Abstract
Voltage-gated potassium (Kv) channels are tetrameric assemblies of transmembrane Kv proteins with cytosolic N- and C-termini. The N-terminal domain of Kv1 proteins binds to β-subunits, but the role of the C-terminus is less clear. Therefore, we studied the role of the C-terminus in regulating Kv1.5 channel and its interactions with Kvβ-subunits. When expressed in COS-7 cells, deletion of the C-terminal domain of Kv1.5 did not affect channel gating or kinetics. Co-expression of Kv1.5 with Kvβ3 increased current inactivation, whereas Kvβ2 caused a hyperpolarizing shift in the voltage-dependence of current activation. Inclusion of NADPH in the patch pipette solution accelerated the inactivation of Kv1.5-Kvβ3 currents. In contrast, NADP+ decreased the rate and the extent of Kvβ3-induced inactivation and reversed the hyperpolarizing shift in the voltage-dependence of activation induced by Kvβ2. Currents generated by Kv1.5ΔC+Kvβ3 or Kv1.5ΔC+Kvβ2 complexes did not respond to changes in intracellular pyridine nucleotide concentration, indicating that the C-terminus is required for pyridine nucleotide-dependent interactions between Kvβ and Kv1.5. A glutathione-S-transferase (GST) fusion protein containing the C-terminal peptide of Kv1.5 did not bind to apoKvβ2, but displayed higher affinity for Kvβ2:NADPH than Kvβ2:NADP+. The GST fusion protein also precipitated Kvβ proteins from mouse brain lysates. Pull-down experiments, structural analysis and electrophysiological data indicated that a specific region of the C-terminus (Arg543-Val583) is required for Kvβ binding. These results suggest that the C-terminal domain of Kv1.5 interacts with β-subunits and that this interaction is essential for the differential regulation of Kv currents by oxidized and reduced nucleotides.
Keywords: Voltage-gated potassium channel, whole-cell patch clamp, Kvβ, pyridine nucleotides, redox, aldo-keto reductase
Introduction
Voltage-gated potassium (Kv) channels generate outward K+ currents that contribute to the resting membrane potential and regulate the frequency and the duration of the action potential [6]. Several families of Kv channels (1-9) have been described. Of these, the Shaker family of Kv channels (Kv1) is the most widely distributed. Members of this family play an important role in several physiological processes, such as hypoxic pulmonary vasoconstriction, oxygen sensing, cell proliferation, and apoptosis [6-8,39]. Abnormal activity of these channels has been linked to the development of pulmonary hypertension, arrhythmias, and epilepsy [35].
Native Kv1 channels are multi-protein aggregates. The conducting pore of these channels is constructed by the assembly of 6 transmembrane regions, while both the N and C termini of the transmembrane monomers are located in the cytosol [16,30,38,7,14]. The T1 domain of the N-terminus of Kv1 channels associates with the auxiliary Kvβ subunits [29,42]. In the mammalian genome these proteins are encoded by three different genes - Kvβ1-3 [27]. The mature Kvβ proteins possess a variable N-terminus and a conserved C-terminal domain. The C-terminus of the Kvβ protein folds into an (α/β)8-TIM barrel that displays high sequence homology with the aldo-keto reductase (AKR) superfamily [22,1]. In heterologous expression systems, the N-terminal domains of Kvβ1 and Kvβ3 proteins impart inactivation to otherwise non-inactivating Kv currents, whereas Kvβ2, which possesses a shorter N-terminus, shifts the voltage-dependence of Kv channels to lower voltages and accelerates inactivation of partially inactivating Kv currents [27].
The Kvβ proteins bind to pyridine nucleotides with high affinity [17] and it has been shown that pyridine nucleotide binding regulates Kvβ-mediated inactivation of Kv current such that reduced pyridine coenzymes (NAD[P]H) support inactivation, whereas the binding of oxidized nucleotides (NAD[P]+) abolishes Kvβ-mediated inactivation of Kv current [34,33]. The differential effects of reduced and oxidized nucleotides suggest that the Kv currents might be sensitive to changes in the cellular redox state induced by variations in metabolic activity or oxygen concentration. Recent work has shown that Kvβ proteins catalyze the reduction of a wide range of aldehydes and ketones [32,40], and that catalysis affects Kvβ-induced inactivation of Kv1 currents [40,26]; however, the physiological significance of Kvβ catalysis in regulating the sensitivity of Kv channels to changes in oxygen concentration or fluctuations in intermediary metabolism remains to be fully assessed.
The Kvβ subunits bind to the N-terminal T1 domain of the α subunit of the Kv1 channel. Crystal structure of the T1-β complex shows that the cytosolic N-termini of Kvα interact with each other to form a docking platform on which the tetramers of the β-subunit are arranged in a fourfold symmetric T14β4 complex [10]. The binding of β-subunits to the T1 domain appears to be essential for α-β interaction as it has been shown that genetic deletion of this domain disrupts the association of the β subunit with Kv channels and prevents Kvβ-induced inactivation of Kv current [10,42]. In contrast, the role of the cytosolic C-terminal domain of Kvα subunits is less clear. It has been suggested that the disordered segments of the C-terminus utilize an inter-molecular fishing rod-like mechanism for binding to scaffold proteins [20,21], however, little is known about the structural features of the C-terminus that mediate its interaction with other proteins. Extant X-ray structures provide no information regarding its position within the α-β complex, although a poorly defined extra density due to the C-terminus overlying the active site of the β-subunit has been observed in electron density maps of Kv1.2-Kvβ2 complex [18]. Moreover, electron microscopic studies and single particle analysis [31] of the Shaker K+ channel bound to Kvβ2 indicate that the C-terminus of the α subunit is in close contact with the β2-tetramer. The C-terminal loop of α changes its position relative to the transmembrane domains when β is bound to the channel and in the α-β complex this loop is located within a crevice close to the pyridine nucleotide binding site of Kvβ. However, the functional implications of this potential interaction have not been examined.
Genetic deletion of the C-terminal loop of Kv1.5 does not abolish the binding of the β-subunits to the T1-domain of the channel [37,10], indicating that simultaneous binding to both the N- and the C-termini of Kv channels is not essential for the tethering of β-subunits to the Kv channel. However, it remains unclear whether the interaction between the C-terminus of Kvα with the β-subunits regulates Kvβ-mediated inactivation of Kv current or its dependence on pyridine nucleotides. The current study was therefore designed to examine the role of the C-terminus of the α subunit of Kv channel in β-mediated inactivation of Kv current. Our results show that deletion of the C-terminus induces minimal changes in Kvβ-mediated inactivation of Kv currents, but it abolishes pyridine nucleotide-dependent changes in channel gating. These observations suggest a unique mechanism by which changes caused by pyridine nucleotide binding are transmitted to a membrane spanning ion channel via interaction with its cytosolic domain.
Materials and methods
Constructs
The rat Kv1.5 (accession # M27158) mammalian expression construct in pIRES-hr-GFP-1α vector (Stratagene) was generated as described previously [34]. To generate Kvα1.5ΔC expression constructs, the last 56, 37, and 18 amino acid residues were deleted from Kvα1.5 by inserting a stop codon after the amino acid position 546, 565, 584 by using the PCR technique. Primer sequences are listed in the Supplemental Material section. The Kvβ3 cDNA (NM_010599) was amplified by RT-PCR from mouse heart cDNA library and also inserted into pIRES-hr-GFP-1α vector (Stratagene). All constructs were sequenced in their entirety to rule out cloning errors.
For bacterial expression, the entire Kvβ3 cDNA was inserted into pET-28a vector in frame with the His tag. The construct for Kvβ2 (AKR part, amino acid residues 39-367) expression was generated as described previously [17] . GST-fusion constructs of 60, 38 or 19 C-terminal amino acid residues of Kvα1.5 attached to the GST at its C-terminus were generated using a pET-49b vector (Novagen). For control, an unrelated peptide in pET-49b vector was used (50 amino acid residues, 315 amino acids total, MW 35,332 Da). The GST-C60 construct in pDEST-26 vector (Invitrogen) was used for quantitative Kvβ2 binding assay using silver staining.
Transfection of COS-7 cells
COS-7 cells were used as a mammalian expression system to study electrophysiological properties of Kvα1.5 WT or ΔC truncated channel protein with or without Kvβ3 or Kvβ2 subunits. Expression constructs were transfected into COS-7 cells using Lipofectamine reagent (Invitrogen) according to the manufacturer’s protocol. In brief, 5μ g of DNA for each subunit were mixed with 11.3 μl of Lipofectamine and incubated with 70% confluent cells in serum-free DMEM for 4 h. The cells were then allowed to grow in 10% FBS DMEM for 48 h before use for Western blotting or electrophysiological recordings. Green fluorescent protein (GFP) expressed by a pIRES-GFP vector served as a reporter to identify cells for patch-clamp recording.
Whole-cell patch clamp
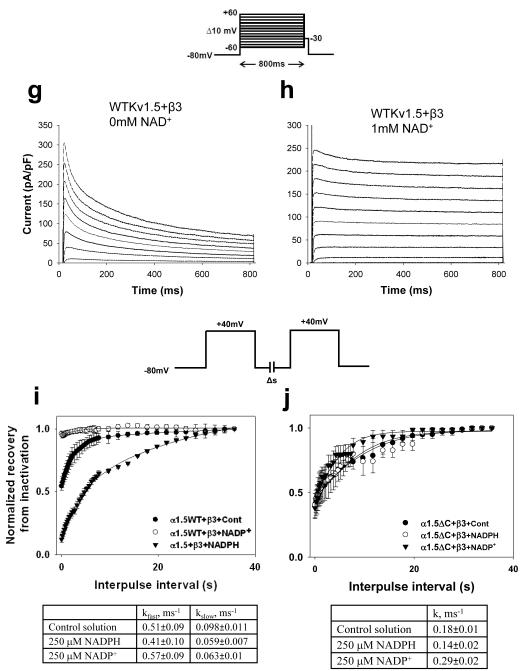
Whole cell currents were recorded and analyzed as described before [34]. The COS-7 cells were placed in a 0.25 ml recording chamber and perfused with normal Tyrode’s solution containing (in mM): NaCl 135, MgCl2 1.1, CaCl2 1.8, KCl 5.4, HEPES 10, glucose 10, pH 7.4 adjusted with NaOH at 2.5 ml/min at room temperature (21-23°C). Voltage-clamp experiments were performed in the whole-cell configuration of the patch-clamp method by using Axopatch-200B patch-clamp amplifier (Axon Instruments, Foster City, CA). Cells were patched with borosilicate glass pipettes (2-4 MΩ) filled with the normal pipette solution containing (in mM): K-aspartate 100, KCl 30, MgCl2 1, HEPES 5, EGTA 5, Mg-ATP 5, Na2-creatine phosphate 5, pH adjusted to 7.2 with KOH (24). The cells were depolarized for 800 ms from a holding potential of −80 mV to potentials ranging from −60 mV to +60 mV in 10mV voltage steps at 0.1 Hz. The current data are expressed as pA/pF by normalizing the current for each cell to the cell capacitance. The extent of current inactivation, voltage-dependence of activation and voltage-dependence of inactivation were quantified as described before [34]. The tail currents were normalized to peak tail current at +50mV and plotted against the potential of the depolarizing step and analyzed using a Boltzmann function. To investigate the voltage-dependence of inactivation, a two-pulse protocol was used. The cell was depolarized to different conditioning potentials from −70 to +60 mV in 10 mV steps for 1s from a holding potential of −80 mV, followed by a 10 ms step to −80 mV and an 800 ms test step to +50 mV. Mean normalized values (+50mV) of peak outward current were plotted against pre-pulse potential and best fits of a single Boltzman function to the data were determined. Recovery from inactivation was recorded by using the two pulse protocol, from a holding potential of −80mV the cell was depolarized to +40 mV for 800 ms and returned to −80mV. The second pulse was applied at varying time intervals from 10-2500 ms following the return to −80 mV holding. For analysis, the peak of the second pulse was measured, normalized and plotted against the inter-pulse interval. Data points were fitted using mono or bi exponential equations for the best fit with correlation coefficients greater than 0.98.
Protein-protein interactions
Animal procedures were approved by the University of Louisville Institutional Animal Care and Use Committee. Male C57BL/6 mice were euthanized by intraperitoneal injection of pentobarbital (50 mg/kg). The brains were harvested, flash frozen in liquid nitrogen, stored at −80°C, and used for GST-pulldown assays. The GST-fusion proteins of 60, 38 or 19 C-terminal amino acid residues of Kvα1.5 or scrambled peptide attached to the GST at its C-terminus (30 μ g each) were mixed with the crude extract of the Kvβ2 or Kvβ3 expressing bacteria (350 μ g) or murine brain (1 mg) in 1 ml of 20 mM potassium phosphate buffer, 1 mM EDTA, pH 6.0 for 30 min, and the resulting protein complexes were pulled down using GSH beads. The beads were washed twice with PBS and the protein bound to the bead was eluted with 10 mM GSH. An equal fraction (by volume) of the eluate from each sample was subjected to SDS-PAGE electrophoresis and transblotted. Antibodies were obtained from the following sources: pan-Kvβ and Kv1.5 N-terminus (Santa-Cruz Biotechnologies), Kv1.5 C-terminus (Alomone Labs), Kvβ2, β1.1, β1.2 (Neuromab), GST (Novagen).
Quantitative GST-C60 - Kvβ2 interaction
Kvβ2 purified from bacteria contains tightly-bound cofactor. Apo-Kvβ2 and the nucleotide-bound forms of Kvβ2 were prepared as described in our previous publications [17,32]. Briefly, to remove the cofactor and obtain Kvβ2 preparation with defined nucleotide content, all bound NADPH was oxidized with 600 μM 4-nitrobenzaldehyde followed by overnight dialysis. This created nucleotide-free enzyme (apo-Kvβ2). This Kvβ2 preparation was incubated with 3-fold molar excess of NADPH and NADP+, and defined amounts of resulting complexes or apo-Kvβ2 were used in a GST-pulldown assay as described in the main text. GST-C60 protein expressed from pDEST-26 vector (Invitrogen) was used in this series of experiments to avoid band overlap due to similar molecular weight of fusion construct from pET-49b vector to that of Kvβ2. Equal aliqouts of the eluate from each incubation were applied on an SDS-PAGE and silver stained; band intensities were quantified using ImageQuant TL (GE Healthcare). Kvβ2 band intensity was normalized by GST-C60 input.
Western blot Analyses
Transfected COS-7 cells were washed with cold phosphate buffer saline and collected by scraping. The cell pellet was disrupted in buffer containing 20 mM potassium phosphate, 1 mM EDTA, pH 7.4 , a protease inhibitor cocktail (Sigma) 1:100 and 1mM DTT and centrifuged at 1000xg to remove cell debris. Enriched membrane fractions were obtained by ultracentrifugation at 100,000xg at 4°C. The pellet containing surface and intracellular membrane was solubilized in 2% SDS containing 20mM Tris, 1mM EDTA, pH 7.4. The cytosol was extracted in the supernatant. Protein was measured using the Lowry’s method [19]. For Western analysis, 10 μ g protein was loaded onto 12% polyacrylamide gels and separated by gel electrophoresis on a Mini-Protean system (BioRad). Immunoreactivity was visualized by chemiluminescence using a Typhoon 9400 detector (Amersham Biosciences), and the intensity of specific bands was analyzed with ImageQuantTL software (Amersham Biosciences). Results are presented as ratios of the intensity of specific bands to actin.
Bioinformatics analysis
Sequence alignment for Drosophila shaker potassium channel protein, rat Kv1.2, and rat Kv1.5 was carried out by using ClustALW [13]. Secondary structure propensity and disorder prediction was done by using the Jpred algorithm and PONDR-FIT-VLXT, respectively [5,28,41]. Identification of Molecular Recognition Feature (MoRF) was completed by the MoRF-II predictor [4]. To identify the binding segments in the C-terminus region of Kv1.5, we used the AIBS (Anchor Identified Binding Sites) strategy in which we utilized Anchor as the analysis software for prediction based analysis [23].
Statistical Analysis
Data are reported as mean + SEM. Data were analyzed using SigmaStat 3.0 with paired or unpaired t test or ANOVA followed by Student-Newman-Keuls (SNK) test for all pairwise comparisons. P values < 0.05 were considered significant.
Results
Expression and intracellular localization of Kv1.5ΔC deletion mutants
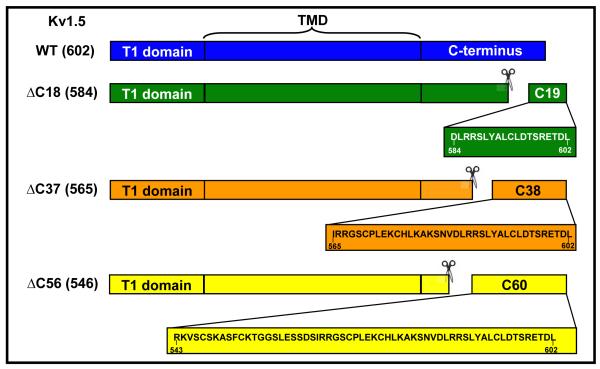
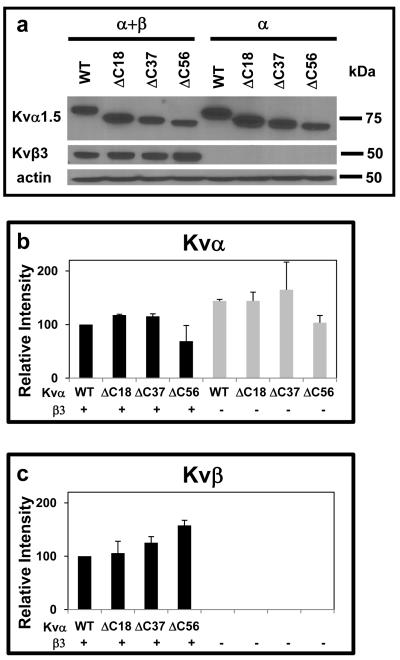
To delineate the function of the C-terminus and its interaction with Kvβ, three different C-terminal deletion constructs were generated, in which nucleotides coding for the terminal 18, 37, and 56 amino acids were deleted (Scheme I). Western analysis of COS-7 cells transfected with WT, Kv1.5ΔC18, ΔC37, and ΔC56 using anti-Kv1.5 antibodies showed no statistically significant differences in the protein abundance of Kv1.5 mutants (Fig. 1a), indicating that deletion of the C-terminus does not prevent Kv1.5 expression.
Scheme I. Construction of Kv1 deletion mutants and GST-fusion proteins.
The full-length Kv protein consists of 3 distinct domains: the N-terminus or the T1-domain, a trans-membrane domain (TMD) which forms the ion-conducting pore, and a C-terminal cytosolic loop. Deletion mutants were generated by successive deletion of 18, 37, and 58 amino acids from C-terminal end as indicated to the left of the drawings. In a separate series of experiments, peptides corresponding to the amino acid sequence spanning the last 19, 38, and 60 amino acids were expressed fused with GST.
Fig. 1. Expression of WT and KvΔC mutants and Kvβ3 in COS-7 cells.
(a) Western blot of the microsomal fraction of COS-7 cell extracts transfected with expression constructs encoding WT Kv1.5 or its deletion mutants either alone or coexpressed with a Kvβ3 expression plasmid. Cells were harvested 48 h after transfection. (b-c) Densitometric analysis of band intensities of Kv1.5 deletion constructs (b), and Kvβ (c). Kv band intensities were normalized by the intensity of the actin band, and plotted as a percentage of the intensity obtained in the Kv1.5WT+Kvβ3 transfection.
During early biogenesis, Kv1 channels co-assemble with Kvβ subunits in the ER [24], hence deletion of the C-terminus of Kv channels could potentially disrupt subunit assembly or maturation. Therefore, to determine how deletion of the C-terminus affects the expression of the α-β complex, COS-7 cells were transfected with Kvβ3 and WT Kv1.5 or Kv1.5ΔC18, ΔC37, or ΔC56. Co-expression with Kvβ3 led to a 1.5-fold decrease in the expression of WT Kv1.5. This is similar to the reported decrease in the expression of Kv1.5 when co-expressed with Kvβ1.3 [34]. A similar decrease in expression of each of the three mutants was observed upon co-expression with Kvβ3. Moreover, Kvα deletion mutants and Kvβ were found in the microsomal, not the cytosolic fraction of the cell (data not shown), however, there is no statistically significant difference between the expression of WT and deletion mutants of Kv1.5 in the presence of Kvβ3 (Fig. 1b). Moreover, deletions in the C-terminus did not affect Kvβ3 expression (p=0.28, ANOVA; Fig. 1c). Together, these data indicate that deletion of different segments of the C-terminus does not significantly affect the expression or the membrane localization of Kv1.5 in COS-7 cells.
Effects of C-terminal deletion on Kv currents
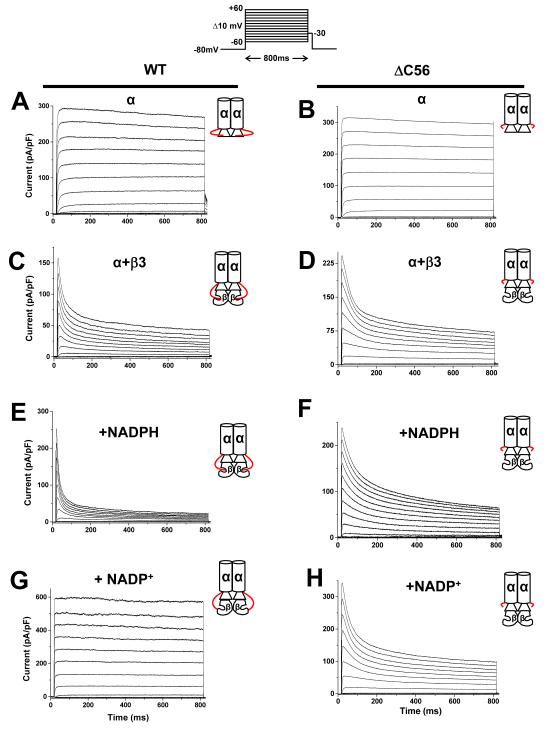
To examine the role of the C-terminus in Kv function, Kv currents were recorded from COS-7 cells transfected with WT and ΔC56 Kv1.5. Cells transfected with Kvα1.5WT cDNA showed rapidly-activating outward currents with the time-to-peak activation of 62±14 ms, and activation time constant of 1.7±0.3 ms (n = 8; Fig 2a). In general, Kv current characteristics of ΔC56 Kv1.5 (Fig. 2b) were similar to WT Kv1.5 (Fig. 2a); however, there was a small shift in the Vh of activation from −6.5±0.2 to −3.2 ±0.6 mV, and a significant decrease in the time-to-peak activation from 62±14 to 22±4.4 ms (Table I). Nevertheless, the extent of inactivation at the end of the pulse of WT Kv1.5 (5.3±1%; at +50mV) was indistinguishable from that of ΔC56 Kv1.5 (4.7±1.5%; at +50mV). These observations suggest that although deletion of the C-terminal domain of Kv1.5 has modest effect on the activation parameters of the channel, it does not affect channel inactivation.
Fig. 2. Effect of Kv1.5 C-terminus deletion on Kvα-β currents.
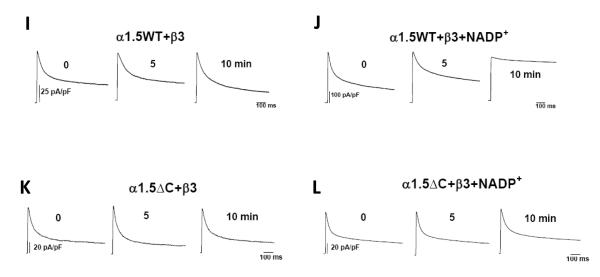
Whole-cell currents recorded from COS-7 cells cotransfected with plasmids encoding WT or ΔC56 Kv1.5 alone or coexpressed with Kvβ3. Outward currents were recorded using the patch-clamp technique (n=6-9 cells each group). Panels show representative current traces recorded with pipette containing control internal solution from cells expressing Kv1.5WT (a); or Kv1.5ΔC56 (b); Kv1.5+β3 (c); Kv1.5ΔC56+β3 (d). Representative outward currents recorded from cells transfected with WT Kv1.5+β3 with patch pipettes containing 250 μ M NADPH (e) or 1mM NADP+(g) or cells expressing ΔC56 Kv1.5+β3 with 250 μM NADPH (f) or 1mM NADP+(h) in the patch pipette. The inset on the top panel shows the pulse protocol in which the membrane potential was held at −80 mV and outward currents were generated by depolarizing the cell from −60 mV to +60mV in 10mV increments for 800 ms. Inset to each trace is depicting the α with or without the β-subunit, The C-terminus of the Kvα-subunit extending from α to the β-subunit is shown in red. Representative traces shown in panels I-L are time course effects depicting the steady-state effects of NADP+. Panels I and K shows the currents from COS-7 cells co-expressing Kv1.5 or KvΔC with Kvβ3 patched with control internal solution. Panels J and L show cells patched with solution containing NADP+ (1mM).
Table 1.
Regulation of Kvα1.5 and Kvα1.5ΔC currents by Kvβ3
| Vh act (mV) |
Time to peak act (ms) |
Act time constant |
Vh inact (mV) |
||
|---|---|---|---|---|---|
| Kvα1.5WT | alone | −6.5 ± 0.26 | 62 ± 14 | 1.7 ± 0.3 | --a |
| +β3 | −19.7 ± 0.6* | 7.1 ± 0.5* | 1.5 ± 0.4 | −22.2 ± 0.9 | |
| Kvα1.5ΔC56 | alone | −3.2 ± 0.6† | 22 ± 4.4† | 1.1 ± 0.2 | --a |
| +β3 | −21.5 ± 1.5* | 6.0 ± 0.6* | 1.1 ± 0.2 | −20.5 ± 0.3 |
Due to minimal inactivation, the kinetics and the voltage-dependence of inactivation of Kvα currents could not be reliably estimated
P<0.05 Kvα1.5 (WT or ΔC) alone versus coexpressed with Kvβ3
P<0.05 Kvα1.5WT versus Kvα1.5ΔC56
Role of Kv C-terminus in interactions between α and β-subunits
To delineate the role of the C-terminus of Kv1.5 in its interaction with β-subunit, we studied how the deletion of this domain would affect the modification of Kv1.5 currents by two Kvβ subunits – Kvβ3, which accelerates the inactivation of Kv1.5 and Kvβ2 which does not affect inactivation but alters the kinetics and voltage-dependence of channel activation [27]. As expected, co-expression with Kvβ3 increased the rate of inactivation of WT Kv1.5 currents. Moreover, the presence of Kvβ3 induced a hyperpolarizing shift in the activation potential and decreased time-to-peak activation for WT Kv1.5 (see below). The nature and the extent of current modification induced by Kvβ3 were similar when the subunit was expressed with Kv1.5ΔC56 (Fig. 2 c and d). The overall magnitude of the current generated in COS-7 cells expressing Kv1.5WT+Kvβ3 or Kv1.5ΔC56+Kvβ3 was similar (248±60 pA/pF; n=8 versus 259±80 pA/pF; n=6). The Vh activation (mV) and the slope (k) shifted from −6.5±0.26; 5.8±0.2 to −19.7±0.6; 4.3±0.6, respectively, when Kvβ3 was coexpressed with WTKvα1.5, and a similar hyperpolarizing shift was observed when Kvβ3 was coexpressed with ΔC56 (Table 1 and Fig. 3). Moreover, Kvβ3 decreased time-to-peak activation of Kvα WT and ΔC56 mutant to a similar extent.
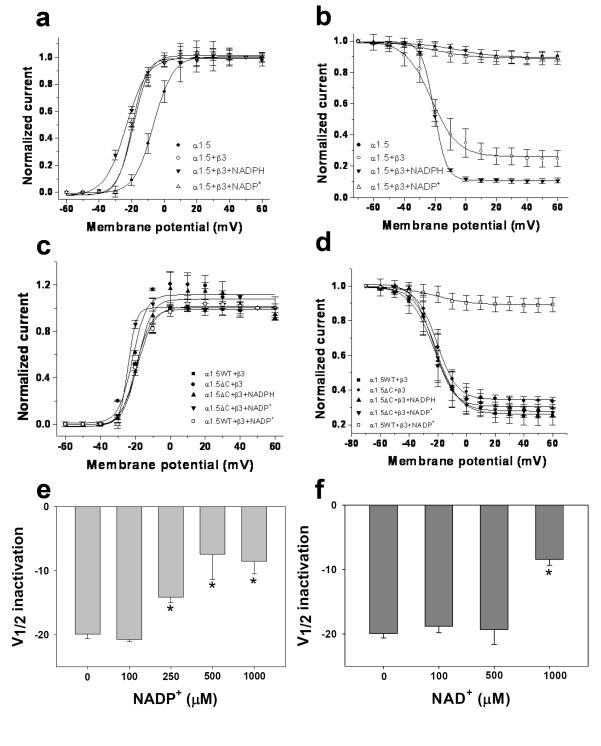
Fig. 3. Regulation of the voltage-dependence of Kv-Kvβ current by pyridine nucleotides.
Outward currents were recorded from COS-7 cells cotransfected with Kv1.5WT or Kv1.5ΔC56 with Kvβ3. (a; c) The voltage-dependence of activation was determined by normalizing outward currents at indicated voltages to +50mV; and (b; d) voltage-dependence of inactivation determined using a two pulse protocol (see Materials and Methods for details). Outward Kv currents for Kv1.5+β3 (filled square), Kvα1.5ΔC+β3 (filled circles) recorded with either the control internal solution or that containing NADPH (250μM; filled triangles) or NADP+ (1mM; inverted filled triangles). Note: In panel d, Kv1.5ΔC+ β3, inclusion of NADP+ (inverted filled triangle) did not cause change in the voltage-dependence of inactivation as compared with Kv1.5WT+ β3+NADP+ (open square). Alteration in voltage dependence of Kv current inactivation (WTKv1.5 +Kvβ3) was determined at different concentrations of NADP+ (e) and NAD+ (f). Compared to inactivation using standard pipette solution of 78% (g), addition of 1mM NAD+ to patch pipette solution decreased inactivation at 800ms in the Kv1.5WT+β3 couple to 12% (h). The recovery from inactivation for Kv1.5WT+β3 (i) and KvΔC56+β3 (j) with or without pyridine nucleotides was plotted against the interpulse interval and fitted parameters are shown in panels below.
Coexpression of Kv1.5 with Kvβ2 did not affect the rate of inactivation of outward currents, however, Kvβ2 induced a hyperpolarizing shift in the voltage-dependence of activation from −6.5±0.2 to −20.6±0.7 mV; n=5; P<0.05 (Fig. 4c). A similar shift in the voltage-dependence of activation was observed when Kvβ2 was expressed with Kv1.5ΔC. Collectively, these data suggest that even in the absence of the C-terminus, Kv1.5 co-assembles with β-subunits, and that deletion of this loop does not abrogate Kvβ3 or Kvβ2-induced changes in the kinetics or the voltage-dependence of Kv1.5 currents.
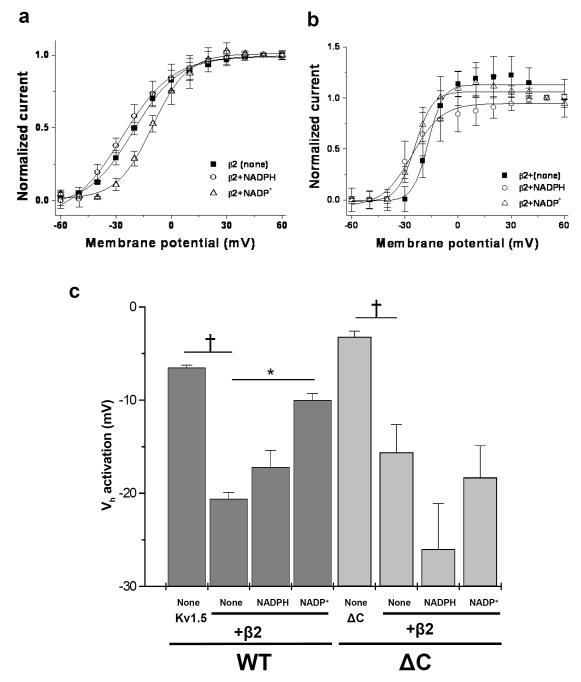
Fig. 4. Effect of C-terminal deletions on regulation of Kv1.5+Kvβ2 currents by pyridine nucleotides.
COS-7 cells were transfected with Kv1.5WT or KvΔC coexpressed with Kvβ2. (a, b) The voltage-dependence of activation was determined by normalizing outward currents at indicated voltages to +50mV. Kv currents were recorded with either control internal solution or solution containing NADPH or NADP+ in the patch pipette. (c) The Vh of activation measured by the deactivating tail currents is plotted for different groups; see Material and Methods for details. *p<0.05 compared with none within each group, †p<0.05 compared with WT (none) or ΔC (none) within each group.
Role of pyridine nucleotides in regulating Kv currents
Given that deletion of the C-terminus of Kv1.5 did not affect its basal interactions with β-subunits, we next examined whether the C-terminal domain modulates the effects of pyridine nucleotides. Previous work from our laboratory has shown that inactivation of Kv1.5 currents induced by Kvβ1.3 is abolished when the β-subunit is bound to NADP+ [34]. Pan et al. have also reported that perfusion with NADP+ reduces inactivation of Kv1.1-Kvβ1 couple [26]; however, the effects of pyridine nucleotides on the regulation of Kv1.5 currents by Kvβ3 or Kvβ2 have not been studied. Hence, to examine whether the Kv1.5 C-terminus participates in nucleotide sensing, we first studied the NADP(H)-induced changes in the Kv1.5-Kvβ3 and Kv1.5-Kvβ2 currents.
Although it has been shown that both Kvβ1 [26] and Kvβ2 [17] bind pyridine nucleotides with high affinity, the binding of Kvβ3 with pyridine nucleotides has not been reported. Therefore, we measured pyridine nucleotide binding affinity to Kvβ3 using fluorescence titrations as described before [17,33]. The dissociation constant (KD) was measured for NADPH and NADP+ for Kvβ3 protein. The KDNADPH for Kvβ3 protein was 5.6±0.26 μ M and the KDNADP+ was 38±9 μ M. These measurements indicate that Kvβ3 binds pyridine nucleotides albeit with a lower affinity than Kvβ2 (K NADPH D = 0.12 μ M; K NADP+ D =0.36 μ M [17]) or Kvβ1 (KDNADPH =1.8 μ M, K NADP+ D = 5.6 μ M [33]).
To examine how pyridine nucleotides affect Kvβ3- and Kvβ2-induced changes in Kv1.5 currents, whole-cell currents were recorded from COS-7 cells transfected with Kv1.5 and Kvβ3 and patched with pipettes containing either NADPH or NADP+ in the internal solution. In cells expressing Kv1.5 and Kvβ3, inclusion of NADPH (250 μ M) in the patch pipette increased the total extent of inactivation and accelerated both the fast and the slow phases (Fig. 2e). In contrast, addition of NADP+ (1mM) markedly attenuated inactivation in which the fast phase was completely removed and decelerated the slow phase (Fig 2g). The characteristics of the currents stayed the same whether the transfection ratio of Kv1.5 to Kvβ3 cDNA was 1:1 or 1:5 (Suppl. Fig. 1) suggesting that the recordings were not subject to variation of Kvα:Kvβ subunit ratio. These results show that NADPH accelerates, whereas NADP+ prevents, Kvβ3-induced inactivation of Kv1.5 currents.
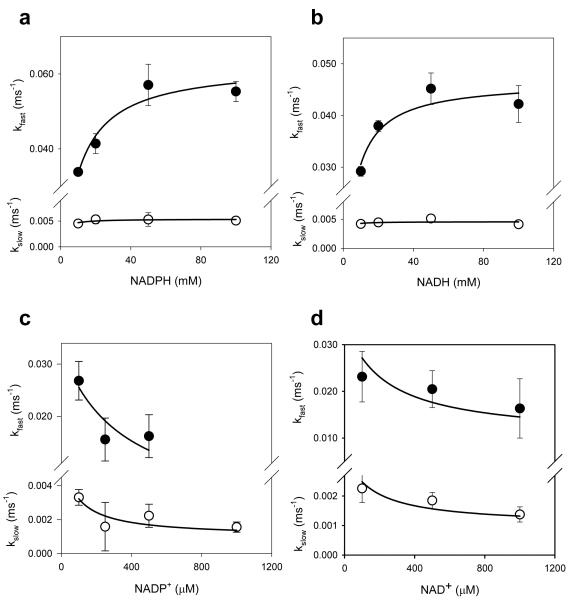
To examine whether pyridine nucleotides regulate Kv1.5-Kvβ3 currents in their physiological range, the concentration-dependence of Kv regulation by pyridine nucleotides was determined using patch pipettes containing varying concentrations of nucleotides. As shown in Fig. 5, plots of the kfast vs NADPH could be described by a simple binding isotherm with an apparent K1/2 of 8.8 ± 2.4 μ M. These values are in excellent agreement with the KDNADPH of recombinant Kvβ3 determined by fluorescence titrations (5.6 μ M). The relationship between kfast and NADH was also hyperbolic with apparent K1/2 value of 5.3 ± 1.6 μ M. Plots of kslow vs NAD(P)H displayed only weak concentration-dependence. Due to competition with endogenous reduced nucleotides, higher levels of oxidized nucleotides were required to affect both kfast and kslow, with apparent K1/2 values of 145 to 275 μ M for NADP+ and 220 to 476 μ M for NAD+. Nevertheless, the complete removal of kfast at high NADP+ concentration indicates robust sensitivity of the currents to oxidized nucleotides. Oxidized nucleotides also induced a concentration-dependent shift in Vh of inactivation of Kv currents to more positive potentials (Fig. 3). No change in recovery from inactivation was observed (rate constants in the presence of 250 μ M NADPH were: kfast 0.41±0.10 ms−1, k 0.059±0.007 ms−1 slow ; and in the presence of 250 μ M NADP+, k 0.57±0.09 ms−1 fast , kslow 0.063±0.01 ms−1, respectively; Fig. 3i). Collectively, these data suggest that Kv1.5-Kvβ3 currents respond to pyridine nucleotides within the concentration range expected in most cells.
Fig. 5. Concentration effects of pyridine nucleotides.
Rate constants for the fast and slow inactivation components are plotted as a function of individual pyridine nucleotide concentrations. Symbols with error bars are experimentally determined values ± SE (n=3 measurements at each nucleotide concentration); lines are best fit of the hyperbolic saturation (panels a and b) or hyperbolic competition (panels c and d) equation to the data.
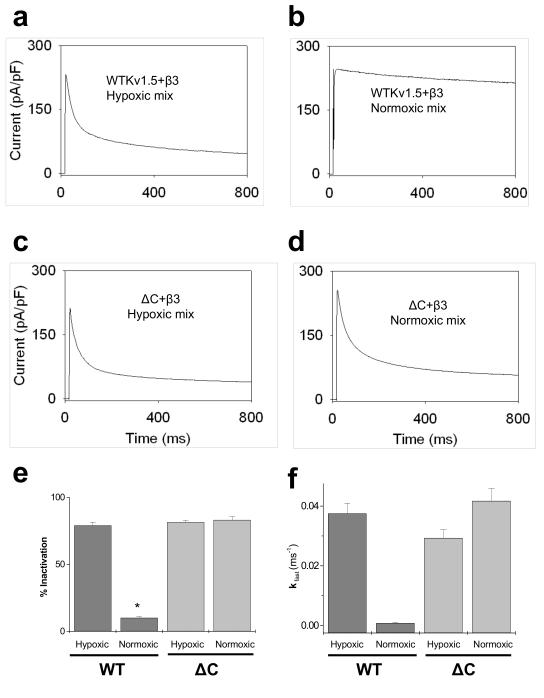
To further assess the physiological significance of pyridine nucleotide-dependent changes in Kv currents, we measured currents from cells expressing Kv1.5-β3 using patch pipettes containing a full set of reduced and oxidized nucleotides expected to be present in vivo under hypoxic and normoxic conditions. Cells patched with a mixture of pyridine nucleotides expected in hypoxic tissue (NADPH 80, NADP+ 50, NADH 1000, and NAD+ 200 μM [2] displayed 79±3% inactivation (Fig. 6a) whereas Kv currents recorded in cells containing the normoxic complement of nucleotides (NADPH 100, NADP+ 30, NADH 50, and NAD+ 1000 μM [2] displayed only 10±1% inactivation (Fig. 6b). Fast inactivation phase was observed only in hypoxic mix ( kfast=0.038±0.0034 ms−1; Fig 6f), whereas it was virtually abolished in normoxic mix. Taken together, these results show that Kv1.5-β3 couple responds not only to changes in individual nucleotide concentrations, but also to nucleotide mixtures reflecting normoxic and hypoxic conditions in the cell.
Fig. 6. Regulation of Kv currents by normoxic and hypoxic complement of pyridine nucleotides.
COS-7 cells co-expressing Kv1.5WT+Kvβ3 or Kv1.5ΔC+Kvβ3 were used for whole-cell patch-clamp recordings (n=5-8 cells). From a holding potential of −80mV the cells were depolarized to +50mV for 800 ms. Whole cell currents were recorded in (a, c) hypoxic and (b, d) normoxic mixture of nucleotides as indicated. The hypoxic complement of pyridine nucleotides consisted of the following: NADPH 80, NADP+ 50, NADH 1000, NAD+ 200 μM and the normoxic complement of pyridine nucleotides consisted of: NADPH 100, NADP+ 30, NADH 50, NAD+ 1000 μM in the patch pipette solution. Percent inactivation was calculated as percentage difference between the peak current and that at 800 ms and depicted as bar graph showing analysis from normoxic and hypoxic complements (e). Panel f shows the kfast of inactivation measured from the currents in hypoxic and normoxic groups. (*P<0.05 Normoxic vs. hypoxic within group)
Like Kvβ3, the effects of Kvβ2 on Kv1.5 were also influenced by pyridine nucleotides. The Vh of activation for Kv1.5+β2 with control internal solution, NADPH or NADP+ was −20.6±0.7, −17.2±1.8 and −10±0.7 mV respectively (Fig. 4); indicating that inclusion of NADP+ causes a significant depolarization shift in the voltage dependence of activation compared with the control internal solution group (p<0.05). Based on the results obtained with both Kvβ2 and Kvβ3, we conclude that both of these β-subunits alter Kv1.5 gating and that their effects are differentially modified by reduced and oxidized nucleotides.
Role of Kv C-terminus in pyridine nucleotide sensing
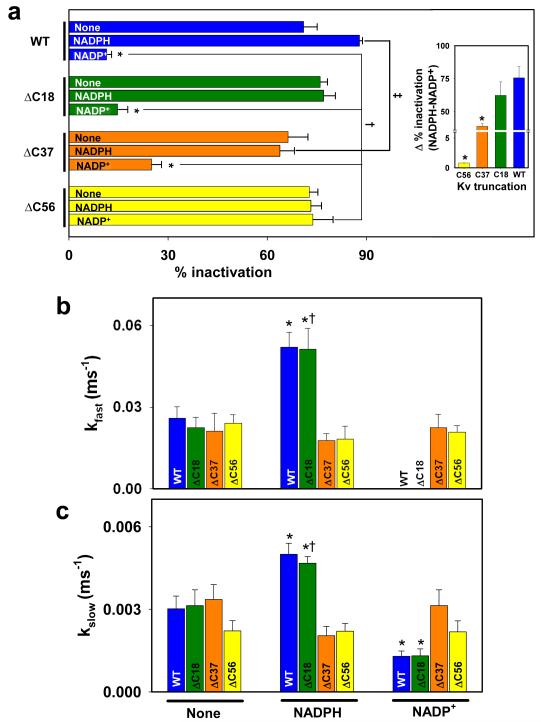
To examine how deletion of the C-terminus affects nucleotide-dependent changes in Kvβ-induced Kv1.5 inactivation, individual deletion mutants were co-expressed with Kvβ3 or Kvβ2 in COS-7 cells; and NADPH or NADP+ were added to the patch pipette. As seen with WT-Kv1.5:Kvβ3, the fast phase of inactivation of Kv1.5ΔC18:Kvβ3 currents were abolished by the addition of NADP+ to the patch pipette, and the total extent of inactivation was decreased. Although NADPH-induced acceleration of both the fast and the slow inactivation rates was preserved, NADPH failed to increase the total inactivation (Fig. 7). These data indicate that deletion of 18 amino acids from the C-terminus of Kv1.5 has only marginal effect on the regulation of Kv1.5 currents by Kvβ bound to either NADPH or NADP+.
Fig. 7. Effect of C-terminal deletions on regulation of Kv currents by pyridine nucleotides.
COS-7 cells were transfected with Kv1.5WT or KvΔC constructs coexpressed with Kvβ3. Kv currents were recorded with either control internal solution or solution containing NADPH or NADP+ in the patch pipette. Percent inactivation was measured from the peak current to the end of 800 ms pulse at +50 mV. (a) % inactivation measured with no additives (none) or NADPH or NADP+ in the patch pipette. Inset shows the range of regulation by pyridine nucleotides calculated as a difference in the overall percent inactivation between NADPH and NADP+ groups. Rate constants of the fast (b) and the slow (c) phases of inactivation, respectively. The decay phase was analyzed by using mono- or bi-exponential equation (see Material and Methods for detail). Bars are color-coded for the deletion construct. *P<0.05 compared with no-additions (none) within each group, †P<0.05 compared with WT (none), ‡P<0.05 compared with WT (NADPH).
In comparison with currents obtained with Kv1.5ΔC18:Kvβ3, those with Kv1.5ΔC38:Kvβ3 were relatively insensitive to pyridine nucleotides. Addition of NADP+ to Kv1.5ΔC38:Kvβ3 couple caused a significantly smaller reduction in the inactivation (25% inactivation remained with 1 mM NADP+) than either WT or ΔC18 Kv1.5 and the total range of regulation by pyridine nucleotides was 23.5% less than ΔC18 (Fig. 7a inset). NADPH did not accelerate inactivation, and NADP+ failed to abolish the fast phase of inactivation and the rate of slow phase was not regulated to the same extent as with WT Kv1.5 (Fig 7 b&c). The largest deletion mutant ΔC56 completely abolished change in inactivation induced by Kvβ3 bound to either reduced or oxidized pyridine nucleotides (see current recordings in Fig. 2 and analysis in Fig. 7). Currents generated by the Kv1.5ΔC56+Kvβ3 exhibited two-phase inactivation with inactivation properties that were very similar to that of the WT Kv1.5+β3, however, neither rate nor extent of inactivation were affected by NADPH or NADP+. In addition, the Kv1.5ΔC+Kvβ3 currents were insensitive to normoxic and hypoxic nucleotide mixtures (Fig. 6 c-d) suggesting that the deletion of the C-terminus abolishes the regulation of the channel by pyridine nucleotides.
To rule out the possibility that the effects of C-terminus deletion are due to delayed changes in inactivation, we examined the time course of NADP+-mediated decline in channel inactivation. As shown in Fig. 2i-j, NADP+ completely abolished inactivation of WT-Kvβ3 currents within 10 min of patch rupture. This reflects the time required for dialysis of the nucleotide from the patch pipette to the cell as well the time required for nucleotide exchange at the Kvβ active site. However, there was no difference in inactivation of Kv1.5ΔC:Kvβ3 current recorded for 10 min with patch pipettes containing NADP+ (Fig. 2 k-l), indicating that that deletion of the C-terminus does not only change the kinetics of the response but that it permanently abolishes the ability of Kv1.5 channel to respond to pyridine nucleotide binding to Kvβ3.
Addition of NADPH did not significantly affect the voltage-dependence of activation or inactivation for any of the Kvα constructs used in this study. NADP+ did not have any effect on the activation parameters (Vh of activation and time to peak), but it induced a depolarizing shift in the Vh of inactivation for Kvβ3: WT, ΔC18, and ΔC37 complexes (Table 2). Significantly, NADP+ failed to induce a depolarizing shift in the Vh of inactivation of the ΔC56 currents (Fig. 3d, circles vs. inverted triangles). Moreover, as shown in Fig. 4, neither NADPH nor NADP+ significantly affected the voltage-dependence of activation of Kv1.5ΔC56-Kvβ2 currents. Based on these observations we conclude that deletion of the C-terminal domain of Kvα1.5 selectively abolishes differential regulation of Kvα-β2 and β3 currents by oxidized and reduced pyridine nucleotides, whereas intermediate deletions have a partial effect.
Table 2.
Regulation of Kv currents by reduced and oxidized pyridine nucleotides
| Vh act (mV) |
Time to peak act (ms) |
Act time constant |
Vh inact (mV) |
||
|---|---|---|---|---|---|
| Kvα1.5WT+Kvβ3 | None | −19.7±0.6 | 7.1±0.5 | 1.5±0.4 | −22.2±0.9 |
| NADPH | −23.5±0.5 | 5.5±0.5 | 1.3±0.2 | −20.4±0.1 | |
| NADP+ | −19.0±0.7 | 7.0±1.0 | 0.5±0.1* | −9.3±2.0*† | |
|
Kvα1.5ΔC18 +Kvβ3 |
None | −21.0±1.0 | 6.0±0.4 | 1.1±0.1 | −24.9±0.8 |
| NADPH | −24.0±1.4 | 4.1±0.9 | 0.9±0.1 | −22.0±0.6 | |
| NADP+ | −18.7±1.8 | 9.1±0.5 | 0.5±0.04‡ | −15.0±3.0* | |
|
Kvα1.5ΔC37 +Kvβ3 |
None | −21.5±0.8 | 8.0±1.5 | 1.4±0.6 | −22.7±0.7 |
| NADPH | −21.7±0.9 | 5.3±0.2 | 1.0±0.1 | −22.9±1.6 | |
| NADP+ | −20.5±0.4 | 9.0±2.5 | 0.6±0.07‡ | −13.1±2.6† | |
|
Kvα1.5ΔC56 +Kvβ3 |
None | −21.5±1.5 | 6.0±0.6 | 1.1±0.2 | −20.5±0.3 |
| NADPH | −18.6±1.2 | 5.4±0.2 | 0.7±0.1 | −22.8±0.5 | |
| NADP+ | −24.1±0.3 | 5.3±0.4 | 0.8±0.04 | −21.2±2.6 |
P<0.05 Within group; none vs. NADPH or NADP+
P<0.05 Within group; NADPH vs. NADP+
P<0.05 Kvα1.5WT+Kvβ3 none vs. all other groups
Pyridine nucleotide dependence of the interactions between Kv C-terminus and Kvβ
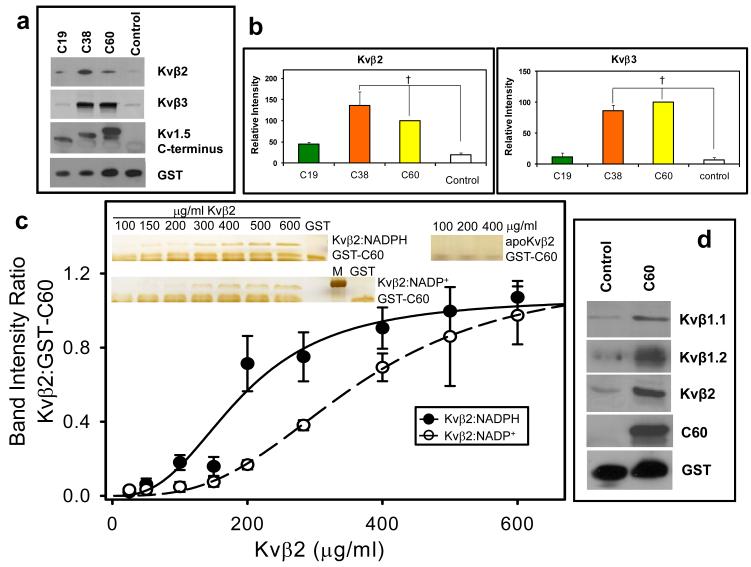
Our electrophysiological studies showed that the C-terminal loop of Kvα is essential for the pyridine nucleotide-dependent changes in Kv inactivation. To examine whether this could be attributed to a direct, physical interaction between the C-terminus of Kv1.5 and Kvβ, we assessed whether the C-terminus by itself binds to β-subunits, and whether this binding is sensitive to the nucleotide bound to Kvβ. To determine the binding of the Kv C-terminus with β-subunits, we generated a series of GST fusion constructs in which the GST sequence was fused with sequences coding for the terminal-most 19 (Asp584-Leu602), 38 (Ile565-Leu602), or 60 (Arg543-Leu602) amino acids of the C-terminal loop of Kv1.5. An unrelated peptide sequence of 50 amino acid residues was fused with the GST protein and used as control. The GST-fusion constructs of the C-terminal fragments of Kv1.5 (GST-C60, C38 or C19) were then incubated with lysates of E. coli expressing the Kvβ2 or the Kvβ3 gene. As shown in Fig. 8a, Kvβ2 and Kvβ3 in bacterial lysate were pulled down by GST-C38 and GST-C60 with equal affinity. Only trace levels of Kvβ3, similar to those with the control peptide, were precipitated by GST-C19, although this peptide did show a weak affinity for Kvβ2 (Fig 8a-b). Coomassie blue staining of the Kvβ3 pull down extracts did not reveal the presence of other proteins in the gel, confirming specificity of interaction (data not shown). These results show that the cytosolic C-terminus of Kv1.5 binds to Kvβ2 and 3 and that this binding is primarily due to an interaction of the C-terminal domain consisting of 41 amino acids (Arg543-Val583) with Kvβ.
Fig. 8. Binding of the C-terminal domain of Kv to Kvβ.
(a) Western blots of Kvβ2 (upper panel) and Kvβ3 (middle panel) pulled down by the GST-Kv1.5 C-terminus fusion peptides. Fusion proteins containing 60, 38, or 19 terminal amino acid peptides from Kvα1.5 C-terminus attached to GST or GST with unrelated peptide (Control; 30μ g each) were incubated with lysate of Kvβ2 or Kvβ3 -expressing E.coli (350 μ g total protein). Protein complexes were pulled down using GST·Bind beads, washed and eluted with 10mM glutathione. The eluate was separated by SDS-PAGE and probed with anti-pan-Kvβ antibody, an antibody directed against the C-terminus of Kv1.5 (bait) or GST; (b) Densitometric analysis of the bands in panel a. The density of the Kvβ band precipitated with GST-C60 was assigned a 100% value. †, P< 0.05 versus Control peptide (n=3-5). (c) Determination of the binding affinity of Kvβ:nucleotide complexes to the C-terminal peptide of Kv1.5. A fixed concentration of GST-C60 (30 μ g/ml) was mixed with variable concentrations of Kvβ2 in a binary complex with NADPH (Kvβ2:NADPH), or NADP+ (Kvβ2:NADP+), or none (apo-Kvβ2) and GST pull down assay was performed. Eluted protein was separated by SDS-PAGE and the gels were silver stained. Band intensities normalized to GST-C60 input were plotted as a function of Kvβ2 concentration. The experiment was repeated 6 times; data points on the graph represent average and standard error, and the lines represent the best fit to experimental data using the Hill equation. Apo:Kvβ2 did not bind GST-C60 and no measurable intensities were found by silver stain. Inset: silver stained gels showing Kvβ2 in complex with NADPH, NADP+ or without nucleotide, pulled down with GST-C60; M, marker, 37 kDa band is shown. (d) Western blots of GST beads incubated with brain lysates. The GST-C60 construct or scrambled construct was used to pull down proteins from mouse brain extract. The eluate was separated on SDS PAGE and protein bands were visualized with antibodies against the indicated Kvβ isoform, Kv1.5 C-terminus, or GST.
To determine whether nucleotides regulate the binding of Kv C-terminus to the β-subunits, a constant amount of GST-C60 was mixed with different concentrations of nucleotide-free Kvβ2 or Kvβ2 bound to either NADPH or NADP+, and the resultant complexes were pulled down with the GSH beads. Kvβ2 stripped of bound nucleotide (apoKvβ2) was not precipitated by GST-C60, suggesting that the C-terminal peptide does not bind Kvβ2 in the absence of the nucleotide (Fig. 8c, inset). However, GST-C60 precipitated Kvβ2 bound to either NADPH or NADP+. The Kvβ2:NADPH complex was precipitated at lower concentrations of the protein than Kvβ2:NADP+ (Fig. 8c). The concentration dependence of the binary complexes of Kvβ2 was sigmoidal in shape and the difference between the NADPH and NADP+-bound forms of Kvβ2 was diminished at high concentrations of Kvβ, indicating that NADPH and NADP+ regulate the affinity for, but not the total extent of binding to, GST-C60. The dissociation constant (KD) calculated by fitting the Hill equation to the data for Kvβ2:NADPH binary complex (5.2 ± 0.8 μ M) was significantly (P=0.02) lower than that calculated for Kvβ2:NADP+ (9.9 ± 1.7 μ M). Taken together, these results suggest that the C-terminus of Kv1.5 does not bind to apoKvβ2 and that it has a higher affinity for Kvβ2:NADPH than Kvβ2:NADP+. Finally, to examine whether the C-terminal domain of Kv1.5 also interacts with native Kvβ proteins, GST-C60 was incubated with murine whole brain homogenates. Western analysis of the pull-down showed strong bands recognized by anti-Kvβ1.1, 1.2, and β2 antibodies (Fig. 8d), indicating that the C-terminus of Kv1.5 interacts with the native β-subunits expressed in the brain.
Structural analysis of Kv C-terminus
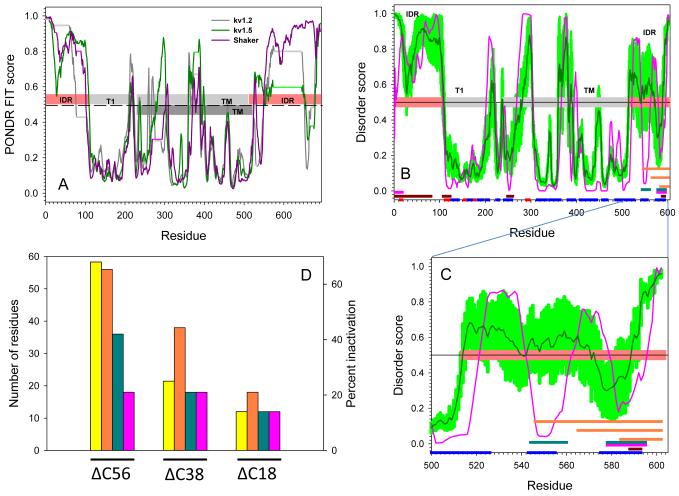
To further examine the propensity of the C-terminal domain of Kv1.5 to bind ancillary subunits, we compared the amino acid sequence of Kv1.5 C-terminus to that of Kv1.2 and the Drosophila Shaker channel (Fig. 9a). In agreement with previous studies [20,21], this analysis showed that both the N- and C-terminal domains of these proteins are located in the disordered zone (disorder score >0.5 threshold) with some downward spikes representing local short segments with increased order propensity within the long disordered region. In contrast, the central regions containing T1 and TM domains are located predominantly in the structured zone with some distinct peaks representing local short disordered/flexible fragments within the long ordered region.
Fig. 9. Evaluation of intrinsic disorder in Kv channels.
(a) Intrinsic disorder propensities of the rat Kv1.2 (dark gray line), rat Kv1.5 (dark green line), and Drosophila Shaker channel (dark pink line). In each protein, per residue disorder propensity was evaluated by PONDR® FIT algorithm. Results are shown for sequences aligned by ClustALW. All residues with disorder score higher than 0.5 (above dashed line) are predicted to be disordered; while residues with disorder score lower than 0.5 (below dashed line) are expected to be structured. Horizontal light gray, light green and light pink lines correspond to the gaps in the sequence alignment between Kv1.2, Kv1.5, and Drosophila Shaker, respectively. Light pink and gray bars in the middle of the plot show localization of the structured domain and disordered regions for Shaker Kv1.2 (PDB id: 3LUT). IDR stands for Intrinsically Disordered Region; T1 is T1 domain; TM refers to the TransMembrane domain. The transmembrane domain of Kv1.5 has been annotated as dark gray bar. Panel (b) shows the Disorder analysis and functionally important sequence features of the Kv1.5 channel. Results of the disorder prediction by PONDR® VLXT and PONDR® FIT are shown by pink and dark green lines, respectively. Light green shadow covers the distribution of errors in evaluation of disorder scores by PONDR® FIT. Three parallel orange bars at the C-terminal are locations of the three segments in deletion mutagenesis experiments. The red and blue bars at the bottom of the plot illustrate the localization of the Jpred-predicted β-strands and α-helices, respectively. Locations of the predicted α-MoRFs, AIBS, and potential VLXT-based binding sites are shown by pink, dark red and dark cyan bars, respectively. Panel (c) is an inset to Panel (b) which represents the extended view for the residue range 500-602 of the disorder distribution and functionally important sequence features in the C-terminal domain of the Kv1.5 channel. Annotations are the same as in (b). (d) Comparison of the % inactivation (yellow bars) with the length of the deleted C-terminal regions (orange bars) and the length of the deleted potential binding regions, MoRFs (pink bars) and VLXT-based binding sites (dark green bars).
To obtain insights into the potential functional roles of the intrinsically disordered regions of Kvα, the distributions of the intrinsic disorder propensities along the protein sequence were generated by two disorder predictors, PONDR® FIT and PONDR® VLXT. The PONDR® FIT analysis, which provides predictions with the highest accuracy [41], was used for the accurate evaluation of disorder propensity, whereas PONDR® VL-XT predictor, which is extremely sensitive to local amino acid composition [28], was used to find potential functional fragments. Being defined as short order-prone motifs that are located within long disordered regions and that undergo a disorder-to-order transition during the binding process, molecular recognition features (MoRFs) are usually more enriched in aliphatic and aromatic amino acids relative to typical intrinsically disordered regions. Due to these sequence peculiarities, MoRFs are frequently observed as sharp dips in the corresponding PONDR®VLXT disorder score plots. Hence, MoRF regions can be reliably identified on the basis of special characteristics of the PONDR® VLXT prediction profiles and several other specific attributes [25,4]. As shown in Fig. 9b, both disordered N- and C-termini of Kvα contain a number of downward order spikes (residues 36-48, 53-62, 71-82, and especially residues 2-10, 542-562, and 578-595), which correspond to the potential binding sites. Two of these sites (residues 1-18 and 578-595) were identified as α-MoRFs by MoRF identifier [4], and several binding sites (residues 1-81, 108-126, 249-260, and 588-592) were also identified by the ANCHOR algorithm [23]. In general there is a good agreement between predictions of the intrinsic disorder propensity and secondary structure (Fig. 9b&c), as many regions with high disorder score typically correspond to protein regions that do not contain ordered secondary structure elements. Furthermore, the potential binding sites located at the C-terminal domain of Kv1.5 were both predicted to possess α-helical propensity. Fig. 9c represents the sequence peculiarities in the C-terminal fragment of Kv1.5 (residues 500-602) which support the idea that deletion mutations at Kv1.5 C-terminus might affect potential binding sites of this protein. Deletion of the last 18 residues is expected to remove a portion of one potential binding site; deletion of the last 37 residues completely removes one C-terminal binding site, whereas deletion of the last 56 residues eliminates both potential C-terminal binding sites observed as sharp dips in the PONDR®VLXT plot and predicted to possess high helical propensity (residues 542-562, and 578-595). These analyses support the notion that the interaction of Kv1.5 with Kvβ subunit is minimally affected by the deletion of C18 (Asp584-Leu602) fragment, whereas the deletion of the C37 (Ile565-Leu602) fragment might cause further decline in this function, and, finally, the removal of the C56 (Arg543-Leu-602) fragment might strongly abrogate functional interaction of these proteins. Collectively, this analysis indicates that the molecular recognition features of the C-terminal domain are consistent with experimental results showing that the residues between 543-584 are involved in binding to β-subunits.
Discussion
The major findings of this study are that the deletion of the cytosolic domain of the C-terminus of Kv1.5 does not significantly alter the gating or the kinetics of the channel, nor does it prevent the interaction of the pore-forming proteins of the channel with β-subunits. Nevertheless, deletion mutants of Kv1.5 lacking the C-terminal domain were unable to respond to changes in the redox state of pyridine nucleotides bound to Kvβ, suggesting that the C-terminus plays an obligatory role in discriminating between the NAD(P)+- and the NADPH-bound forms of the β-subunit and in mediating the effects of these nucleotides on channel gating.
Our observation that deletion of the C-terminus does not affect membrane targeting or the electrophysiological properties of Kv1.5 is consistent with previous work showing that deletion of the last 57 amino acids of channel subunit does not affect whole cell currents in mouse L-cells [37]. However, it has been reported that the deletion of the C-terminus increases the surface expression of Kv1.5 in transfected HEK [9]. No such increase in the expression of the channel was observed in our study; however, such changes may be masked in our overexpression studies in which channel proportion and current density appear to saturate before cell surface expression. We also found that deletion of the C-terminus does not affect the association of Kvα with Kvβ: both Kvβ2 and β3 induced similar changes in channel gating in WT and Kv1.5ΔC, suggesting that functional interactions between the α and the β subunits were preserved in the absence of the Kv C-terminus. Crystal structures of Kvα-Kvβ complexes show that these proteins form a tetrameric assembly around the T1 or the N-terminal domain of Kv channels [29,10], hence it is not surprising that the binding of the β-subunits does not depend on the presence of the C-terminus. However, electron microscopic studies suggest that the C-terminus of Kvα is disordered and in the absence of Kvβ, it is closely association with the portion of the T1-domain closer to the membrane surface; whereas, in the presence of the β-subunit the C-terminus moves away from the T1 domain [31] and grazes the surface of the β-subunits. Consistent with previous observation [27], our results showed that co-expression with Kvβ3 or Kvβ2 shifts the voltage for half-maximal activation of Kv1.5 currents to more negative potentials, giving rise to shallower slopes of peak conductance-voltage relationship [15,36,34]. Because this shift in voltage-dependence of activation was not affected by the deletion of the C-terminus, we suggest that this is due to the binding of the β-subunits to the T1 domain of Kv1.5 and that movement of the C-terminus away from the T1 domain upon binding of this domain to β-subunits is an electrically silent event.
The most significant change upon deletion of the C-terminus was the loss of the ability of the channel to discriminate between reduced and oxidized nucleotides bound to the β-subunit. Our previous studies with Kvβ1.3 [34] and those of Pan et al. with Kvβ1.1 [26] indicate that these proteins bind pyridine nucleotides with high affinity and that the binding of oxidized nucleotides removes β-induced inactivation of Kv currents. In extension of these studies, we report here that Kvβ3, like Kvβ1 and Kvβ2, binds to pyridine nucleotide with high affinity and responds to physiological changes in pyridine nucleotide levels that would be expected to prevail in cells under normoxic and hypoxic conditions. From our measurements of the nucleotide-dependence of Kv1.5 inactivation by Kvβ3, we found that channel inactivation is saturated at low levels of NADPH (K1/2 = 5 μ M), therefore we expect that in most metabolically-active tissues such as the heart, brain, liver and skeletal muscle (which maintain 100 to 300 μ M NADPH in the cytosol [11,3,12] Kv1.5α-β complexes will be maximally saturated with NADPH under aerobic non-stressed conditions. However, metabolic inhibition or hypoxia, which leads to a drastic decrease in NADPH, could significantly impact Kvβ-mediated inactivation of Kv currents. In these circumstances, the extent to which changes in pyridine nucleotides affect Kv currents would depend not only on the absolute concentration of NADPH but on the relative concentration of oxidized nucleotides [17], which in contrast to reduced nucleotides abolish channel inactivation. Thus the overall response will be a summation of the changes in the redox ratios of pyridine nucleotides and the set of Kvβ proteins expressed in the cell. Because of differences in nucleotide affinity, different subunits are likely to respond differently to changes in cellular nucleotide levels. Thus, the presence of multiple β-subunits within the same tissue might enable responses to a wider range of changes in nucleotide concentrations than possible with a single β-subunit. For instance, we have found that increasing NADPH accelerates Kv inactivation by Kvβ3 (Fig. 2), but not by Kvβ1 [34]. Thus Kvβ3 is likely to be more responsive to an increase in NADPH, whereas Kvβ1 and β2, because of their higher affinity nucleotide binding, are likely to be more responsive to NAD(P)H concentrations.
The observation that the deletion of the C-terminus leads to the loss of nucleotide sensitivity of Kv1.5-Kvβ3 complexes suggests that the C-terminus is a sensory domain of Kvα essential for distinguishing whether reduced or oxidized pyridine nucleotide is bound to Kvβ. The C-terminus, however, does not seem to be required for basal, pyridine nucleotide-insensitive inactivation of Kvα, which is mediated by the N-terminal inactivation gate of Kvβ3. The most straightforward explanation for this observation is that in the absence of the Kvα C-terminus, Kvβ is unable to bind to pyridine nucleotides or that the C-terminus regulates the nucleotide affinity of Kvβ. However, we consider this unlikely because Kvβ proteins avidly bind pyridine nucleotides in solution in the absence of the C-terminus. Another possibility is that the C-terminus selectively increases K NADP+d without affecting NADPH binding (since basal inactivation was preserved) thereby preventing NADP+-mediated removal of inactivation, but not NADPH-mediated increase in inactivation. This also appears unlikely because the effects of both NADPH and NADP+ were abolished by the deletion of the Kv C-terminus, indicating that this domain of the Kv channel is required for enhancing inactivation by NADPH as well as for removing inactivation in the presence of NADP+. However, the ability to discriminate between NADPH and NADP+ bound forms of Kvβ appears to be a unique property of the C-terminus of Kv1.5, because deletion of the C-terminal domain of Kv1.1 does not affect nucleotide sensing [26]. Our structural analysis shows that the molecular recognition features are absent from the C-terminus of Kv1.2, suggesting that the mechanism by which Kv channels other than Kv1.5 respond to nucleotide binding to Kvβ may be different. This difference may enable specific Kv channels to respond differently to distinct regulatory influences. For instance, binding of other proteins to the C-terminus might regulate pyridine coenzyme-dependent changes in potassium current generated by Kv1.5, but not other Kv channels.
Changes in the voltage-dependence of activation and inactivation clearly suggest that channel gating could be regulated by pyridine nucleotides, which could directly affect the movement of the N-terminus of the β-subunit, independent of the C-terminus. However, in cells expressing Kvβ2 (which lacks the inactivating N-terminus) and Kv1.5, the voltage-dependence of activation was altered by NADP+ and abolished by deletion of Kv C-terminus, indicating that the effects of nucleotides could be transmitted independent of change in the movement of the N-terminal inactivating peptide and dependent upon the interaction of the Kv C-terminus. Similarly, NADP+ caused a shift in the inactivation potential of Kv1.5-Kvβ3 currents and this shift was absent in the ΔC mutant. With WT Kv1.5-Kvβ3, addition of NADP+ caused a depolarization shift in the half-inactivation potential (from −20 to −8 mV), with little or no change in the voltage-dependence of activation (which shifted from −20 to −18 mV). Significantly, addition of NADP+ did not cause a depolarizing shift in the voltage-dependence of inactivation of Kv1.5ΔC+Kvβ3 couple (Vh inactivation = −19 mV). Hence, it appears that activation-independent changes in the voltage-dependence of inactivation due to pyridine-nucleotide bound states of Kvβ are transmitted via the C-terminal domain of Kv1.5. This is consistent with the model in which the movement of a charged particle in the voltage sensor region is away from the ball-and-chain movement imparted by Kvβ-subunit at the channel pore. As pointed out by Long et al. [18], binding of the C-terminus near the active site of the β-subunit, with an affinity that depends upon whether it is bound to NADP+ or NADPH, could affect channel gating because the C-terminus is connected directly to the inner helices which must undergo large movements to open or close the channel. Our biochemical experiments showing that the C-terminus has a higher affinity for β2-NADPH than for β2-NADP+ support the concept that binding of the β-subunit to NADP+ weakens the interaction of the β-subunit with Kv C-terminus and the movement of this peptide away from Kvβ removes inactivation and shifts the voltage-dependence of activation to more positive potentials.
The results obtained from progressive deletions of the C-terminus indicate that amino acid residues between positions 565 and 584 are required for binding Kvβ. This location is consistent with our biochemical, electrophysiological and structural analyses. Our biochemical analysis showed that the 584-602 peptide displayed little (β2) or no (β3) binding, whereas the 565-602 peptide bound Kvβ as strongly as the entire C-terminus (543-602), localizing the Kvβ site between residues 565-584. This localization is in agreement with electrophysiological findings. Deletion of amino acids from 585-602 had only minor effects on the channel inactivation, whereas deletion of the 566-602 had a much larger impact on the range of regulation by pyridine nucleotides, indicating that the 565-602 loop of the C-terminus is sufficient for binding to Kvβ, residues 543-565 play an additional role in nucleotide recognition. This may be attributed to the fact that the region between amino acids 543 and 584 is rich in positively charged residues. It contains 9 positively charged, but only 3 negatively charged amino acid residues. Therefore, electrostatic repulsion between this domain and the positive charge of NADP+ could account for the lower affinity of the C-terminus for Kvβ bound to NADP+ than that bound to NADPH. Finally, our analysis of the intrinsic disorder propensity of the C-terminal domain is also consistent with the notion that molecular recognition features of the 564-602 peptide support the binding of the C-terminus to Kvβ. Altogether, these findings suggest the presence of a unique structural domain within the cytosolic C-terminal domain of Kv1.5 which interacts with the β-subunits and that the effects of this interaction on the channel depend upon whether the β-subunits are bound to NADPH or NADP+. Additional studies are required to determine how the interaction of the C-terminus of Kv1.5 with Kvβ might be affected by other proteins (protein kinases, phosphatases and cytoskeletal elements) that are associated with the α-β assembly in native Kv channels.
Supplementary Material
Acknowledgements
The authors thank Dr. Maiying Kong for her advice on statistical analysis and Joshua Salabei and Ermin Villa for their help in cloning experiments. This work was partly supported by NIH grants RR024489, HL-55477, HL-59378 (to A.B.), HL-089372 (to O.A.B.), 0865466D American Heart Beginning-grant-in-aid, HL-102171, Deans Research Funding USF-COP (to S.M.T), and the Program of the Russian Academy of Sciences for the “Molecular and Cellular Biology” (to V.N.U).
Abbreviations
- Kvβ
β-subunit of the voltage-gated potassium channel
- Kv
voltage-gated potassium channel
- KvΔC
deletion of the last C-terminal amino acids of Kv channel
- GST
Glutathione-S-transferase fusion protein
- Vh act
voltage at which half of the channel are activation
- Vhinact
voltage at which half of the channel are inactivated
References
- 1.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40(4):553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceconi C, Bernocchi P, Boraso A, Cargnoni A, Pepi P, Curello S, Ferrari R. New insights on myocardial pyridine nucleotides and thiol redox state in ischemia and reperfusion damage. Cardiovasc Res. 2000;47(3):586–594. doi: 10.1016/s0008-6363(00)00104-8. [DOI] [PubMed] [Google Scholar]
- 3.Ceconi C, Bernocchi P, Boraso A, Cargnoni A, Pepi P, Curello S, Ferrari R. New insights on myocardial pyridine nucleotides and thiol redox state in ischemia and reperfusion damage. Cardiovascular research. 2000;47(3):586–594. doi: 10.1016/s0008-6363(00)00104-8. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deal KK, England SK, Tamkun MM. Molecular physiology of cardiac potassium channels. Physiol Rev. 1996;76(1):49–67. doi: 10.1152/physrev.1996.76.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Dubois JM, Rouzaire-Dubois B. Role of potassium channels in mitogenesis. Prog Biophys Mol Biol. 1993;59(1):1–21. doi: 10.1016/0079-6107(93)90005-5. [DOI] [PubMed] [Google Scholar]
- 8.Ekhterae D, Platoshyn O, Zhang S, Remillard CV, Yuan JX. Apoptosis repressor with caspase domain inhibits cardiomyocyte apoptosis by reducing K+ currents. Am J Physiol Cell Physiol. 2003;284(6):C1405–1410. doi: 10.1152/ajpcell.00279.2002. [DOI] [PubMed] [Google Scholar]
- 9.Eldstrom J, Doerksen KW, Steele DF, Fedida D. N-terminal PDZ-binding domain in Kv1 potassium channels. FEBS Lett. 2002;531(3):529–537. doi: 10.1016/s0014-5793(02)03572-x. [DOI] [PubMed] [Google Scholar]
- 10.Gulbis JM, Zhou M, Mann S, MacKinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science. 2000;289(5476):123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- 11.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klaidman LK, Leung AC, Adams JD., Jr. High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem. 1995;228(2):312–317. doi: 10.1006/abio.1995.1356. [DOI] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Lee TE, Philipson LH, Kuznetsov A, Nelson DJ. Structural determinant for assembly of mammalian K+ channels. Biophys J. 1994;66(3 Pt 1):667–673. doi: 10.1016/s0006-3495(94)80840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leicher T, Bahring R, Isbrandt D, Pongs O. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium channel. J Biol Chem. 1998;273(52):35095–35101. doi: 10.1074/jbc.273.52.35095. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science. 1992;257(5074):1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 17.Liu SQ, Jin H, Zacarias A, Srivastava S, Bhatnagar A. Binding of pyridine nucleotide coenzymes to the beta-subunit of the voltage-sensitive K+ channel. J Biol Chem. 2001;276(15):11812–11820. doi: 10.1074/jbc.M008259200. [DOI] [PubMed] [Google Scholar]
- 18.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 19.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 20.Magidovich E, Fleishman SJ, Yifrach O. Intrinsically disordered C-terminal segments of voltage-activated potassium channels: a possible fishing rod-like mechanism for channel binding to scaffold proteins. Bioinformatics. 2006;22(13):1546–1550. doi: 10.1093/bioinformatics/btl137. [DOI] [PubMed] [Google Scholar]
- 21.Magidovich E, Orr I, Fass D, Abdu U, Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K+ channel modulates its interaction with scaffold proteins. Proc Natl Acad Sci U S A. 2007;104(32):13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79(7):1133–1135. doi: 10.1016/0092-8674(94)90004-3. doi:0092-8674(94)90004-3 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5(5):e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaya N, Papazian DM. Potassium channel alpha and beta subunits assemble in the endoplasmic reticulum. J Biol Chem. 1997;272(5):3022–3027. doi: 10.1074/jbc.272.5.3022. [DOI] [PubMed] [Google Scholar]
- 25.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 26.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283(13):8634–8642. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90(2):755–796. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- 28.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Sewing S, Roeper J, Pongs O. Kv beta 1 subunit binding specific for shaker-related potassium channel alpha subunits. Neuron. 1996;16(2):455–463. doi: 10.1016/s0896-6273(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 30.Shen NV, Chen X, Boyer MM, Pfaffinger PJ. Deletion analysis of K+ channel assembly. Neuron. 1993;11(1):67–76. doi: 10.1016/0896-6273(93)90271-r. [DOI] [PubMed] [Google Scholar]
- 31.Sokolova O, Accardi A, Gutierrez D, Lau A, Rigney M, Grigorieff N. Conformational changes in the C terminus of Shaker K+ channel bound to the rat Kvbeta2-subunit. Proc Natl Acad Sci U S A. 2003;100(22):12607–12612. doi: 10.1073/pnas.2235650100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tipparaju SM, Barski OA, Srivastava S, Bhatnagar A. Catalytic mechanism and substrate specificity of the beta-subunit of the voltage-gated potassium channel. Biochemistry. 2008;47(34):8840–8854. doi: 10.1021/bi800301b. doi:10.1021/bi800301b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tipparaju SM, Liu SQ, Barski OA, Bhatnagar A. NADPH binding to beta-subunit regulates inactivation of voltage-gated K(+) channels. Biochem Biophys Res Commun. 2007;359(2):269–276. doi: 10.1016/j.bbrc.2007.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tipparaju SM, Saxena N, Liu SQ, Kumar R, Bhatnagar A. Differential regulation of voltage-gated K+ channels by oxidized and reduced pyridine nucleotide coenzymes. Am J Physiol Cell Physiol. 2005;288(2):C366–376. doi: 10.1152/ajpcell.00354.2004. [DOI] [PubMed] [Google Scholar]
- 35.Tristani-Firouzi M, Chen J, Mitcheson JS, Sanguinetti MC. Molecular biology of K(+) channels and their role in cardiac arrhythmias. Am J Med. 2001;110(1):50–59. doi: 10.1016/s0002-9343(00)00623-9. [DOI] [PubMed] [Google Scholar]
- 36.Uebele VN, England SK, Gallagher DJ, Snyders DJ, Bennett PB, Tamkun MM. Distinct domains of the voltage-gated K+ channel Kv beta 1.3 beta-subunit affect voltage-dependent gating. Am J Physiol. 1998;274(6 Pt 1):C1485–1495. doi: 10.1152/ajpcell.1998.274.6.C1485. [DOI] [PubMed] [Google Scholar]
- 37.Uebele VN, Yeola SW, Snyders DJ, Tamkun MM. Deletion of highly conserved C-terminal sequences in the Kv1 K+ channel sub-family does not prevent expression of currents with wild-type characteristics. FEBS Lett. 1994;340(1-2):104–108. doi: 10.1016/0014-5793(94)80181-9. [DOI] [PubMed] [Google Scholar]
- 38.VanDongen AM, Frech GC, Drewe JA, Joho RH, Brown AM. Alteration and restoration of K+ channel function by deletions at the N- and C-termini. Neuron. 1990;5(4):433–443. doi: 10.1016/0896-6273(90)90082-q. [DOI] [PubMed] [Google Scholar]
- 39.Weir EK, Lopez-Barneo J, Buckler KJ, Archer SL. Acute oxygen-sensing mechanisms. N Engl J Med. 2005;353(19):2042–2055. doi: 10.1056/NEJMra050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281(22):15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu W, Xu J, Li M. NAB domain is essential for the subunit assembly of both alpha-alpha and alpha-beta complexes of shaker-like potassium channels. Neuron. 1996;16(2):441–453. doi: 10.1016/s0896-6273(00)80062-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.