Abstract
We performed high-throughput sequencing of DNA from fossilized faeces to evaluate this material as a source of information on the genome and diet of Pleistocene carnivores. We analysed coprolites derived from the extinct cave hyena (Crocuta crocuta spelaea), and sequenced 90 million DNA fragments from two specimens. The DNA reads enabled a reconstruction of the cave hyena mitochondrial genome with up to a 158-fold coverage. This genome, and those sequenced from extant spotted (Crocuta crocuta) and striped (Hyaena hyaena) hyena specimens, allows for the establishment of a robust phylogeny that supports a close relationship between the cave and the spotted hyena. We also demonstrate that high-throughput sequencing yields data for cave hyena multi-copy and single-copy nuclear genes, and that about 50 per cent of the coprolite DNA can be ascribed to this species. Analysing the data for additional species to indicate the cave hyena diet, we retrieved abundant sequences for the red deer (Cervus elaphus), and characterized its mitochondrial genome with up to a 3.8-fold coverage. In conclusion, we have demonstrated the presence of abundant ancient DNA in the coprolites surveyed. Shotgun sequencing of this material yielded a wealth of DNA sequences for a Pleistocene carnivore and allowed unbiased identification of diet.
Keywords: ancient DNA, Cervus elaphus, Crocuta crocuta, Hyaena hyaena, mitochondrial genome
1. Introduction
The first analyses of ancient DNA were initiated in 1984 [1], but it was not until 2001 that the first complete mitochondrial genome for an extinct species was reported [2,3]. With the release of ‘next generation’ DNA sequencing in 2006, large ancient DNA datasets have rapidly accumulated. These have delivered complete mitochondrial and nuclear genomes for the woolly mammoth [4,5] and a number of extinct species, including Neandertals [6]. In addition to providing ancient human and animal genomes with unprecedented speed, high-throughput DNA sequencing makes global (as opposed to targeted) analyses of genetic material feasible. This is of interest for the unbiased characterization of poorly-defined archaeological specimens [7,8], as well as the simultaneous sequencing of several genomes. Fossilised faeces or coprolites are of particular interest for such studies, as they contain DNA from the defecator [9,10] and its diet [9]. However, high-throughput DNA sequencing has not been undertaken on Pleistocene coprolites, limiting the information available from such samples to a few hundred base pairs of DNA.
In the present study, we undertook the analysis of coprolites that are abundant in European cave sites and are ascribed to the cave hyena. This animal was widespread across mid-latitude northern Eurasia during the Middle and Upper Pleistocene [11]. It is considered a vanished animal, and has been referred to as Crocuta crocuta spelaea [12]. The only extant member of the same genus, the spotted hyena (Crocuta crocuta), is confined to Sub-Saharan Africa. The cave hyena was considerably larger than the extant spotted hyena, and also displayed different body proportions [11]. The cave and spotted hyena are nevertheless so closely related that a phylogenetic tree constructed using a fragment of the mitochondrial cytochrome b (cytb) gene did not show a taxonomic delineation [13].
The spotted hyena is both a hunter and a scavenger, and the majority of its food consists of hunted medium- and large-bodied ungulates. The foraging behaviour of the cave hyena was also varied as indicated by the different cracked and chewed bones found in Pleistocene hyena dens [14]. This suggests that analyses of cave hyena coprolites might offer a rich window into the genetics of prey and carrion items, providing that the DNA from consumed species survives in the fossilized faecal material. To investigate this, we have assayed a series of cave hyena coprolites collected from a site in the French Pyrenees.
2. Material and methods
(a). Authenticity of ancient DNA sequences
The key measures implemented to avoid contamination are described in the electronic supplementary material. The efficiency of these procedures was verified through the consistent negative results obtained from mock extracts. In addition, sequence data demonstrated that the libraries mainly contained DNA from animal species that had not been previously studied by our group, or by the high-throughput DNA sequencing facility where the sequence data were generated.
To limit nucleotide misidentifications caused by post-mortem DNA damage, which mostly consist of cytosine to uracil deamination [15], we used Phusion DNA polymerase (that cannot replicate through uracil) to produce the libraries [16]. The extensive coverage of the cave hyena mitochondrial genome allowed us to calculate a sequencing accuracy greater than 99 per cent. For sequence validation studies, we performed two to four PCR replicates, cloned the amplicons and sequenced both DNA strands of eight clones to deduce a reliable consensus sequence.
(b). DNA extraction
For DNA extraction (electronic supplementary material), we used 0.8–1 g of material retrieved from the coprolite core, and a solubilization buffer consisting of 0.45 M EDTA, 10 mM Tris–HCl (pH 8.0), 0.1 per cent SDS, 65 mM DTT and 0.5 mg ml−1 proteinase K. The DNA extract was recovered as a 200 µl sample volume.
Coprolite samples were screened by PCR using primers (electronic supplementary material, table S1) designed from the partial cytb sequence of the cave hyena [13]. Initially, all samples were amplified using primer pair 1, which yields an 84 bp fragment. This was performed using a high number of PCR cycles (45 cycles) in order to ensure the detection of even trace amounts of DNA. Following this initial test, and in order to detect better differences between samples, we used primer pair 2, which yields a 127 bp DNA fragment, and performed only 33 PCR cycles.
Amplification was carried out as described [17] using a single round of 33 or 45 PCR cycles. Sequence analysis was carried out on eight clones for each amplicon (electronic supplementary material).
(c). Generating and sequencing coprolite DNA libraries
Libraries of DNA fragments suitable for single-pass sequencing with the Illumina procedure [18] were generated following the manufacturer's recommendations (San Diego, CA, USA), except for the following modifications that were introduced for the purpose of analysing ancient DNA. First, we omitted the DNA fragmentation step, owing to the already fragmented nature of the ancient DNA. An additional benefit of this is that high-molecular weight DNA, derived from modern contaminants, will be unlikely to enter the sequencing pipeline, thus reducing contamination sources. Second, the quantity of library adapters introduced in the ligation reaction was reduced by a factor of three to 10, when compared with the level recommended for libraries generated from 5 µg of modern DNA. Third, the adapter-ligated material was amplified using 40 per cent of the ligation reaction and 12 PCR cycles. This number of cycles compares favourably to that used for generating libraries from 0.5–5 µg of modern DNA (range: 10–12 PCR cycles) and was found high enough to provide robust amplification.
DNA sequencing was performed on the Illumina Genome Analyser IIx platform, and data acquisition rested on SCS2.4/RTA1.6 software. For the CC8 coprolite specimen, sequencing yielded 67.3 million high-quality DNA reads, which after trimming the adapter and the removal of sequences of less than 10 nucleotides provided 66.7 million unique fragments. For the CC9 specimen, we obtained 25.0 million high-quality reads, 24.2 million of which corresponded to unique fragments. Duplicate reads were removed, and data analysis was carried out using sequence reads greater than or equal to 20 nucleotides. This corresponds to 65.3 million reads for the CC8 sample, and to 23.6 million of reads for the CC9 sample.
(d). Sequence assembly and phylogenetic analysis
Contigs were generated from unique DNA reads using SOAP2 software [19] with a perfect match identity over 23 nucleotides.
Cave hyena mitochondrial genome and 18S gene sequences were reconstructed using both contigs and DNA reads. For cave hyena single-copy nuclear genes and for red deer mitochondrial genomes, contigs were scarce and we only used DNA reads to characterize the sequences. Full details of genes and genomes reconstruction are available in the electronic supplementary material.
For phylogenetic analysis, DNA sequences were aligned with ClustalW, and trees were constructed with maximum likelihood and Bayesian phylogenetic inference. DNA sequences and programmes used for phylogenetic analysis are described in the electronic supplementary material, tables S2 and S3.
3. Results and discussion
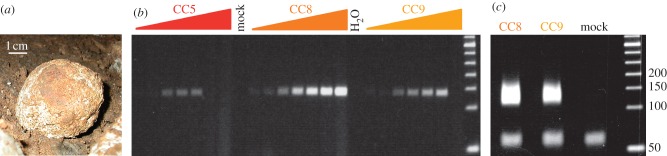
We collected nine cave hyena coprolites from the ground surface in the Coumère Cave (figure 1a, and electronic supplementary material, figure S1), which is located at 680 m above sea level in the Balaguères area of Ariège (France). To date, the cave has yielded 30 such coprolites, a cave hyena skull, a single red deer (Cervus elaphus) tooth and a number of cave bear (Ursus spelaeus) bones. We tested the samples for the presence of amplifiable ancient DNA using PCR primers targeting a fragment of the cave hyena cytb gene. Successful amplification was obtained from all coprolites using a moderate number of PCR cycles. Two samples (CC8 and CC9), which stood out from the others for the amount of DNA and were almost devoid of PCR inhibitors (figure 1b) were selected for further studies.
Figure 1.
Selecting coprolites. (a) Coumère cave coprolite. (b) PCR amplification of a fragment of the cave hyena cytb gene. Amplification (33 PCR cycles) was carried out on 0.04 to 2.5% of each DNA extract. (c) Production of libraries of DNA fragments for high-throughput sequencing. Coprolite and mock extracts ligated to oligonucleotide adapters were amplified using 12 PCR cycles. The upper band (specific for coprolite samples) was recovered for Illumina sequencing.
We prepared libraries of DNA fragments (figure 1c), and sequenced them. Read lengths were scaled to 51 or 76 nucleotides and yielded 67.3 million and 25.0 million DNA sequences for CC8 and CC9 libraries, respectively. We obtained libraries of high complexity (greater than 96% unique fragments), which as expected for ancient DNA mostly contained very short fragments (electronic supplementary material, figure S2).
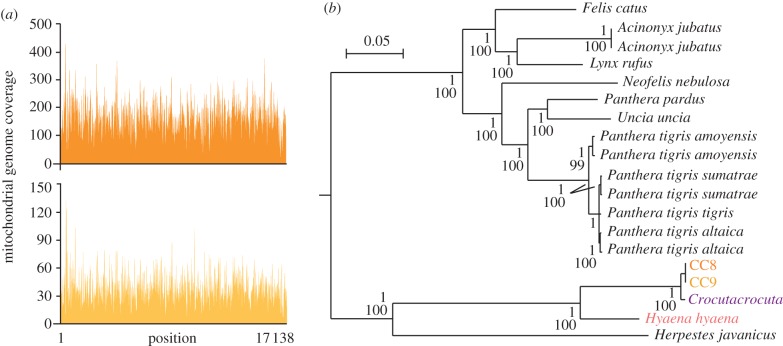
Our data mining procedure included both the analysis of individual reads and contigs obtained by de novo assembly of the reads. We aligned the reads to sequences recorded in GenBank nt and wgs using MegaBlast [20]. Bacterial DNA accounted for 0.8 per cent of the sequences. In human fresh faeces, 7.6 per cent of Illumina reads corresponded to bacterial genomes deposited in GenBank [21]. In our dataset, the largest number of hits (6.4% of the reads) corresponded with the domestic cat (Felis catus). This observation is consistent with the fact that the domestic cat is the closest species to the cave hyena (a Feliformia) for which a nuclear genome sequence is available. However, considering that these two species belong to different families (Hyaenidae versus Felidae) and that only 65 per cent of the euchromatin cat genome sequence has been deciphered [22], the number of cave hyena sequences is probably underestimated by database search. Further evidence of abundant Hyaenidae DNA in the coprolites was obtained by de novo assembly of the reads, which yielded a series of contigs of up to 8.3 kbp aligning with high confidence to the mitochondrial genome of extant hyenas. We assembled overlapping contigs into a provisional cave hyena sequence that was used to retrieve all DNA reads aligning to it with a maximum of one mismatch and one indel. This strategy yielded complete circular mitochondrial genomes of 17 138 bp with an average unique read depth of 158x and 35x for CC8 and CC9, respectively (figure 2a). The high redundancy and close similarity between individual reads and the consensus sequence (electronic supplementary material, figure S3) show that we were able to recover reliable cave hyena mitochondrial genome sequences. This conclusion is supported by open reading frames in all 13 protein coding genes of the mitochondrial genome that predict polypeptides whose size is similar to that reported in other Feliformia species.
Figure 2.
Cave hyena mitochondrial genome. (a) Number of reads for each position of the cave hyena mitochondrial genome for CC8 (top) and CC9 (bottom) coprolites. (b) Phylogenetic analysis of the cave hyena with complete mitochondrial genome sequences. Tree construction was performed by Bayesian phylogenetic inference using Caniformia sequences as an outgroup. The posterior probability and bootstrap values that support the nodes are indicated for Bayesian phylogenetic inference and maximum-likelihood analysis, respectively. Coloured characters indicate mitochondrial genomes provided by this study. The scale indicates the genetic distance.
The two cave hyena mitochondrial genome sequences differ from each other by two substitutions located in the repetitive motifs of the control region, which may be of little significance since such motifs are difficult to sequence and align in ancient specimens and are not taken into account for the phylogenetic analyses. These two mitochondrial genomes, together with those for the extant spotted and striped hyena, were used to perform a comprehensive phylogeny of Feliformia (figure 2b). The topology of the tree is robustly supported and establishes unequivocally the close evolutionary relationship between the spotted and the cave hyena. The cave hyena genome only displays 115 differences with the spotted hyena sequence. We screened some of these differences by PCR analysis and systematically confirmed them (electronic supplementary material, figure S4), which further supports the accuracy of our cave hyena genome sequence. The genetic distance between the cave and spotted hyena is markedly lower than that recorded between different species from the Hyaenidae family, as shown by the comparison with the striped hyena genome. Further comparison with other Feliformia (e.g. Panthera tigris) indicate that the genetic distance between the cave and the spotted hyena is in the range of that exhibited between sub-species or even different specimens from the same sub-species (electronic supplementary material, table S4). The close phylogenetic position of the cave and spotted hyena mitochondrial genome sequences is in line with the observation that one of the four Crocuta clades characterized using a fragment of the cytb gene included cave as well as extant spotted hyena specimens [13]. The cave and spotted hyena cytb sequences of the current study belong to this clade. They correspond to the A2 and A3 haplotype, respectively.
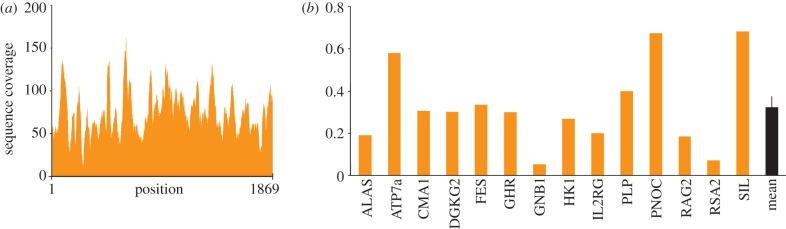
We estimated the coverage of the cave hyena nuclear genome using two strategies. First, we analysed the 18S gene, which displays approximately 200 copies in mammalian genomes [23]. The full-length sequence of this gene (1869 nucleotides) was obtained for both samples, with an average unique read depth of 75x for CC8 (figure 3a) and 31x for CC9. The ratios of the 18S read depth to the number of copies for this gene (CC8: 75/200; CC9: 31/200) provide estimates of the genome coverage of 0.38x (CC8) and 0.16x (CC9). Second, to evaluate independently the genome coverage, we selected a set of 14 phylogenetically informative genes that have been characterized in a number of Feliformia species [24]. This was performed using the most complete sequenced specimen (CC8). We obtained DNA reads for all 14 genes (figure 3b), and the average unique read depth for these single-copy genes (0.32 ± 0.05x) predicted a nuclear genome coverage very close to that deduced for the same sample from the 18S gene data (0.38x). The phylogeny deduced from the 14 nuclear genes again highlighted the close evolutionary relationship between the cave and the spotted hyena (electronic supplementary material, figure S5). However, the branch leading to the cave hyena was much longer than that leading to the spotted hyena. This may be explained by the fact that the information gathered on most nucleotides consisted of single-pass sequencing. We tested this hypothesis in a series of PCR experiments by sequencing several clones from different amplicons to achieve a high redundancy for each nucleotide. The consensus sequence deduced from PCR analysis revealed a single difference between the cave and the spotted hyena among 604 nuclear positions surveyed (electronic supplementary material, figure S6). This extensive similarity supports the notion that, from a genetic point of view, the cave hyena is the Eurasian representative of the Pleistocene spotted hyena rather than a distinct species.
Figure 3.
Cave hyena nuclear sequence data for CC8 coprolite. (a) Number of reads for each position of the 18S gene sequence. (b) Individual sequence coverage of 14 cave hyena nuclear genes. The dark column indicates the mean read depth ± s.e.m. for the 14 genes.
The cave hyena nuclear genome coverage calculated from the 18S data mainly varied from one specimen to another because we sequenced a different number of DNA fragments from these samples (CC8, 2,39 Gb; CC9, 0,86 Gb). Using these data, we estimated (electronic supplementary material) that 43 per cent of CC8, and 50 per cent of CC9 DNA can be ascribed to the cave hyena. These values compare favourably with the amount of animal DNA in Pleistocene specimens from frozen environments, which reached 40 per cent in a polar bear tooth [25], 55 per cent in a mammoth bone [4] and 58–90% in mammoth hair shafts [5]. By contrast, Pleistocene tooth or bone samples from cave sites usually contain less than 6 per cent of animal or human DNA [6,7]. One noticeable exception is a phalanx from the Denisova Cave that yielded 70 per cent of hominid DNA [8].
Several attempts were performed for radiocarbon dating of the coprolites by accelerator mass spectrometry (AMS). Initially, stringent protein extraction procedures did not yield a sufficient amount of organic carbon for dating (J. van der Plicht 2012, personal communication). Further attempts returned radiocarbon ages of 9170 ± 50 (Beta-281961) and 13 060 ± 90 (Beta-281962) years before present (BP) for CC8 and CC9 samples, respectively. These ages are much younger than the date assumed for the disappearance of the cave hyena from Europe, which can be traced back to at least 15 000 BP or even as old as 26 000 BP [26]. Therefore, we explored further the reliability of the radiocarbon ages by dating a cave hyena coprolite retrieved from a well-defined stratigraphic unit that contains cave bear bones of 27 440–30 220 BP [27]. This coprolite sample was AMS-dated to 22 590 ± 110 BP (Beta-300724). Thus, these data demonstrate that the actual age of the cave hyena coprolites was underestimated.
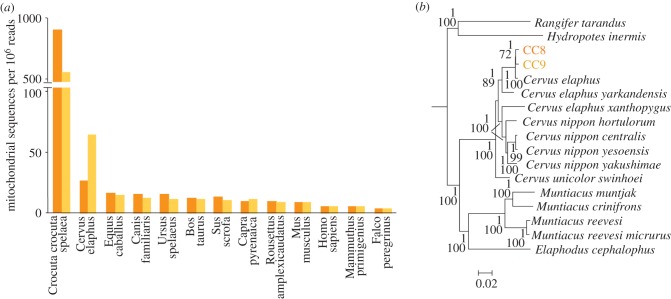
To gain a molecular insight into the cave hyena diet, we searched for DNA reads that may indicate reliably additional animal species. Mitochondrial genomes are well suited for this purpose, since they are available for a number of extant and extinct species. Apart from the cave hyena, we only observed a high number of reliable matches for the red deer (figure 4a). As outlined in the electronic supplementary material, methods, hits for other species consisted of evolutionary conserved portions of mammalian mitochondrial genomes. To analyse further the Cervidae sequences, we retrieved all DNA reads that aligned with one or the other of the available deer mitochondrial genomes. This enabled the characterization of mitochondrial genomes with 3.5x and 3.8x coverage for CC8 and CC9, respectively. Phylogenetic analysis demonstrated that these mitochondrial sequences correspond to Cervus elaphus DNA (figure 4b). We also explored the possibility that the deer sequences could correspond to the extinct giant deer (Megaloceros giganteus). In the absence of a complete mitochondrial genome for this species, we performed a phylogenetic analysis based on the cytb sequence. This analysis confirmed the presence of red deer DNA in the samples (electronic supplementary material, figure S7).
Figure 4.
Identifying Cervus elaphus DNA in coprolites. (a) Number of reads that display a perfect match to the indicated mitochondrial genomes. (b) Phylogenetic analysis of the Cervidae mitochondrial sequences retrieved from cave hyena coprolites. Tree construction was performed by Bayesian phylogenetic inference using Bovidae sequences for delineating an outgroup. The posterior probability and bootstrap values that support the nodes are indicated for Bayesian phylogenetic inference and maximum-likelihood analysis, respectively. Yellow bars, CC9 coprolite; orange bars, CC8 coprolite.
In conclusion, this work provides, to our knowledge, the first genomic dataset for the extinct cave hyena. The high-throughput sequencing strategy also allowed unbiased identification of the cave hyena diet thus highlighting the interaction between two animal species. Altogether these data show that cave hyena coprolites and possibly those of other carnivore species deserve special attention for palaeogenomic studies.
Acknowledgements
We thank B. Mulot (Beauval Zoo, France) and F. Huyghe (Cerza Zoo, France) for the extant hyena samples, A. Martel for help in data analysis, and N. Griffiths for language correction. This work was supported by grants from the CEA. C.B. received PhD funding from the CEA. The DNA reads of this study have been deposited at EBI under accession number ERA030882 and ERA030883. GenBank entries for annotated sequences are JF894376-JF894380.
References
- 1.Higuchi R., Bowman B., Freiberger M., Ryder O. A., Wilson A. C. 1984. DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282–284 10.1038/312282a0 (doi:10.1038/312282a0) [DOI] [PubMed] [Google Scholar]
- 2.Cooper A., Lalueza-Fox C., Anderson S., Rambaut A., Austin J., Ward R. 2001. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409, 704–707 10.1038/35055536 (doi:10.1038/35055536) [DOI] [PubMed] [Google Scholar]
- 3.Haddrath O., Baker A. J. 2001. Complete mitochondrial DNA genome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proc. R. Soc. Lond. B 268, 939–945 10.1098/rspb.2001.1587 (doi:10.1098/rspb.2001.1587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poinar H. N., et al. 2006. Metagenomics to paleogenomics, large-scale sequencing of mammoth DNA. Science 311, 392–394 10.1126/science.1123360 (doi:10.1126/science.1123360) [DOI] [PubMed] [Google Scholar]
- 5.Miller W., et al. 2008 Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456, 387–390 10.1038/nature07446 (doi:10.1038/nature07446) [DOI] [PubMed] [Google Scholar]
- 6.Green R. E., et al. 2010. A draft sequence of the Neandertal genome. Science 328, 710–722 10.1126/science.1188021 (doi:10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blow M. J., Zhang T., Woyke T., Speller C. F., Krivoshapkin A., Yang D. Y., Derevianko A., Rubin E. M. 2008. Identification of ancient remains through genomic sequencing. Genome Res. 18, 1347–1353 10.1101/gr.076091.108 (doi:10.1101/gr.076091.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reich D., et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 10.1038/nature09710 (doi:10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poinar H. N., Hofreiter M., Spaulding W. G., Martin P. S., Stankiewicz B. A., Bland H., Evershed R. P., Possnert G., Pääbo S. 1998. Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281, 402–406 10.1126/science.281.5375.402 (doi:10.1126/science.281.5375.402) [DOI] [PubMed] [Google Scholar]
- 10.Gilbert M. T., et al. 2008. DNA from pre-Clovis human coprolites in Oregon, North America. Science 320, 786–789 10.1126/science.1154116 (doi:10.1126/science.1154116) [DOI] [PubMed] [Google Scholar]
- 11.Kurtén B. 1968. Pleistocene mammals of Europe. London, UK: Weidenfeld & Nicholson [Google Scholar]
- 12.Goldfuss G. A. 1823. Osteologische Beiträge zur Kenntnis verschiedener Säugethiere der Vorwelt. VI. Ueber die Hölen-Hyäne (Hyaena spelaea). Nova Acta Physico-Medica Academiea Caesarae Leopoldino-Carolinae Naturae Curiosorum 3, 456–490 [Google Scholar]
- 13.Rohland N., Pollack J. L., Nagel D., Beauval C., Airvaux J., Pääbo S., Hofreiter M. 2005. The population history of extant and extinct hyenas. Mol. Biol. Evol. 22, 2435–2443 10.1093/molbev/msi244 (doi:10.1093/molbev/msi244) [DOI] [PubMed] [Google Scholar]
- 14.Fosse P. 1997. Variabilité des assemblages osseux créés par l'hyène des caverns. Paléo 9, 15–54 10.3406/pal.1997.1227 (doi:10.3406/pal.1997.1227) [DOI] [Google Scholar]
- 15.Stiller M., et al. 2006. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA 103, 13 578–13 584 10.1073/pnas.0605327103 (doi:10.1073/pnas.0605327103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen M., et al. 2010. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762 10.1038/nature08835 (doi:10.1038/nature08835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bon C., et al. 2008. Deciphering the complete mitochondrial genome and phylogeny of the extinct cave bear in the Paleolithic painted cave of Chauvet. Proc. Natl Acad. Sci. USA 105, 17 447–17 452 10.1073/pnas.0806143105 (doi:10.1073/pnas.0806143105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley D. R., et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 10.1038/nature07517 (doi:10.1038/nature07517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 10.1093/bioinformatics/btp336 (doi:10.1093/bioinformatics/btp336) [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z., Schwartz S., Wagner L., Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214 10.1089/10665270050081478 (doi:10.1089/10665270050081478) [DOI] [PubMed] [Google Scholar]
- 21.Qin J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 10.1038/nature08821 (doi:10.1038/nature08821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pontius J. U., et al. 2007. Initial sequence and comparative analysis of the cat genome. Genome Res. 17, 1675–1689 10.1101/gr.6380007 (doi:10.1101/gr.6380007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paule M. R. 1998. Class I genes. In Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I (ed. Paule M. R.), pp. 1–8 New York, NY: Springer [Google Scholar]
- 24.Johnson W. E., Eizirik E., Pecon-Slattery J., Murphy W. J., Antunes A., Teeling E., O'Brien S. J. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311, 73–77 10.1126/science.1122277 (doi:10.1126/science.1122277) [DOI] [PubMed] [Google Scholar]
- 25.Lindqvist C., et al. 2010. Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc. Natl Acad. Sci. USA 107, 5053–5057 10.1073/pnas.0914266107 (doi:10.1073/pnas.0914266107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart A. J., Lister A. M. 2007. Patterns of Late Quaternary megafaunal extinctions in Europe and northern Asia. Cour. Forsch. Inst. Senckenberg 259, 287–297 [Google Scholar]
- 27.Bon C., Berthonaud V., Fosse P., Gély B., Maksud F., Vitalis R., Philippe M., van der Plicht J., Elalouf J. M. 2011. Low regional diversity of late cave bears mitochondrial DNA at the time of Chauvet Aurignacian paintings. J. Archaeol. Sci. 38, 1886–1895 10.1016/j.jas.2011.03.033 (doi:10.1016/j.jas.2011.03.033) [DOI] [Google Scholar]