brucei, metabolites are still transported across the MOM in the TbVDAC knockout [4]. Thus, the p1 sequence (Tb11.01.255) is a promising candidate for a second VDAC isoform, because it is more closely related to VDAC than p2 and also its sequence length is VDAC-like (TbVDAC: 270 amino acids, p1: 279 amino acids; electronic supplementary material, table S1).
A putative functional assignment for the p2 sequence (Tb927.4.1610), however, is difficult. As the protein translocase in the MOM of Trypanosoma has recently been identified [26], a VDAC-like function of Tb927.4.1610 in solute transport is likely. The latter is also supported by VDAC-like features in the N-terminal helical region. Additionally, a function in tRNA transport of p1 and/or p2 sequences is conceivable as it was shown that TbVDAC is not involved in tRNA import [4].
Thus, based on sequence analyses, we present evidence for the existence of two additional porin-like proteins in Kinetoplastida, which have already been confirmed to reside in mitochondria. The functional prediction has now to be challenged by biochemical approaches.
Acknowledgements
We thank Dennis Gessmann and Stephan Nussberger for discussions. We apologize to our colleagues from fold recognition as we could only cite their work in the electronic supplementary material due to space restrictions. N.F. was funded by a fellowship from CMP and TRAM. The project was funded by Volkswagenstiftung and Deutsche Forschungsgemeinschaft in the frame of CEF and SFB807 project 17 to E.S.
References
- 1.Cavalier-Smith T. 2010. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345 10.1098/rsbl.2009.0948 (doi:10.1098/rsbl.2009.0948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burri L., Williams B. A., Bursac D., Lithgow T., Keeling P. J. 2006. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc. Natl Acad. Sci. USA 103, 15 916–15 920 10.1073/pnas.0604109103 (doi:10.1073/pnas.0604109103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredemeier R., Schlegel T., Ertel F., Vojta A., Borissenko L., Bohnsack M. T., Groll M., von Haeseler A., Schleiff E. 2007. Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J. Biol. Chem. 282, 1882–1890 10.1074/jbc.M609598200 (doi:10.1074/jbc.M609598200) [DOI] [PubMed] [Google Scholar]
- 4.Pusnik M., Charrière F., Mäser P., Waller R. F., Dagley M. J., Lithgow T., Schneider A. 2009. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 26, 671–680 10.1093/molbev/msn288 (doi:10.1093/molbev/msn288) [DOI] [PubMed] [Google Scholar]
- 5.Gessmann D., Flinner N., Pfannstiel J., Schlösinger A., Schleiff E., Nussberger S., Mirus O. 2011. Structural elements of the mitochondrial preprotein-conducting channel Tom40 dissolved by bioinformatics and mass spectrometry. Biochim. Biophys. Acta 1807, 1647–1657 10.1016/j.bbabio.2011.08.006 (doi:10.1016/j.bbabio.2011.08.006) [DOI] [PubMed] [Google Scholar]
- 6.Naveed H., Jackups R., Jr, Liang J. 2009. Predicting weakly stable regions, oligomerization state, and protein-protein interfaces in transmembrane domains of outer membrane proteins. Proc. Natl Acad. Sci. USA 106, 12 735–12 740 10.1073/pnas.0902169106 (doi:10.1073/pnas.0902169106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565, 308–317 10.1016/S0005-2736(02)00577-1 (doi:10.1016/S0005-2736(02)00577-1) [DOI] [PubMed] [Google Scholar]
- 8.Ujwal R., Cascio D., Colletier J. P., Faham S., Zhang J., Toro L., Ping P., Abramson J. 2008. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl Acad. Sci. USA 105, 17 742–17 747 10.1073/pnas.0809634105 (doi:10.1073/pnas.0809634105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salinas T., Duchêne A. M., Maréchal-Drouard L. 2008. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 33, 320–329 10.1016/j.tibs.2008.04.010 (doi:10.1016/j.tibs.2008.04.010) [DOI] [PubMed] [Google Scholar]
- 10.Jackson A. P., Quail M. A., Berriman M. 2008. Insights into the genome sequence of a free-living Kinetoplastid: Bodo saltans (Kinetoplastida: Euglenozoa). BMC Genomics 9, 594. 10.1186/1471-2164-9-594 (doi:10.1186/1471-2164-9-594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 10.1093/bioinformatics/btl158 (doi:10.1093/bioinformatics/btl158) [DOI] [PubMed] [Google Scholar]
- 13.Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 10.1093/nar/gkp985 (doi:10.1093/nar/gkp985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frickey T., Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 10.1093/bioinformatics/bth444 (doi:10.1093/bioinformatics/bth444) [DOI] [PubMed] [Google Scholar]
- 15.Katoh K., Kuma K., Toh H., Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 10.1093/nar/gki198 (doi:10.1093/nar/gki198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D. T., Taylor W. R., Thornton J. M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 17.Likic V. A., Dolezal P., Celik N., Dagley M., Lithgow T. 2010. Using hidden Markov models to discover new protein transport machines. Methods Mol. Biol. 619, 271–284 10.1007/978-1-60327-412-8_16 (doi:10.1007/978-1-60327-412-8_16) [DOI] [PubMed] [Google Scholar]
- 18.Singha U. K., Sharma S., Chaudhuri M. 2009. Downregulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell 8, 1418–1428 10.1128/EC.00132-09 (doi:10.1128/EC.00132-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutik S., et al. 2008. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 132, 1011–1024 10.1016/j.cell.2008.01.028 (doi:10.1016/j.cell.2008.01.028) [DOI] [PubMed] [Google Scholar]
- 20.Imai K., Fujita N., Gromiha M. M., Horton P. 2011. Eukaryote-wide sequence analysis of mitochondrial β-barrel outer membrane proteins. BMC Genomics 12, 79. 10.1186/1471-2164-12-79 (doi:10.1186/1471-2164-12-79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young M. J., Bay D. C., Hausner G., Court D. A. 2007. The evolutionary history of mitochondrial porins. BMC Evol. Biol. 7, 31. 10.1186/1471-2148-7-31 (doi:10.1186/1471-2148-7-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38, 4946–4957 10.1093/nar/gkq237 (doi:10.1093/nar/gkq237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veitch N. J., Johnson P. C., Trivedi U., Terry S., Wildridge D., MacLeod A. 2010. Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genomics 11, 124. 10.1186/1471-2164-11-124 (doi:10.1186/1471-2164-11-124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acestor N., Panigrahi A. K., Ogata Y., Anupama A., Stuart K. D. 2009. Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics 9, 5497–5508 10.1002/pmic.200900354 (doi:10.1002/pmic.200900354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panigrahi A. K., Ogata Y., Zíková A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450 10.1002/pmic.200800477 (doi:10.1002/pmic.200800477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pusnik M., Schmidt O., Perry A. J., Oeljeklaus S., Niemann M., Warscheid B., Lithgow T., Meisinger C., Schneider A. 2011. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 10.1016/j.cub.2011.08.060 (doi:10.1016/j.cub.2011.08.060) [DOI] [PubMed] [Google Scholar]


