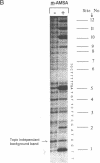
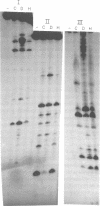
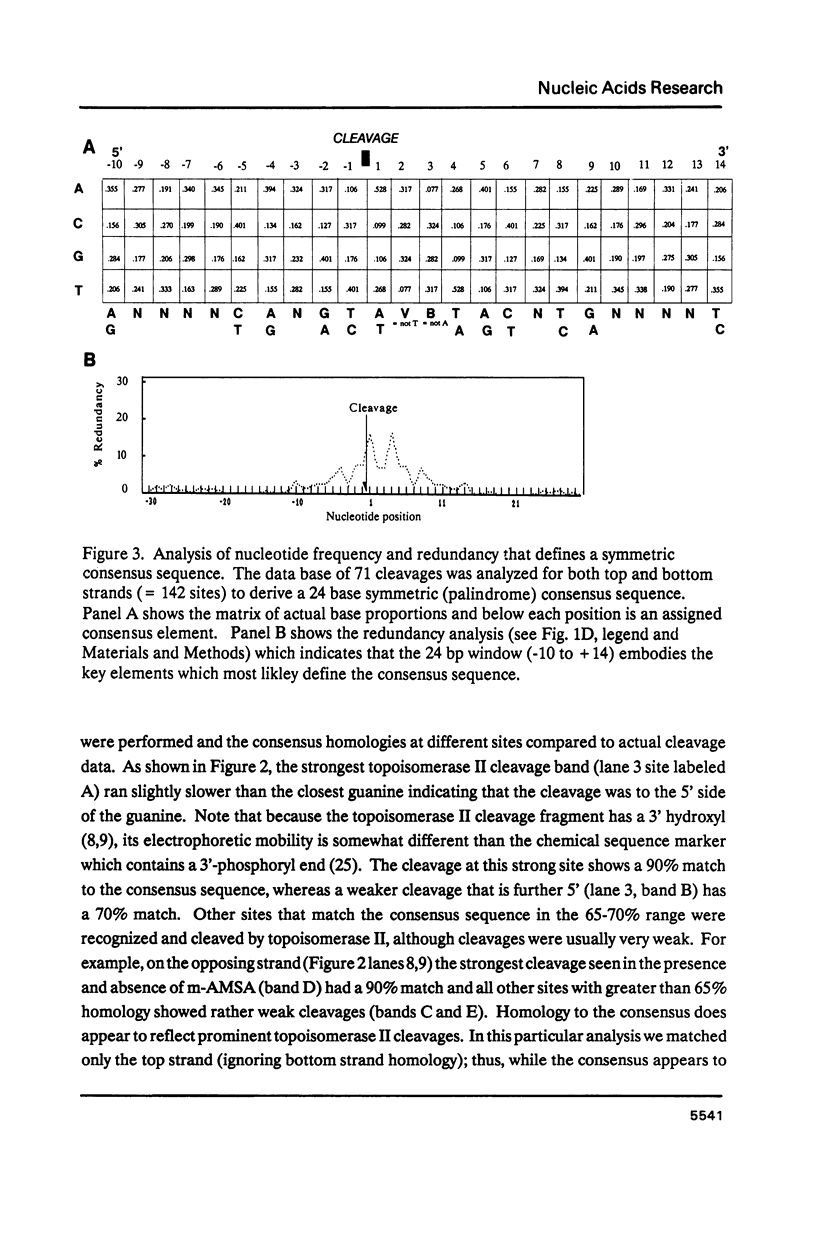
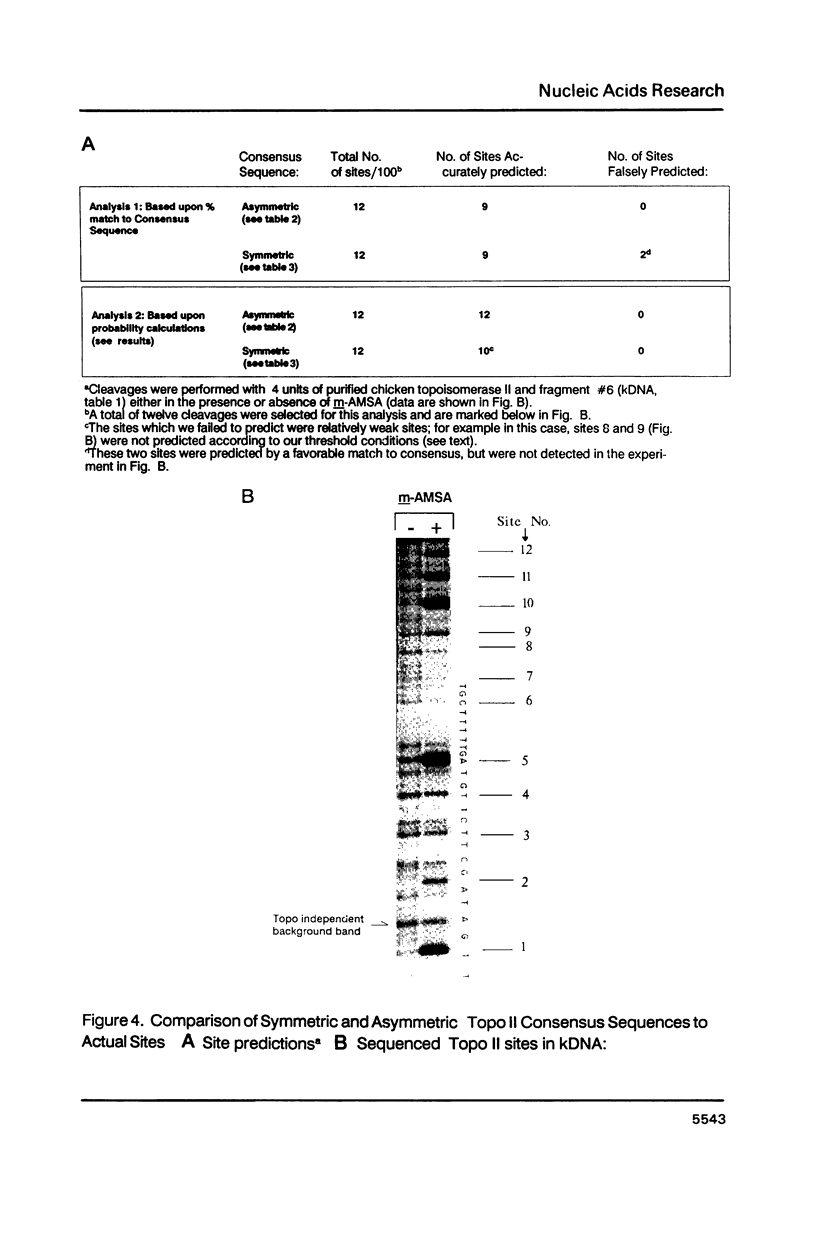
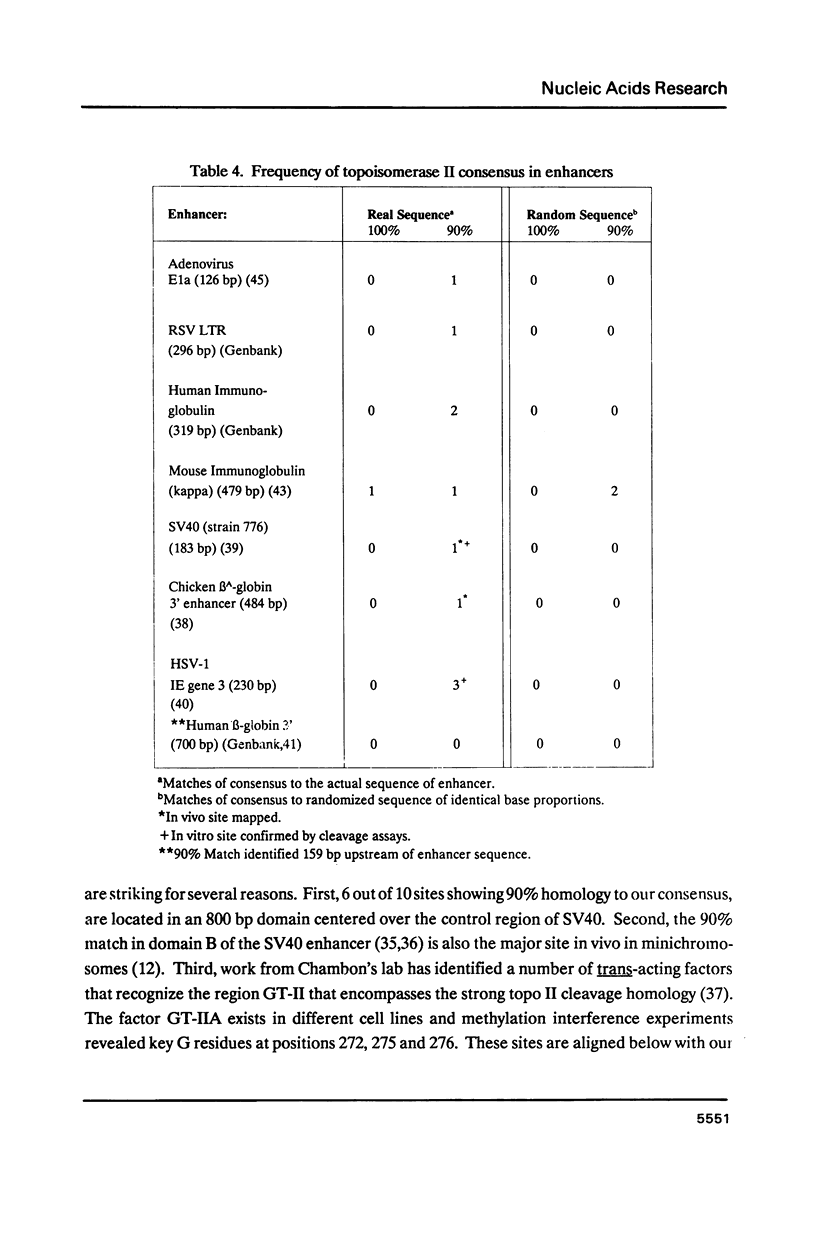
Abstract
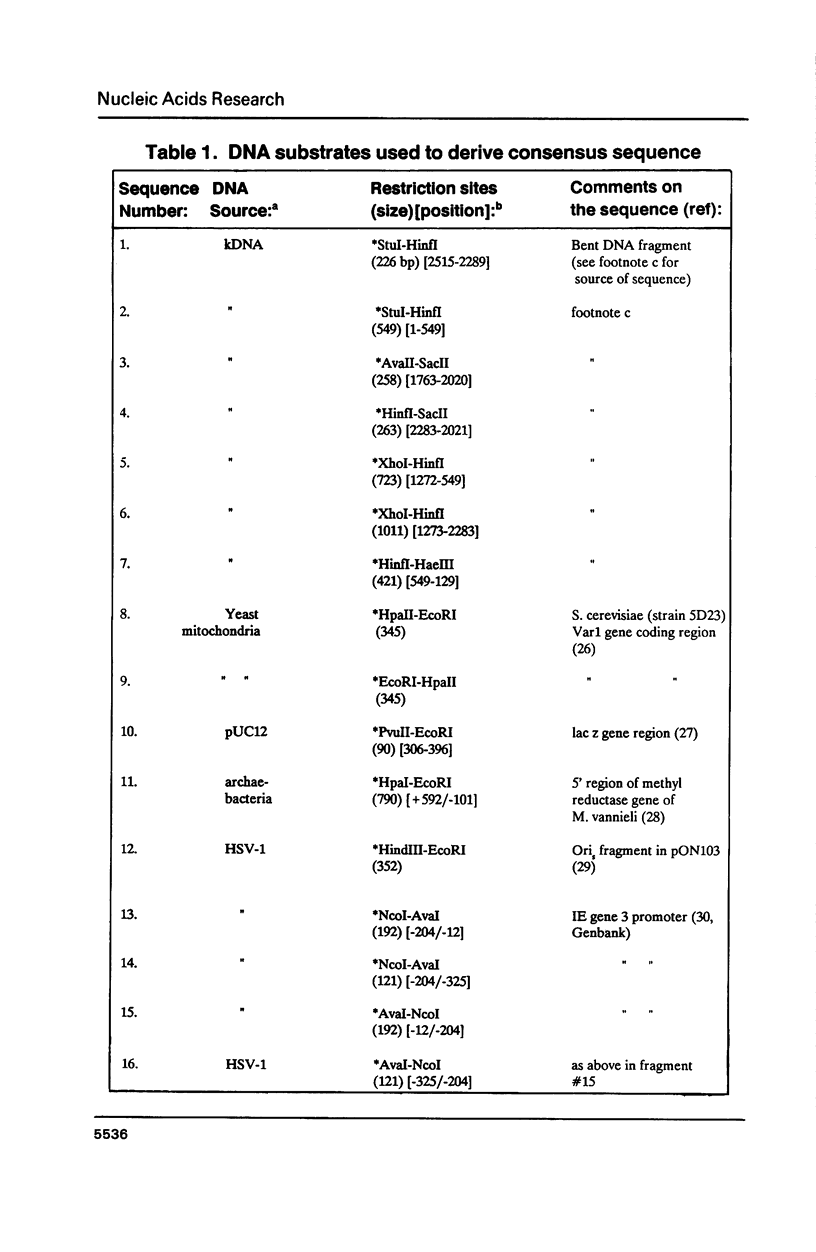
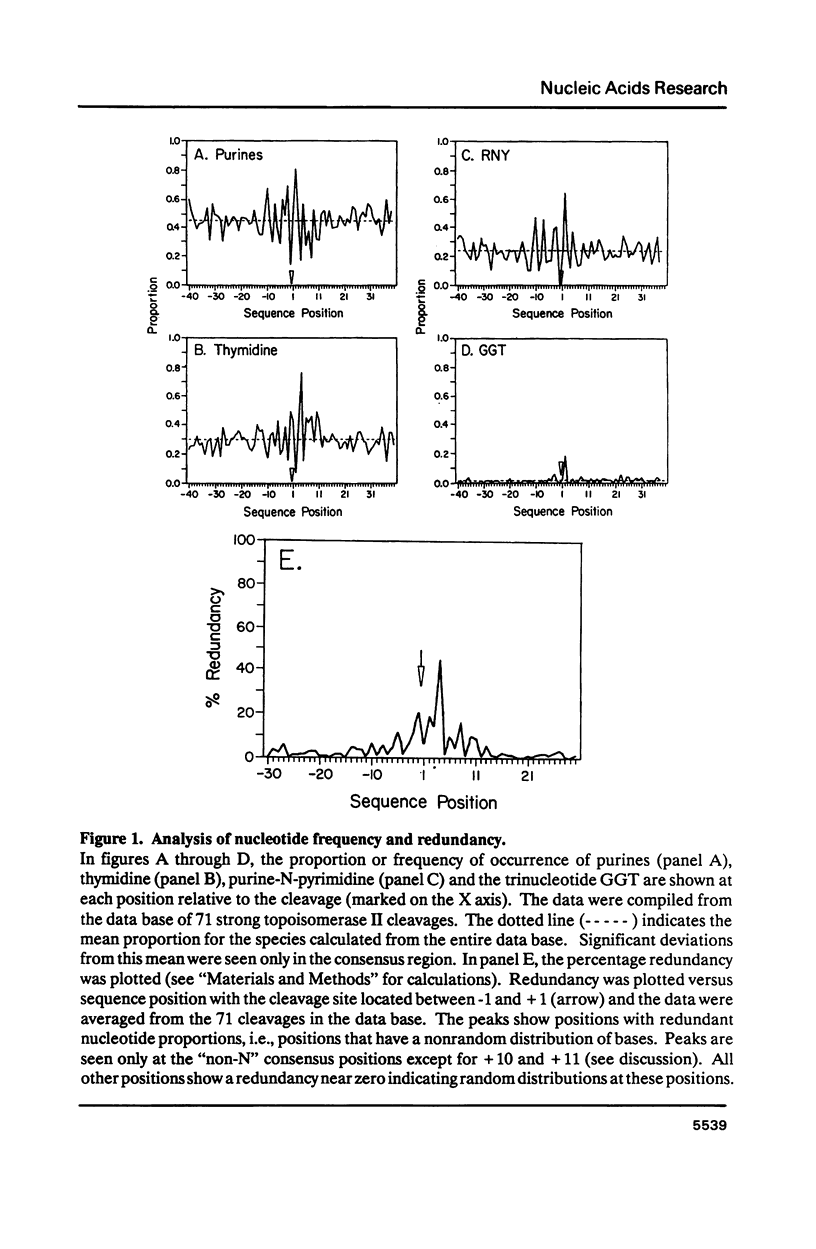
Topoisomerase II, purified from chicken erythrocytes, was reacted with a large number of different DNA fragments and cleavages were catalogued in the presence and absence of drugs that stabilize the cleavage intermediate. Cleavages were sequenced to derive a consensus for topoisomerase II that predicts catalytic sites. The consensus is: (sequence; see text) where N is any base and cleavage occurs at the indicated mark between -1 and +1. The consensus accurately predicts topoisomerase II sites in vitro. This consensus is not closely related to the Drosophila consensus sequence, but the two enzymes show some similarities in site recognition. Topoisomerase II purified from human placenta cleaves DNA sites that are essentially identical to the chicken enzyme, suggesting that vertebrate type II enzymes share a common catalytic sequence. Both viral and tissue specific enhancers contain sites sharing strong homology to the consensus and endogenous topoisomerase II recognizes some of these sites in vivo.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman P., Glover C. V., Osheroff N. Phosphorylation of DNA topoisomerase II by casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc Natl Acad Sci U S A. 1985 May;82(10):3164–3168. doi: 10.1073/pnas.82.10.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti P., Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Purification and characterization of Xenopus laevis topoisomerase II. EMBO J. 1983;2(8):1303–1308. doi: 10.1002/j.1460-2075.1983.tb01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios M., Osheroff N., Fisher P. A. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cram D. S., Sherf B. A., Libby R. T., Mattaliano R. J., Ramachandran K. L., Reeve J. N. Structure and expression of the genes, mcrBDCGA, which encode the subunits of component C of methyl coenzyme M reductase in Methanococcus vannielii. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3992–3996. doi: 10.1073/pnas.84.12.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby M. K., Herrera R. E., Vosberg H. P., Nordheim A. DNA topoisomerase II cleaves at specific sites in the 5' flanking region of c-fos proto-oncogenes in vitro. EMBO J. 1986 Sep;5(9):2257–2265. doi: 10.1002/j.1460-2075.1986.tb04493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douc-Rasy S., Kayser A., Riou J. F., Riou G. ATP-independent type II topoisomerase from trypanosomes. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7152–7156. doi: 10.1073/pnas.83.19.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet M., Lavenot C., Harper F., Mirambeau G., De Recondo A. M. DNA topoisomerases from rat liver: physiological variations. Nucleic Acids Res. 1983 Feb 25;11(4):1059–1075. doi: 10.1093/nar/11.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Heck M. M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., O'Donnell M. E., Mocarski E. S., Lehman I. R. A DNA binding protein specific for an origin of replication of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6322–6326. doi: 10.1073/pnas.83.17.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D. DNA sequence elements required for regulated expression of the HSV-1 glycoprotein D gene lie within 83 bp of the RNA capsites. Nucleic Acids Res. 1983 Oct 11;11(19):6647–6666. doi: 10.1093/nar/11.19.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986 Apr 20;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Goto T., Wang J. C. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J Biol Chem. 1982 May 25;257(10):5866–5872. [PubMed] [Google Scholar]
- Harr R., Häggström M., Gustafsson P. Search algorithm for pattern match analysis of nucleic acid sequences. Nucleic Acids Res. 1983 May 11;11(9):2943–2957. doi: 10.1093/nar/11.9.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hesse J. E., Nickol J. M., Lieber M. R., Felsenfeld G. Regulated gene expression in transfected primary chicken erythrocytes. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4312–4316. doi: 10.1073/pnas.83.12.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Vincent R. D., Perlman P. S., Shumard D. S., Treisman L. O., Grossman L. I. Expandable var1 gene of yeast mitochondrial DNA: in-frame insertions can explain the strain-specific protein size polymorphisms. Proc Natl Acad Sci U S A. 1984 May;81(10):3148–3152. doi: 10.1073/pnas.81.10.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias G., Hurst J., deBoer E., Grosveld F. The human beta-globin gene contains a downstream developmental specific enhancer. Nucleic Acids Res. 1987 Jul 24;15(14):5739–5747. doi: 10.1093/nar/15.14.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J. C., Spandidos D. A., Wilkie N. M. Transcriptional regulation of a herpes simplex virus immediate early gene is mediated through an enhancer-type sequence. EMBO J. 1984 Feb;3(2):389–395. doi: 10.1002/j.1460-2075.1984.tb01817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Rowe T. C., Yang L., Tewey K. M., Chen G. L. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem. 1983 Dec 25;258(24):15365–15370. [PubMed] [Google Scholar]
- Mackem S., Roizman B. Differentiation between alpha promoter and regulator regions of herpes simplex virus 1: the functional domains and sequence of a movable alpha regulator. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4917–4921. doi: 10.1073/pnas.79.16.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Liu L. F., Englund P. T. A homogeneous type II DNA topoisomerase from HeLa cell nuclei. J Biol Chem. 1981 Sep 10;256(17):9334–9339. [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E. M., Tewey K. M., Liu L. F. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4'-(9-acridinylamino)-methanesulfon-m-anisidide. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Covey J., Kerrigan D., Mattes W., Markovits J., Kohn K. W. Role of DNA intercalation in the inhibition of purified mouse leukemia (L1210) DNA topoisomerase II by 9-aminoacridines. Biochem Pharmacol. 1987 Oct 15;36(20):3477–3486. doi: 10.1016/0006-2952(87)90329-7. [DOI] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Rowe T. C., Wang J. C., Liu L. F. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986 Apr;6(4):985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983 Jul 10;258(13):8421–8428. [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Shelton E. R., Osheroff N., Brutlag D. L. DNA topoisomerase II from Drosophila melanogaster. Purification and physical characterization. J Biol Chem. 1983 Aug 10;258(15):9530–9535. [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D. Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem. 1988;17:241–263. doi: 10.1146/annurev.bb.17.060188.001325. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Novel partitioning of DNA cleavage sites for Drosophila topoisomerase II. Cell. 1985 Apr;40(4):933–941. doi: 10.1016/0092-8674(85)90353-8. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Topoisomerase II cleavage in chromatin. J Mol Biol. 1986 Sep 20;191(2):231–246. doi: 10.1016/0022-2836(86)90260-3. [DOI] [PubMed] [Google Scholar]
- Uemura T., Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986 May;5(5):1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984 Aug;3(8):1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Ferrandon D., Rosales R., Vigneron M., Macchi M., Ruffenach F., Chambon P. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987 Oct;6(10):3005–3013. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Rowe T. C., Nelson E. M., Liu L. F. In vivo mapping of DNA topoisomerase II-specific cleavage sites on SV40 chromatin. Cell. 1985 May;41(1):127–132. doi: 10.1016/0092-8674(85)90067-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]