Abstract
The Wnt pathways contribute to many processes in cancer and development, with β-catenin being a key canonical component. p120-catenin, which is structurally similar to β-catenin, regulates the expression of certain Wnt target genes, relieving repression conferred by the POZ- and zinc-finger-domain-containing transcription factor Kaiso. We have identified the kinase Dyrk1A as a component of the p120-catenin–Kaiso trajectory of the Wnt pathway. Using rescue and other approaches in Xenopus laevis embryos and mammalian cells, we found that Dyrk1A positively and selectively modulates p120-catenin protein levels, thus having an impact on p120-catenin and Kaiso (and canonical Wnt) gene targets such as siamois and wnt11. The Dyrk1A gene resides within the Down's syndrome critical region, which is amplified in Down's syndrome. A consensus Dyrk phosphorylation site in p120-catenin was identified, with a mutant mimicking phosphorylation exhibiting the predicted enhanced capacity to promote endogenous Wnt-11 and Siamois expression, and gastrulation defects. In summary, we report the biochemical and functional relationship of Dyrk1A with the p120-catenin–Kaiso signaling trajectory, with a linkage to canonical Wnt target genes. Conceivably, this work might also prove relevant to understanding the contribution of Dyrk1A dosage imbalance in Down's syndrome.
Key words: Dyrk1A, Hipk, Siamois, Wnt11, p120-catenin
Introduction
The canonical Wnt signaling pathway is fundamental to embryo development and tumor progression, contributing to embryonic gene expression programs as well as later homeostasis (Behrens and Lustig, 2004; Huelsken and Birchmeier, 2001; Klaus and Birchmeier, 2008; Lai et al., 2009; Moon et al., 2004; van Amerongen and Nusse, 2009; Wend et al., 2010). Numerous studies have demonstrated that disruption of Wnt signaling results in a variety of developmental defects and adult disease. β-catenin is a key component in Wnt signaling (Grigoryan et al., 2008; MacDonald et al., 2009; Moon, 2005; Mosimann et al., 2009). Metabolically stabilized β-catenin, which has an unphosphorylated N-terminus, is associated with pathway activation, leading to the relief of TCF–LEF-mediated repression (i.e. activation) of Wnt target genes (Liu et al., 2002; MacDonald et al., 2009; Verheyen and Gottardi, 2010).
p120-catenin (also known as catenin-δ1) is a catenin family member recently proposed to act coordinately with β-catenin in certain vertebrate Wnt signaling contexts (Hong et al., 2010; Johnson et al., 2010; Park et al., 2006; Park et al., 2005; Spring et al., 2005; van Roy and McCrea, 2005). p120-catenin binds Kaiso (Daniel and Reynolds, 1999; Kim et al., 2002) and its targets, although this is still the subject of some debate (Iioka et al., 2009; Ruzov et al., 2009a; Ruzov et al., 2009b), include certain Wnt target genes such as siamois and wnt11 (Hong et al., 2010; Kim et al., 2004; Park et al., 2005). Kaiso is a vertebrate-specific transcriptional repressor of the BTB (POZ)- and zinc-finger-domain-containing protein family, and the Kaiso repressor has bi-modal DNA binding activity, associating with both sequence-specific Kaiso binding sites (KBS) and with methylated DNA (Daniel, 2007; Daniel et al., 2002; Prokhortchouk et al., 2001). Through the competitive displacement of Kaiso from DNA, p120-catenin is thought to relieve Kaiso-mediated repression at KBS (the role of p120-catenin is unclear at methylated sites) (Daniel, 2007; Kelly et al., 2004; Park et al., 2005; Spring et al., 2005).
Work in our laboratory recently found that the signaling pool of p120-catenin isoform-1 is subject to modulation by the canonical Wnt pathway, in a manner similar to that of β-catenin (Hong et al., 2010). Additional biochemical and function evidence supports the involvement of p120-catenin in vertebrate developmental Wnt signaling (Hong et al., 2010; Park et al., 2006; Park et al., 2005; Spring et al., 2005), with work from an independent group using cell lines also pointing to the positive role of p120-catenin in the canonical Wnt pathway, albeit by a different mechanism (Casagolda et al., 2010).
Aside from the role of vertebrate p120-catenin in the nucleus, p120-catenin binds the cytoplasmic tails of cadherins, thereby contributing to cadherin stabilization and the functional state of cell–cell contacts (Davis et al., 2003; Reynolds and Carnahan, 2004; Xiao et al., 2007). Additionally, p120-catenin directly or indirectly modulates small GTPases such as RhoA and Rac1, resulting in downstream effects that prominently include cytoskeletal rearrangements (Anastasiadis, 2007; Anastasiadis and Reynolds, 2001; Bernards and Settleman, 2005; Wildenberg et al., 2006). Because of the role of p120-catenin in the nucleus and its interaction with cadherin and small-GTPases, it (and/or other p120-catenin subfamily members) is believed to be pertinent to the progression of cancer and other pathologies (Bartlett et al., 2010; Davis and Reynolds, 2006; Dohn et al., 2009; Perez-Moreno et al., 2006; Perez-Moreno et al., 2008; Reynolds and Roczniak-Ferguson, 2004).
Dual-specificity tyrosine-regulated kinase 1A (Dyrk1A) belongs to the DYRK subfamily of protein kinases, and maps to chromosome 21 (21q22.2), within the Down's syndrome critical region (DSC) (Cheon et al., 2003; Shindoh et al., 1996; Song et al., 1996). Dyrk1A is highly conserved at the protein level across vertebrates and invertebrates, being most prominently expressed in developing brain and heart (Okui et al., 1999). Evidence is accumulating that DYRK1A, one of approximately 20 genes located in this region, is important in the neurobiological alterations of Down's syndrome (Altafaj et al., 2001; Fotaki et al., 2002; Park et al., 2009; Smith et al., 1997; Tejedor and Hammerle, 2010; Wegiel et al., 2010). Recently, a related subfamily member, Hipk2, was found to phosphorylate and relieve TCF3-mediated repression in Xenopus laevis (Hikasa et al., 2010). There is also some evidence of Hipk2 and Hipk1 kinase activity either increasing or decreasing β-catenin stability (Hikasa et al., 2010; Kim et al., 2010; Lee et al., 2009; Louie et al., 2009), and thus having roles in canonical Wnt signaling.
Here we report the identification of Dyrk1A as a positive modulator of p120-catenin. Because Dyrk1A did not modulate β-catenin levels in our studies, Dyrk1A might preferentially or specifically act upon p120-catenin. Dyrk1A expression results in increased p120-catenin levels, whereas depletion has the opposite effect. We further determined that Dyrk1A modulates the expression of target genes previously shown to be regulated by the p120-catenin–Kaiso pathway, and that appropriate manipulation of p120-catenin or Kaiso levels rescues developmental and transcriptional (gene target) effects of Dyrk1A. Consistent with an impact upon certain Wnt-target genes (presumably occurring through stabilization of p120-catenin), Dyrk1A expression promoted duplicate axis formation in Xenopus laevis embryos when coexpressed with β-catenin. Taken together, our results suggest that one means by which Dyrk1A participates in embryogenesis is through modulation of the p120-catenin–Kaiso pathway. Upstream control of Dyrk1A remains an area of uncertainty, with questions remaining about whether it is responsive to Wnt or other ligands.
Results and Discussion
Dyrk1A specifically increases p120-catenin protein levels
To gain insight into the mechanisms by which p120-catenin is regulated, we performed a small-scale kinase screen. Various kinases were transfected into HEK293T cells, and p120-catenin levels monitored by immunoblotting (data not shown). Dyrk1A was selected because of its robust impact on p120-catenin levels (see below). Coexpression of either Dyrk1A1 or Dyrk1A2 with p120-catenin resulted in heightened p120-catenin protein in Xenopus embryo extracts (Fig. 1A). This was in contrast to Dyrk1A kinase dead (KD), which had no such effect (Fig. 1B). Consistent with our Xenopus data, Dyrk1A transfected into HEK293T cells increased endogenous p120-catenin levels, while β-catenin remained unaffected (Fig. 1C). Likewise, we determined that depletion of Dyrk1A, through the use of two independent Dyrk1A siRNAs, strikingly lowered endogenous p120-catenin but not β-catenin levels (Fig. 1D). Previous reports from our lab and others have shown that p120-catenin relieves Kaiso-mediated repression of certain Wnt target genes such as siamois and wnt-11 (Hong et al., 2010; Kim et al., 2004; Park et al., 2005). Upon binding, p120-catenin is thought to somehow displace or compete Kaiso from its sequence-specific sites and facilitate the exit of Kaiso from the nucleus. We thus tested whether Dyrk1A expression likewise recruits Kaiso from the nucleus. Consistent with such a model, exogenous Dyrk1A expression resulted in a substantially greater proportion of Kaiso appearing in the cytoplasm (supplementary material Fig. S1A).
Fig. 1.
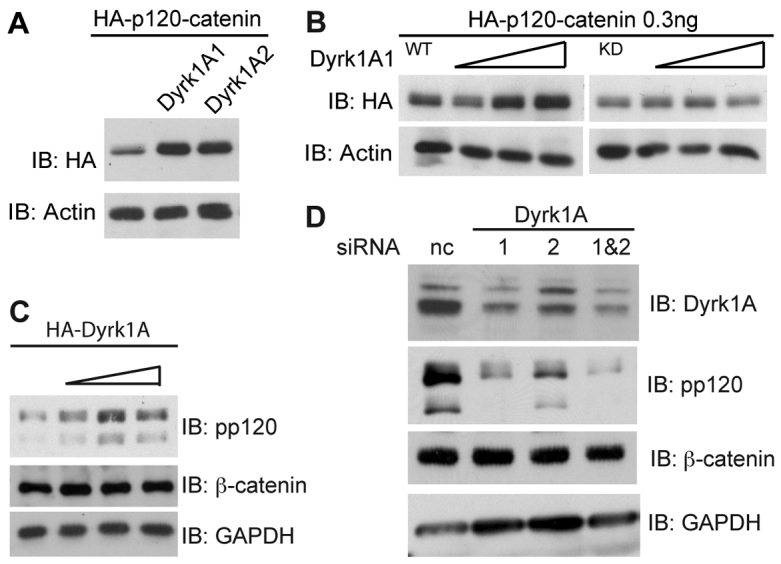
Dyrk1A modulates p120-catenin levels and the intracellular localization of Kaiso. (A) Either Dyrk1A1 or -1A2 was co-injected with HA-tagged p120-catenin (0.25 ng) into Xenopus embryos at the one-cell stage, followed by immunoblotting for HA–p120-catenin (12CA5) or actin (negative control). (B) HA–p120-catenin (0.25 ng) was microinjected into Xenopus embryos with wild-type or kinase-dead (KD-K188R) Dyrk1A. Embryos were harvested as early gastrulas (stage 10–11) and immunoblotted for HA–p120-catenin, with actin serving as an internal loading control. (C) Increasing doses of HA–Dyrk1A were transfected into 293T cells, and endogenous p120-catenin, β-catenin and GAPDH monitored by immunoblotting. (D) HEK293T cells were transfected with one or both Dyrk1A siRNAs (50 pmol), as indicated, for 48 hours. Endogenous p120-catenin, β-catenin, Dyrk1A and GAPDH levels were monitored by immunoblotting (pp120, BD Transduction; Dyrk1A, ab71464, Abcam). Representative outcomes of experiments repeated three or more times with consistent results are shown.
Association of Dyrk1A with p120-catenin
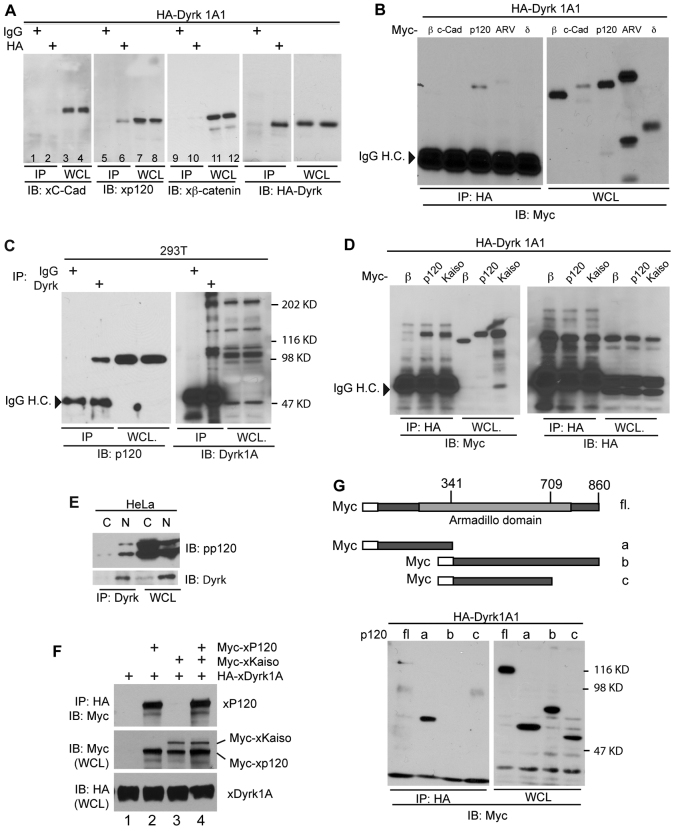
Given the effect of Dyrk1A on p120-catenin, we next examined whether Dyrk1A interacts with p120-catenin. A modest association of the two proteins was detected (Fig. 2A, lane 6), but there was no apparent co-precipitation of Dyrk1A and C-cadherin or β-catenin (negative control lanes 2 and 10). Because p120-catenin belongs to the p120-catenin subfamily, other members such as ARVCF and δ-catenin were examined (δ-catenin is also known as NPRAP and catenin δ2). Interestingly, only p120-catenin associated with Dyrk1A (Fig. 2B, lane 3). However, we cannot exclude the possibility that other p120-catenin family members are phosphorylated and stabilized by Dyrk1A, because δ-catenin, ARVCF and Pkp3 possess multiple potential Dyrk1A phosphorylation sites (results not shown). To assess the interaction of endogenous Dyrk1A with endogenous p120-catenin, we immunoprecipitated Dyrk1A from HEK293T cell extracts and employing the 6H11 antibody to resolve p120-catenin (Fig. 2C, lane 2). Immunoprecipitation was also performed using an antibody directed against Dyrk1A (Fig. 2E), which was used in conjunction with the pp120 antibody in immunoblot assays to determine the association of Dyrk1a with both long and short p120-catenin isoforms. Interestingly, an association of Dyrk1A with Kaiso, presumably through p120-catenin or other p120-catenin family members, was resolved when hemagglutinin epitope tagged (HA)–Dyrk1A was immunoprecipitated from Xenopus embryo extracts (Fig. 2D, lane 3). Because Dyrk1A is mainly present in the nucleus (Hammerle et al., 2003; Hammerle et al., 2008), we assayed where Dyrk1A interacts with p120-catenin to stabilize it, by performing co-immunoprecipitations on nuclear and cytoplasmic extracts. Interestingly, despite p120-catenin being more abundant in the cytoplasm, it predominately associated with Dyrk1A in the nucleus (Fig. 2E). Using immunofluorescence we also demonstrated that nuclear p120-catenin levels increased upon Dyrk1A overexpression (supplementary material Fig. S1B). This suggests that Dyrk associates with p120-catenin in the nucleus to phosphorylate and/or stabilize it, and thereby to increase its effectiveness in relieving Kaiso-mediated repression. Because Dyrk1A predominantly interacts with p120-catenin in the nucleus, we wondered if Dyrk1A might evidence a lesser impact upon cadherin levels or cell–cell adhesion. We found that cadherin levels do not respond in a consistent fashion following Dyrk1A depletion versus overexpression (although in individual experiments responses were at times noted; supplementary material Fig. S2D), and that cell–cell adhesion was not obviously affected when tested using an in vitro adhesion assay (Gu et al., 2009) (data not shown). Thus, these data suggest that Dyrk1A is more directly involved in the nuclear functions of p120-catenin, or that additional effects upon cadherin were difficult to discern using our experimental conditions.
Fig. 2.
Association of Dyrk1A with p120-catenin. (A) Embryos were injected with HA–Dyrk1A and harvested as early gastrulas (stage 10.5). Lysates were immunoprecipitated for HA–Dyrk1A, and the association with endogenous p120-catenin resolved by immunoblotting. β-catenin and C-cadherin served negative controls. (B) HA–Dyrk1A (0.5 ng) was co-injected with Myc-tagged β-catenin, C-cadherin, p120-catenin, ARVCF or δ-catenin (1 ng) into both blastomeres of two-cell embryos. HA–Dyrk1A immunoprecipitates were immunoblotted with anti-Myc antibody to detect co-associating proteins. (C) Endogenous Dyrk1A was immunoprecipitated from HEK293T cells, and endogenous p120-catenin monitored using anti-p120-catenin antibody (6H11, Santa Cruz). (D) HA–Dyrk1A was co-injected with Myc–p120-catenin, Myc–β-catenin (β) or Myc–Kaiso into early Xenopus embryos. Embryos lysates were immunoprecipitated for HA–Dyrk1A. Anti-Myc or -HA immunoblotting was used to test association (versus none) of β-catenin, p120-catenin or Kaiso. (E) Nuclear (N) and cytoplasmic (C) extracts were made from HeLa cells, followed by Dyrk1A immunoprecipitation and immunoblotting using anti-p120-catenin or Dyrk1A antibodies. WCL, whole cell lysate. (F) Myc–p120-catenin, Myc–Kaiso and HA–Dyrk1A were in vitro transcribed as described previously (Hong et al., 2010). Differing combinations of the proteins were mixed as indicated, and immunoprecipitated for Dyrk1A (using anti-HA). Co-precipitating p120-catenin and Kaiso were then monitored (using anti-Myc). (G) Depiction of Myc–p120-catenin deletion constructs (a–f). Panels to the right show immunoblotting of the Myc–p120-catenin constructs (a–f; 0.5 ng mRNA), co-precipitating (versus not) with co-injected HA–Dyrk1A1 (0.5 ng mRNA).
Given that Dyrk1A also associates with Kaiso (Fig. 2D), we next used in vitro binding assays to test whether Dyrk1A associates with Kaiso indirectly or directly. Using in-vitro-translated Dyrk1A, p120-catenin and Kaiso, we found that Dyrk1A associates directly with p120-catenin but not with Kaiso (Fig. 2F). Initially, we expected that p120-catenin might bridge Kaiso to Dyrk1A. However, coexpression of p120-catenin and Kaiso did not increase Kaiso association with Dyrk1A in vitro, suggesting that Dyrk1A interacts with Kaiso by another means, conceivably involving other p120-catenin family members, or Frodo or Dapper, which are known to associate with and stabilize p120-catenin (Park et al., 2006). To map the region of p120-catenin that interacts with Dyrk1A, we expressed various p120-catenin deletion constructs (a–f) generated previously (Hong et al., 2010), and performed co-immunoprecipitations. Full-length, but more prominently the isolated N-terminal region of p120-catenin (construct a) associated with Dyrk1A (Fig. 2G, lane 2), whereas p120-catenin constructs lacking this region (constructs b and c), did not. These findings indicate that the Dyrk1A kinase is capable of associating with the N-terminal region of p120-catenin, presumably accounting for the noted impact of Dyrk1A on p120-catenin protein levels (Fig. 1).
Dyrk1A relieves Kaiso repressor activity through p120-catenin
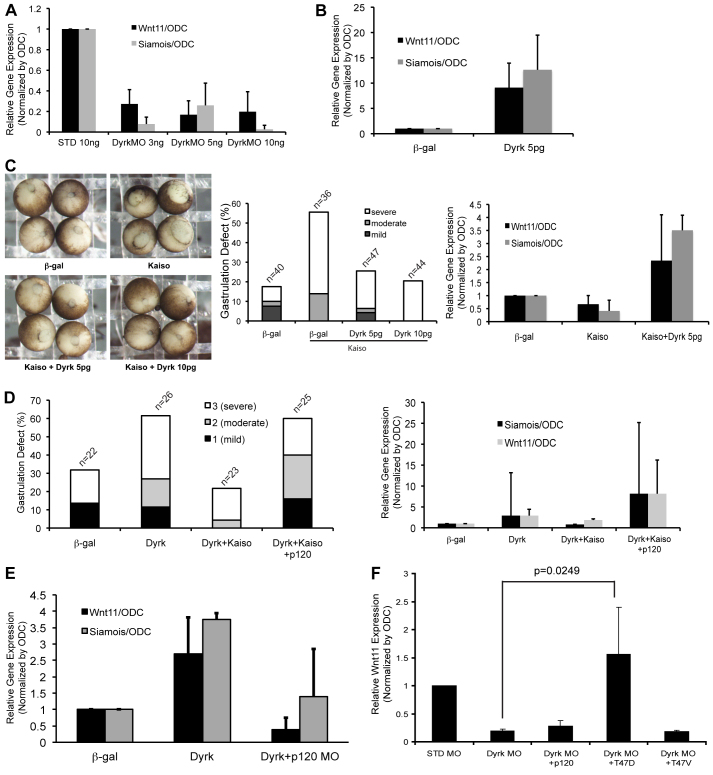
At the functional level, p120-catenin has a number of in vivo activities, including the capacity to relieve Kaiso-mediated repression of target genes containing sequence-specific Kaiso consensus sites (KCS). Given that Dyrk1A associates with and phosphorylates p120-catenin (supplementary material Fig. S3), we tested whether Dyrk1A expression has an impact upon established endogenous p120-catenin–Kaiso target genes such as wnt11 and siamois. Indeed, as seen with real-time PCR analysis, the expression of exogenous Dyrk1A in early Xenopus embryos (resulting in p120-catenin stabilization and thus relief of Kaiso-mediated repression) enhanced wnt-11 and siamois gene transcription (Fig. 3B). Complimenting such overexpression data, the morpholino-directed knockdown of Dyrk1A dramatically decreased both wnt11 and siamois expression (Fig. 3A). Characterization of the Dyrk1A morpholino is summarized in the supplementary material Fig. S2B,D,E. Although variable, the embryonic depletion of Dyrk1A in some instances led to increased or decreased C-cadherin protein levels (supplementary material Fig. S2B). Despite such observations, in vitro cadherin adhesion assays and ConA precipitations (to resolve the extent of cadherin-bound p120-catenin) suggested that Dyrk1A levels are not obviously linked to effects upon cadherin levels or adhesive function (results not shown). This leaves open the possibility that Dyrk1A predominantly affects the signaling pool of p120-catenin. p120-catenin levels are generally correlated with cadherin levels, so this might need to be evaluated in future work. In all cases, the knockdown of Dyrk1A in embryos resulted in gastrulation failure (supplementary material Fig. S2E). A previous report from our laboratory showed that the Kaiso repressor is required for Xenopus gastrulation, with its depletion or overexpression resulting in heightened or lowered expression of Wnt11, respectively, and thus failed gastrulation movements (Kim et al., 2004). We therefore tested whether Kaiso overexpression phenotypes could be rescued by co-expression of Dyrk1A. As anticipated, exogenous Kaiso alone resulted in a reduction in normal gastrulation. However, when Kaiso was coexpressed with a minimal amount of Dyrk1A (previously titrated to produce a small phenotypic consequence), substantial rescues were observed (Fig. 3C). Consistent with such phenotypic rescues, real-time PCR showed, at the molecular level, that exogenous Dyrk1A relieved gene repression by exogenous Kaiso acting upon Wnt11 and Siamois (Fig. 3C). To further test the functional link between Dyrk1A and Kaiso (presumably taking place through p120-catenin), we found that exogenous Dyrk1A produced gastrulation delays in Xenopus embryos. Thus, we next tested if exogenous Kaiso could rescue Dyrk1A gastrulation delays. Although gastrulation delays produced by Dyrk1A are subtle, we observed substantial rescue with the appropriately titrated expression of Kaiso (supplementary material Fig. S4A). Likewise, the Wnt11 and Siamois gene expression data supported our model of the functional interrelationship between Dyrk1A and the p120-catenin–Kaiso pathway. We next tested whether p120-catenin overexpression could interfere with rescue of Dyrk1 expression by Kaiso. Indeed, the co-injection of p120-catenin with Kaiso and Dyrk1A re-elevated the level of gastrulation failure, as well raising the expression of wnt11 and siamois (Fig. 3D). To evaluate the effect of p120-catenin on the interaction between Dyrk1A and Kaiso, we depleted p120-catenin after overexpression of Dyrk1A. As expected from earlier findings, Dyrk1A overexpression resulted in increased levels of Wnt11 and Siamois. If a morpholino to deplete p120-catenin was injected together with Dyrk1A, wnt11 and siamois expression were instead reduced (Fig. 3E). Together, these data suggest that the biological effects of Dyrk1A occur predominantly through its interactions within the p120-catenin–Kaiso pathway.
Fig. 3.
Dyrk1A modulates p120-catenin–Kaiso-dependent gene expression. (A) The indicated amounts of Dyrk1A morpholino were injected into embryos at the 1-cell stage. Gastrula cDNA was assayed by real-time RT-PCR for Wnt11 and Siamois transcript levels. Embryos were harvested at stage 9–10, which proved best for tests involving Siamois expression. For Wnt 11, embryos were collected mainly at stage 10.5–12. Gene expression levels were normalized to ODC (ornithine decarboxylase). (B) Dyrk1A (5 pg) was injected into embryos at the 1-cell stage. Wnt11 and Siamois transcript levels were analyzed by real-time PCR as described in A. (C) Gastrulation (blastopore closure) failures followed the expression of Kaiso (0.5 ng), whereas the co-expression of Dyrk1A (5 pg or 10 pg), partially rescued the effects of Kaiso. Under similar experimental conditions, gastrula cDNA was assayed by real-time RT-PCR for wnt11 and siamois transcript levels, normalized to ODC (right panel). (D) Gastrulation failures resulting from Dyrk1A expression are rescued by co-expression of Kaiso. The coexpression of a third component, p120-catenin, once again results in increased gastrulation failures, presumably in part by relieving the repression and/or rescue conferred by Kaiso. (E) Dyrk1A (5 pg) or β-galactosidase (negative control) were co-injected or not with p120-catenin morpholino (20 ng). Gastrula cDNA was analyzed by real-time PCR for expression of wnt11 and siamois and normalized to ODC. (F) Standard or Dyrk1A morpholinos (10 ng) were microinjected into Xenopus embryos. To test for rescue of morpholino-directed Dyrk1A depletion, p120-catenin wild type, p120-catenin T47D or p120-catenin were co-injected. The gene expression levels of Wnt11 were monitored using real-time PCR.
Dyrk1A overexpression facilitates canonical Wnt signaling
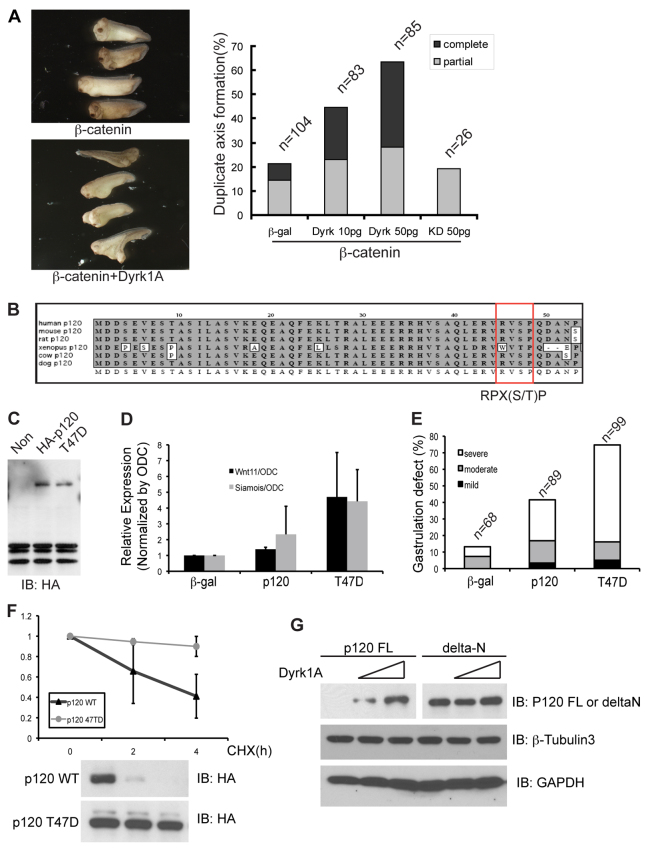
Previously, our group determined that exogenous Kaiso represses canonical Wnt signaling as evaluated using a classic in vivo assay, the suppression of ectopic β-catenin-dependent axis duplication (Park et al., 2005). Given that Dyrk1A stabilizes p120-catenin, which in turn transcriptionally modulates siamois, a key gene product involved in axis specification, we tested whether Dyrk1A has an impact upon β-catenin-mediated axis duplication. By co-injecting mRNA encoding Dyrk1A and β-catenin into one cell of four-cell (as opposed to one-cell) embryos (ventral-vegetal region), we largely avoided gastrulation delays or failures resulting from Dyrk1A overexpression. As predicted, co-expression of Dyrk1A with an intentionally modest (almost sub-phenotypic) dose of β-catenin enhanced secondary axis formation in a dose-dependent manner (Fig. 4A). Kinase-dead Dyrk1A did not produce such effects (Dyrk1A KD, negative control).
Fig. 4.
Phosphorylation-dependent Dyrk1A in embryonic development. (A) Duplicate axis formation following the expression of a sub-maximal dose of β-catenin, is enhanced upon the co-expression of the indicated doses of Dyrk1A, as evaluated in tailbud embryos (stage 27–29). Kinase-dead (KD) Dyrk1A serves as a negative control. (B) Cross-species p120-catenin sequence alignment of the conserved predicted Dyrk1A phosphorylation-site region (box). (C) Expression of HA–p120-catenin wild type, versus the phosphomimic mutant (T47D), was detected by immunoblotting of Xenopus embryo lysates. (D) p120-catenin wild type (WT) or the p120-catenin point-mutant (T47D), was injected into Xenopus embryos at the one-cell stage, followed later by cDNA isolation and real-time PCR, to monitor increased wnt11 and siamois transcript levels. (E) Gastrulation failures were more severe in embryos expressing p120-cateninT47D relative to p120-catenin WT. (F) HA–p120-catenin WT or p120-cateninT47D was expressed in HeLa cells. Cells transfected with each construct were treated with cyclohexamide (CHX) for the times indicated. Each HA-tagged construct was detected by immunoblotting of the corresponding cell extracts (right panel), followed by densitometer quantification of the band intensities (left panel). These data were collected from two independent experiments. (G) Full-length p120-catenin (p120 FL), or an N-terminal-deleted construct of p120-catenin (delta-N), were transfected into HEK293T cells and increasing amounts of Dyrk1A co-transfected. The levels of p120-catenin FL and delta-N protein were monitored (using an anti-Myc antibody). β-tubulin3 and GAPDH served as negative controls.
A phosphomimic mutant of p120-catenin enhances target gene expression
As a first step in assessing how Dryk1A acts we searched for potential Dyrk phosphorylation sites in p120-catenin. Consistent with our earlier Dyrk1A–p120-catenin association results (Fig. 2), one evolutionarily conserved Dyrk1A phosphorylation site was identified in the N-terminal region of p120-catenin. This R45VSP48 motif was comparable with the Dyrk canonical consensus (RPX(S/T)P; Fig. 4B), satisfying the requirement of proline-directed kinases and the requirement of arginine for substrate recognition (Himpel et al., 2000). To approximate the impact of Dyrk1A phosphorylation at this site, we generated a phosphomimic point mutant (p120-catenin T47D). p120-catenin wild-type or p120-catenin T47D was expressed in Xenopus embryos (Fig. 4C). Because wild-type p120-catenin over-expression relieves Kaiso-mediated gene repression and thus contributes to failed gastrulation, we suspected that p120-catenin T47D might be yet more active in relieving Kaiso-mediated repression, possibly resulting from its increased protein levels. As expected, p120-catenin T47D proved more effective than the wild-type protein in promoting expression of the known p120-catenin–Kaiso gene targets siamois and wnt11 (Fig. 4D). Correspondingly, p120-catenin T47D caused more gastrulation failure than corresponding doses of wild-type p120-catenin (Fig. 4E). We further tested whether p120-catenin overexpression might rescue Dyrk1A depletion. By measuring Wnt11 transcript levels, we found that exogenous wild-type p120-catenin did not rescue Dyrk1A depletion, nor did an unstable mutant form of p120-catenin (unphosphorylated form, p120-catenin T47V), whereas, stabilized p120-catenin (phosphomimic, p120-catenin T47D) did (Fig. 3F). The half-life of p120-catenin wild-type versus the p120-catenin T47D mutant was then tested in the presence of cyclohexamide (Fig. 4F). As anticipated, p120-catenin T47D had a notably prolonged half-life. To assess whether threonine 47 of p120-catenin is directly phosphorylated by Dyrk1A, we conducted an in vitro kinase assay using the N-terminal region of p120-catenin versus the same region of p120-catenin T47D. Phosphorylation of p120-catenin by Dyrk1A was faint but reproducible, but did not occur with p120-catenin T47D, nor when kinase-dead Dyrk1A was used (supplementary material Fig. S2). These data suggest that p120-catenin phosphorylation by Dyrk1A is direct, and that such modification is relevant to its functional role in embryonic development.
p120-catenin has several isoforms resulting from the use of four distinct transcriptional start sites and alternative RNA splicing. There are functional differences between some of these isoforms (Hong et al., 2010; Liu et al., 2009). Interestingly, one Dyrk1A phosphorylation site (T47) exists in an N-terminal region present only in the longest isoform, p120-catenin isoform-1, which is generated starting at the most upstream translational start site. Thus, we suspected that Dyrk1A might preferentially regulate p120-catenin isoform-1. We expressed Xenopus p120-catenin isoform-1 in HEK293T cells, and a form of p120-catenin corresponding to human isoform-4 that lacks most of the N-terminus. Confirming our expectation, the N-terminal-deleted p120-catenin (isoform-4) did not appear to respond to exogenous Dyrk1A, whereas isoform-1 levels were increased (Fig. 4G, top panels are low exposure blots, such that p120-catenin FL was not observed in lane 1). Isoform-3 of p120-catenin, however, still associated with Dyrk1A and remained somewhat responsive to Dyrk1A expression (resolved using antibody pp120). This could have arisen from indirect effects, or from an additional phosphorylation site between the translational start site of isoform-3 and isoform-4, but further study is needed to clarify this.
In summary, we report that the Down's-syndrome-related kinase, Dyrk1A, associates with p120-catenin and increases p120-catenin levels, especially p120-catenin isoform-1, which in turn has an impact upon certain Wnt–p120-catenin–Kaiso pathway target genes, such as wnt11 and siamois. We show that exogenous Kaiso rescues the effect of Dyrk1A on target genes, suggesting that Dyrk1A transmits signals through the p120-catenin–Kaiso trajectory of the Wnt signaling pathway. The data presented here provide new insights into the role of Dyrk1A in development and perhaps disease. Although Dyrk1A is recognized as a crucial component of Down's syndrome pathology, molecular analysis of Dyrk1A is at an early stage. Until now, to our knowledge, there have been no reports indicating a functional relationship between Dyrk1A and the Wnt–p120-catenin–Kaiso pathway. Understanding such an interaction could assist in addressing the contribution of Dyrk1A to Down's syndrome, possibly, for example, through its effects on p120-catenin and thus Wnt signaling in central nervous system development. Alternatively, it is conceivable that Dyrk1A alters p120-catenin effects on small GTPases such as RhoA and Rac1, perhaps thereby regulating dendritic spine and synapse formation in the developing brain. Likewise, there are gaps in the understanding of upstream signals relevant to Dyrk1A activity. Intriguingly, a previous report from our lab identified Frodo as an upstream binding-mate and modulator of p120-catenin, but not of β-catenin (Hong et al., 2010; Park et al., 2006). The depletion of Frodo decreases p120-catenin levels, seemingly similar to the effect of Dyrk1A. Thus, in the context of addressing p120-catenin levels, and thereby the p120-catenin–Kaiso signaling trajectory, Frodo and Dyrk1A might functionally interact. Given that Frodo is involved in the Wnt pathway, it is possible that canonical Wnt signals reside upstream of Dyrk within the (Frodo)–p120-catenin–Kaiso trajectory, just as we and others recently revealed is the case for p120-catenin, and probably for ARVCF and δ-catenin (Casagolda et al., 2010; Hong et al., 2010; Oh et al., 2009; Park et al., 2006; Park et al., 2005; Spring et al., 2005). We have preliminarily findings that Dyrk1A increases the levels of coexpressed ARVCF and δ-catenin in addition to p120-catenin (data not shown). Thus, it will be very interesting to investigate the effects of Dyrk1A (and Wnt) on other p120-catenin family members. Given that Dyrk1A dosage imbalance is correlated with Down's syndrome, the study of Dyrk1A-mediated effects on p120-catenin, and thereby upon nuclear Wnt target genes and cytoplasmic small-GTPases, warrants further examination.
Materials and Methods
Embryo culture, microinjections, in vitro transcription and antisense oligonucleotides
Fertilization, embryo culture, in vitro transcription and microinjections were performed as described previously (Fang et al., 2004). We employed a Xenopus Dyrk1A morpholino (Dyrk1A-MO: 5′-ATGAGACTTGAAAGAGGACGATGCA-3′) and a standard control morpholino (STD MO).
Transfection and RNA interference
siRNA oligonucleotide sequences directed against the transcripts of Dyrk1A1 and A2 were gathered from published reports, and synthesized by Applied Biosystems (Seifert et al., 2008; Shi et al., 2008). Duplex oligonulceotides were directed against the target sequences: Dyrk1A-1, 5′-TTAAGGATGCTTGATTATGAC-3′; Dyrk1A-2, 5′-AAACTCGAATTCAACCTTATT-3′. Transfections of siRNA or DNA were performed as described (Hong et al., 2010).
Analysis and gene expression using real-time RT-PCR
Total RNA was prepared as described (Hong et al., 2010). Primers for RT-PCR were: Siamois, 5′-CCCAACTTCAGAAGGACCTAGATC-3′ and 5′-TGGGTAGGGCTGTGTATTTGA-3′; Wnt11, 5′-TGACAGCTGCAACCTCATGT-3′ and 5′-ACAGAGGGCTGTCAGTGCTT-3′; ornithine decarboxylase, 5′-CGAGCGGATTATCTATGCA-3′ and 5′-GCGTATTTGATCTGGGAAA-3′.
Supplementary Material
Acknowledgements
We thank Savrina A. Stratton (UT-MD Anderson) and Eun-Ah Kim (UT-MD Anderson) for providing advice for Real-time PCR.
Footnotes
Funding
This work was supported by the National Institute of Health [grant number RO1GM52112 to P.D.M.]; a Texas Advanced Research Project Grant [grant number 003657-0008-2006 to P.D.M.]; a University of Texas M.D. Anderson Cancer Center National Cancer Institute Core Grant [grant number CA-16672]; a William Randolf Hearst Foundation Student Research Award to J.Y.H.; and a Developmental Award from the University of Texas Specialized Programs of Research Excellence (SPORE) in Lung Cancer [grant number P50 CA070907 to P.D.M.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.086173/-/DC1
References
- Altafaj X., Dierssen M., Baamonde C., Marti E., Visa J., Guimera J., Oset M., Gonzalez J. R., Florez J., Fillat C., et al. (2001). Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 10, 1915-1923 [DOI] [PubMed] [Google Scholar]
- Anastasiadis P. Z. (2007). p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta. 1773, 34-46 [DOI] [PubMed] [Google Scholar]
- Anastasiadis P. Z., Reynolds A. B. (2001). Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 13, 604-610 [DOI] [PubMed] [Google Scholar]
- Bartlett J. D., Dobeck J. M., Tye C. E., Perez-Moreno M., Stokes N., Reynolds A. B., Fuchs E., Skobe Z. (2010). Targeted p120-catenin ablation disrupts dental enamel development. PLoS One 5, pii:e12703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J., Lustig B. (2004). The Wnt connection to tumorigenesis. Int. J. Dev. Biol. 48, 477-487 [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J. (2005). GAPs in growth factor signalling. Growth Factors 23, 143-149 [DOI] [PubMed] [Google Scholar]
- Casagolda D., Del Valle-Perez B., Valls G., Lugilde E., Vinyoles M., Casado-Vela J., Solanas G., Batlle E., Reynolds A. B., Casal J. I., et al. (2010). A p120-catenin-CK1epsilon complex regulates Wnt signaling. J. Cell Sci. 123, 2621-2631 [DOI] [PubMed] [Google Scholar]
- Cheon M. S., Shim K. S., Kim S. H., Hara A., Lubec G. (2003). Protein levels of genes encoded on chromosome 21 in fetal Down syndrome brain: challenging the gene dosage effect hypothesis (Part IV). Amino Acids 25, 41-47 [DOI] [PubMed] [Google Scholar]
- Daniel J. M. (2007). Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim. Biophys. Acta. 1773, 59-68 [DOI] [PubMed] [Google Scholar]
- Daniel J. M., Reynolds A. B. (1999). The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell Biol. 19, 3614-3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Spring C. M., Crawford H. C., Reynolds A. B., Baig A. (2002). The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30, 2911-2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Reynolds A. B. (2006). Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell 10, 21-31 [DOI] [PubMed] [Google Scholar]
- Davis M. A., Ireton R. C., Reynolds A. B. (2003). A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163, 525-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn M. R., Brown M. V., Reynolds A. B. (2009). An essential role for p120-catenin in Src- and Rac1-mediated anchorage-independent cell growth. J. Cell Biol. 184, 437-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D., Sater A. K., Ji H., Cho K., Clark M., Stratton S. A., Michelle B. C., Lu Q., McCrea P. D. (2009). Xenopus delta-catenin is expressed widely across developing and adult tissues, is essential in early embryogenesis and is functionally linked to cadherins and small GTPases. J. Cell Sci. 122, 4049-4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Ji H., Kim S. W., Park J. I., Vaught T. G., Anastasiadis P. Z., Ciesiolka M., McCrea P. D. (2004). Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. J. Cell Biol. 165, 87-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki V., Dierssen M., Alcantara S., Martinez S., Marti E., Casas C., Visa J., Soriano E., Estivill X., Arbones M. L. (2002). Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell. Biol. 22, 6636-6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan T., Wend P., Klaus A., Birchmeier W. (2008). Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 22, 2308-2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle B., Carnicero A., Elizalde C., Ceron J., Martinez S., Tejedor F. J. (2003). Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur. J. Neurosci. 17, 2277-2286 [DOI] [PubMed] [Google Scholar]
- Hammerle B., Elizalde C., Tejedor F. J. (2008). The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur. J. Neurosci. 27, 1061-1074 [DOI] [PubMed] [Google Scholar]
- Hikasa H., Ezan J., Itoh K., Li X., Klymkowsky M. W., Sokol S. Y. (2010). Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev. Cell 19, 521-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. (2000). Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 275, 2431-2438 [DOI] [PubMed] [Google Scholar]
- Hong J. Y., Park J. I., Cho K., Gu D., Ji H., Artandi S. E., McCrea P. D. (2010). Shared molecular mechanisms regulate multiple catenin proteins: canonical Wnt signals and components modulate p120-catenin isoform-1 and additional p120 subfamily members. J. Cell Sci. 123, 4351-4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Birchmeier W. (2001). New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 11, 547-553 [DOI] [PubMed] [Google Scholar]
- Iioka H., Doerner S. K., Tamai K. (2009). Kaiso is a bimodal modulator for Wnt/beta-catenin signaling. FEBS Lett. 583, 627-632 [DOI] [PubMed] [Google Scholar]
- Johnson E., Seachrist D. D., DeLeon-Rodriguez C. M., Lozada K. L., Miedler J., Abdul-Karim F. W., Keri R. A. (2010). HER2/ErbB2-induced breast cancer cell migration and invasion require p120 catenin activation of Rac1 and Cdc42. J. Biol. Chem. 285, 29491-29501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. F., Spring C. M., Otchere A. A., Daniel J. M. (2004). NLS-dependent nuclear localization of p120ctn is necessary to relieve Kaiso-mediated transcriptional repression. J. Cell Sci. 117, 2675-2686 [DOI] [PubMed] [Google Scholar]
- Kim S. W., Fang X., Ji H., Paulson A. F., Daniel J. M., Ciesiolka M., van Roy F., McCrea P. D. (2002). Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J. Biol. Chem. 277, 8202-8208 [DOI] [PubMed] [Google Scholar]
- Kim S. W., Park J. I., Spring C. M., Sater A. K., Ji H., Otchere A. A., Daniel J. M., McCrea P. D. (2004). Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat. Cell Biol. 6, 1212-1220 [DOI] [PubMed] [Google Scholar]
- Kim E. A., Kim J. E., Sung K. S., Choi D. W., Lee B. J., Choi C. Y. (2010). Homeodomain-interacting protein kinase 2 (HIPK2) targets beta-catenin for phosphorylation and proteasomal degradation. Biochem. Biophys. Res. Commun. 394, 966-971 [DOI] [PubMed] [Google Scholar]
- Klaus A., Birchmeier W. (2008). Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387-398 [DOI] [PubMed] [Google Scholar]
- Lai S. L., Chien A. J., Moon R. T. (2009). Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 19, 532-545 [DOI] [PubMed] [Google Scholar]
- Lee W., Swarup S., Chen J., Ishitani T., Verheyen E. M. (2009). Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development 136, 241-251 [DOI] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002). Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell, 108, 837-847 [DOI] [PubMed] [Google Scholar]
- Liu Y., Dong Q. Z., Zhao Y., Dong X. J., Miao Y., Dai S. D., Yang Z. Q., Zhang D., Wang Y., Li Q. C., et al. (2009). P120-catenin isoforms 1A and 3A differently affect invasion and proliferation of lung cancer cells. Exp. Cell Res. 315, 890-898 [DOI] [PubMed] [Google Scholar]
- Louie S. H., Yang X. Y., Conrad W. H., Muster J., Angers S., Moon R. T., Cheyette B. N. (2009). Modulation of the beta-catenin signaling pathway by the dishevelled-associated protein Hipk1. PLoS One 4, e4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R. T. (2005). Wnt/beta-catenin pathway. Sci. STKE 2005, 271, p. cm1 [DOI] [PubMed] [Google Scholar]
- Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004). WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691-701 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. (2009). Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol. Cell Biol. 10, 276-286 [DOI] [PubMed] [Google Scholar]
- Oh M., Kim H., Yang I., Park J. H., Cong W. T., Baek M. C., Bareiss S., Ki H., Lu Q., No J., et al. (2009). GSK-3 phosphorylates delta-catenin and negatively regulates its stability via ubiquitination/proteosome-mediated proteolysis. J. Biol. Chem. 284, 28579-28589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okui M., Ide T., Morita K., Funakoshi E., Ito F., Ogita K., Yoneda Y., Kudoh J., Shimizu N. (1999). High-level expression of the Mnb/Dyrk1A gene in brain and heart during rat early development. Genomics 62, 165-171 [DOI] [PubMed] [Google Scholar]
- Park J. I., Kim S. W., Lyons J. P., Ji H., Nguyen T. T., Cho K., Barton M. C., Deroo T., Vleminckx K., Moon R. T., et al. (2005). Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell 8, 843-854 [DOI] [PubMed] [Google Scholar]
- Park J. I., Ji H., Jun S., Gu D., Hikasa H., Li L., Sokol S. Y., McCrea P. D. (2006). Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev. Cell 11, 683-695 [DOI] [PubMed] [Google Scholar]
- Park J., Song W. J., Chung K. C. (2009). Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol. Life Sci. 66, 3235-3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M., Davis M. A., Wong E., Pasolli H. A., Reynolds A. B., Fuchs E. (2006). p120-catenin mediates inflammatory responses in the skin. Cell 124, 631-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M., Song W., Pasolli H. A., Williams S. E., Fuchs E. (2008). Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc. Natl. Acad. Sci. USA 105, 15399-15404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A., Hendrich B., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E. (2001). The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15, 1613-1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B., Carnahan R. H. (2004). Regulation of cadherin stability and turnover by p120ctn: implications in disease and cancer. Semin. Cell Dev. Biol. 15, 657-663 [DOI] [PubMed] [Google Scholar]
- Reynolds A. B., Roczniak-Ferguson A. (2004). Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23, 7947-7956 [DOI] [PubMed] [Google Scholar]
- Ruzov A., Hackett J. A., Prokhortchouk A., Reddington J. P., Madej M. J., Dunican D. S., Prokhortchouk E., Pennings S., Meehan R. R. (2009a). The interaction of xKaiso with xTcf3: a revised model for integration of epigenetic and Wnt signalling pathways. Development 136, 723-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A., Savitskaya E., Hackett J. A., Reddington J. P., Prokhortchouk A., Madej M. J., Chekanov N., Li M., Dunican D. S., Prokhortchouk E., et al. (2009b). The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development 136, 729-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A., Allan L. A., Clarke P. R. (2008). DYRK1A phosphorylates caspase 9 at an inhibitory site and is potently inhibited in human cells by harmine. FEBS J. 275, 6268-6280 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhang T., Zhou C., Chohan M. O., Gu X., Wegiel J., Zhou J., Hwang Y. W., Iqbal K., Grundke-Iqbal I., et al. (2008). Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J. Biol. Chem. 283, 28660-28669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindoh N., Kudoh J., MAeda H., Yamaki A., Minoshima S., Shimizu Y., Shimizu N. (1996). Cloning of a human homolog of the Drosophila Minibrain/rat Dyrk gene from “the Down syndrome critical region of chromosome 21.”. Biochem. and Biophys. Res. Commun. 225, 92-99 [DOI] [PubMed] [Google Scholar]
- Smith D. J., Stevens M. E., Sudanagunta S. P., Bronson R. T., Makhinson M., Watabe A. M., O'Dell T. J., Fung J., Weier H. U., Cheng J. F., et al. (1997). Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 16, 28-36 [DOI] [PubMed] [Google Scholar]
- Song W. J., Sternberg L. R., Kasten-Sportes C., Keuren M. L., Chung S. H., Slack A. C., Miller D. E., Glover T. W., Chiang P. W., Lou L., et al. (1996). Isolation of human and murine homologues of the Drosophila minibrain gene: human homologue maps to 21q22.2 in the Down syndrome “critical region”. Genomics 38, 331-339 [DOI] [PubMed] [Google Scholar]
- Spring C. M., Kelly K. F., O'Kelly I., Graham M., Crawford H. C., Daniel J. M. (2005). The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp. Cell Res. 305, 253-265 [DOI] [PubMed] [Google Scholar]
- Tejedor F. J., Hammerle B. (2010). MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 278, 223-235 [DOI] [PubMed] [Google Scholar]
- van Amerongen R., Nusse R. (2009). Towards an integrated view of Wnt signaling in development. Development 136, 3205-3214 [DOI] [PubMed] [Google Scholar]
- van Roy F. M., McCrea P. D. (2005). A role for Kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer 5, 956-964 [DOI] [PubMed] [Google Scholar]
- Verheyen E. M., Gottardi C. J. (2010). Regulation of Wnt/beta-catenin signaling by protein kinases. Dev. Dyn. 239, 34-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J., Gong C. X., Hwang Y. W. (2010). The role of DYRK1A in neurodegenerative diseases. FEBS J. 278, 236-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend P., Holland J. D., Ziebold U., Birchmeier W. (2010). Wnt signaling in stem and cancer stem cells. Semin. Cell Dev. Biol. 21, 855-863 [DOI] [PubMed] [Google Scholar]
- Wildenberg G. A., Dohn M. R., Carnahan R. H., Davis M. A., Lobdell N. A., Settleman J., Reynolds A. B. (2006). p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 127, 1027-1039 [DOI] [PubMed] [Google Scholar]
- Xiao K., Oas R. G., Chiasson C. M., Kowalczyk A. P. (2007). Role of p120-catenin in cadherin trafficking. Biochim. Biophys. Acta. 1773, 8-16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.