Abstract
Glioma cell lines are an important tool for research in basic and translational neuro-oncology. Documentation of their genetic identity has become a requirement for scientific journals and grant applications to exclude cross-contamination and misidentification that lead to misinterpretation of results. Here, we report the standard 16 marker short tandem repeat (STR) DNA fingerprints for a panel of 39 widely used glioma cell lines as reference. Comparison of the fingerprints among themselves and with the large DSMZ database comprising 9 marker STRs for 2278 cell lines uncovered 3 misidentified cell lines and confirmed previously known cross-contaminations. Furthermore, 2 glioma cell lines exhibited identity scores of 0.8, which is proposed as the cutoff for detecting cross-contamination. Additional characteristics, comprising lack of a B-raf mutation in one line and a similarity score of 1 with the original tumor tissue in the other, excluded a cross-contamination. Subsequent simulation procedures suggested that, when using DNA fingerprints comprising only 9 STR markers, the commonly used similarity score of 0.8 is not sufficiently stringent to unambiguously differentiate the origin. DNA fingerprints are confounded by frequent genetic alterations in cancer cell lines, particularly loss of heterozygosity, that reduce the informativeness of STR markers and, thereby, the overall power for distinction. The similarity score depends on the number of markers measured; thus, more markers or additional cell line characteristics, such as information on specific mutations, may be necessary to clarify the origin.
Keywords: glioblastoma cell lines, misidentification, similarity score, STR fingerprint
Glioma cell lines are a major tool to uncover molecular mechanisms relevant for malignant behavior of gliomas and are used as in vitro or in vivo models to identify and test novel targets for therapy. The research community has become aware that cross-contamination of cell lines is common and a major problem leading to misinterpretation of results.1,2 Recently, the mix up of cell lines in the brain tumor field has received wide coverage.3,4 New standards for the authentication of human cell lines using short tandem repeat (STR) profiling have been proposed,1,2 and many major journals and research agencies now require authentication of cell lines for publication or grant applications, respectively.
Here, we provide 16 marker DNA fingerprints as reference for 39 widely used glioma cell lines in accordance with worldwide database recommendations of identity testing (15 STR markers plus the amelogen sex-determining marker).5 Moreover, we propose a simulation procedure to better differentiate between identical or just similar cell lines.6,7 In fact, similarity scores are confounded by the notorious genetic instability of tumor cell lines with frequent loss of heterozygosity, reducing the informativeness and, thereby, the complexity of the DNA fingerprints, thus lowering the power for discrimination.
Material and Methods
Glioma Cell Lines and Glioma Derived Sphere Lines
Thirty-six permanent glioma cell lines were cultured as described previously.8 Of these, 33 cell lines have been characterized previously for common genetic alterations, including TP53, PTEN, and p16/ARF, and their potential to form tumors in the flanks of nude mice.8 The same reference8 details the origin of each of these cell lines. The glioma cell line BS-153 was kindly provided by Adrian Merlo.9 Three lines are glioma-derived sphere lines (LN-2540GS, LN-2683GS, LN-2826GS) kept under stem cell conditions, as described elsewhere.10 For 2 new adherent cell lines (LN-2207, LN-2669), we have also respective glioma-derived spheres lines (LN-2207GS, LN-2669GS). The 24 cell lines with the prefix “LN” have been established in our laboratory.
STR Fingerprinting
DNA was isolated from cell lines using a standard DNA isolation kit and from paraffin embedded tissue sections using the Ex-Wax DNA extraction kit (Millipore). The DNA fingerprinting was performed by STR profiling. DNA amplifications were made using the PowerPlex 16 HS kit (Promega) according to the manufacturer's recommendations. The primers of the kit amplify 15 tetranucleotide repeat loci plus the amelogenin (AMEL) sex-determining marker. The combination of this set of markers is in accordance with worldwide database recommendations of identity testing.5 A genetic analyzer ABI 3100 (Applied Biosystems) was used to separate and identify the alleles using standard procedures. The results were confirmed in an independent experiment. For comparisons, STR-fingerprints from cell lines were downloaded from the German Collection of Microorganisms and Cell Cultures (DSMZ) database (http://www.dsmz.de/fp/cgi-bin/str.html), which comprises 9 marker profiles (8 STR markers plus the AMEL marker) of 2289 cell lines from DSMZ, American Type Cell Culture (ATCC), Japanese Collection of Research Bioresources (JCRB), and RIKEN. In addition, we obtained the 16 marker (15 STR + AMEL) profiles of the NCI-60 cell line panel that has been published recently.5 These profiles were established with the same standard marker set used in this study.
Gene Analysis
The cDNA of the TP53 gene was sequenced using Sanger sequencing with previously published primers8 (Microsynth). PTEN mutation analysis was performed using Sanger sequencing of the coding sequence (exons 1–9) including intron/exon boundaries, and gene dosage analysis was performed using multiplex ligation-dependent primer amplification (MLPA assay P158-B1, lot 0509, MRC Holland). Primer sequences are available on request. Determination of p16/ARF deletions in the sphere lines are based on array CGH data.10 B-raf mutation analysis of codon 600 was evaluated using diagnostic pyro-sequencing in the laboratory of Molecular Pathology at the Lausanne University Hospital (Lausanne, Switzerland).
Statistics
The fingerprint profiles were summarized by binary variables in which the values one and zero correspond to the presence or absence of a signal (or peak), respectively. To determine pairwise similarity between profiles, we used the Sørensen index,11,12 which corresponds to the similarity index for DNA fingerprinting described by Lynch et al.13 and the evaluation value used in Tanabe et al.7 We used asymmetrical coefficients to limit the effect of double zeros (double absences). The similarity score between 2 profiles can be defined as follows:
where nxy, nx, and ny correspond to the number of peaks common to both samples x and y, the number of peaks of sample x, and the number of peaks of the sample y. All details on their proprieties and implementation have been described elsewhere.13,14
To analyze the robustness of thresholds proposed in the literature,6,7 we performed data resampling to simulate the distribution of similarity indices for unrelated cell lines in each dataset separately. The simulation consisted of 3 steps (Supplementary material, Fig. S1): (1) for each marker, a genotype was randomly sampled, with repetition from the set of observed genotypes from the same collection; (2) the procedure was repeated to obtain n random profiles, where n corresponds to number of cell lines in the dataset; and (3) the similarity index was computed for each pair of the random profiles, providing (n × n− n)/2 values. The maximal simulated similarity (MSS) was defined as the upper limit of the simulated similarity values. Graphical representations, such as histogram and quantile-quantile representation (QQ-plot),15 were used to illustrate the comparison between the distributions of the observed and the simulated similarity index values.
For the glioma cell line panel and the NCI-60 dataset, a second resampling procedure was used to analyze how discrimination improves by increasing the number of markers and to determinate saturation curves for the MSS value. The simulation was done using the aforementioned procedure. After 100 repetitions, MSS empirical distributions were summarized by medians, means, and 95% confidence intervals for MSS by using the percentile method.16,17 Standard deviation (SD) and median absolute deviation (MAD) were used to evaluate the accuracy of the MSS estimation. Analyses and graphical representations were performed using R-2.13.1 and the R package MASS.18
Results
Pairwise Comparisons of 39 Established Glioma Cell Lines Using 16 Marker Fingerprinting
The DNA fingerprint profiles of 39 glioma cell lines are shown in Table 1. For 5 previously uncharacterized cell lines, information on mutations in TP53 and PTEN, p16/ARF copy number status, and tumorigenicity in nude mice is available in Supplementary material, Table S1. The pairwise comparison of fingerprints depicted in Fig. 1 revealed 4 matched pairs with similarity scores >0.9. Of the 24 cell lines established in our laboratory (LN lines), 4 lines were actually 2 pairs. Analysis of original tumor tissues available established that LN-319 is a tumorigenic subclone of LN-992, which is not tumorigenic in nude mice.8,19 In accordance, both lines carry the same TP53 hotspot mutation in codon 175 (CGC to CAC) and the same PTEN mutation in codon 15 (AGA to AGT),8 reconfirmed in the present study. Similarly, cell line LN-443 is a subline of LN-444, and accordingly, both lines contain the same PTEN mutation (splice deletion exon 5) and are wild-type for TP53.8 One cell line, LN-751, exhibits several markers with 3 alleles that may reflect microsatellite instability, which is found in <10% of glioblastoma, usually associated with pediatric glioblastoma.20 Contamination with another glioma cell line is unlikely, because in this series, it was the only cell line with a homozygous TP53 CGT to TGT mutation in codon 273 and a silent mutation in codon 128 of p16.8
Table 1.
Sixteen marker STR genotypes of the glioblastoma cell lines
| Cell lines | AMELOa | CSF1POa | D13S317a | D16S539a | D18S51 | D21S11 | D3S1358 | D5S818a |
|---|---|---|---|---|---|---|---|---|
| LN-18 | X, Y | 12 | 12, 13 | 11, 13 | 17, 19 | 28 | 15, 16 | 11, 13 |
| LN-71 | X | 11, 12 | 12, 13 | 11, 9 | 13 | 29, 31.2 | 18 | 12 |
| LN-215 | X | 10, 11 | 14 | 12, 9 | 14, 16 | 29 | 14, 17 | 12 |
| LN-229 | X | 12 | 10, 11 | 12 | 13, 15 | 29, 30 | 16, 17 | 11, 12 |
| LN-235 | X, Y | 11, 12 | 9 | 11 | 13, 16 | 29, 32.2 | 15, 17 | 11, 12 |
| LN-Z308 | X, Y | 11, 12 | 11, 13 | 12, 9 | 13, 19 | 31, 31.2 | 15 | 12 |
| LN-319 | X, Y | 10 | 12 | 11, 12 | 14, 19 | 30, 31 | 16, 17 | 11 |
| LN-340 | X, Y | 11 | 11, 12 | 11, 14 | 14, 18 | 28, 32.2 | 15 | 11, 12 |
| LN-382T | X | 10, 11 | 13 | 12, 9 | 15, 16 | 30, 31.2 | 14, 18 | 11 |
| LN-401 | X, Y | 11 | 12, 13 | 9 | 18 | 31.2 | 16, 17 | 12, 13 |
| LN-405 | X | 10, 11 | 8 | 10 | 12, 15 | 29, 31.2 | 14 | 11, 12 |
| LN-427 | X, Y | 11, 12 | 12 | 11, 12 | 12, 15 | 28 | 15, 16 | 10, 12 |
| LN-428 | X, Y | 10 | 8 | 9 | 13, 17 | 30, 31 | 16, 17 | 11, 13 |
| LN-443 | X | 10, 12 | 8 | 10, 11 | 15, 16 | 28, 30 | 15, 17 | 12, 9 |
| LN-444 | X | 10, 12 | 8 | 10, 11 | 15, 16 | 28, 30 | 15, 17 | 12, 9 |
| LN-464 | X | 11 | 11, 13 | 11 | 16 | 28 | 16 | 12 |
| LN-751 | X, Y | 10, 11 | 11, 12 | 11, 12 | 12, 14 | 30, 32.2, 33.2 | 17 | 11, 9 |
| LN-827 | X | 10, 11 | 11 | 12, 13 | 12 | 28, 32 | 15, 17 | 11, 12 |
| LN-992 | X, Y | 10 | 12 | 11, 12 | 14, 19 | 30 | 17 | 11 |
| U87MG | X | 10, 11 | 11, 8 | 12 | 13 | 28, 32.2 | 16, 17 | 11, 12 |
| U118MG | X | 11, 12 | 11, 9 | 12, 13 | 13 | 27, 32.2 | 15 | 11 |
| U138MG | X, Y | 12 | 11, 9 | 12, 13 | 13 | 27, 32.2 | 15 | 11 |
| U178MG | X, Y | 10, 12 | 11 | 10, 13 | 14, 15 | 28, 30 | 17 | 12, 13 |
| U251MG | X, Y | 11, 12 | 10, 11 | 12 | 13 | 29 | 16 | 11, 12 |
| U343MG | X, Y | 10, 12 | 13, 9 | 12, 9 | 23 | 31, 33.2 | 15, 17 | 12, 13 |
| U373MG | X, Y | 11, 12 | 10, 11 | 12 | 13 | 29, 30 | 16, 17 | 11, 12 |
| D247MG | X | 11, 9 | 10, 8 | 12, 9 | 15, 17 | 30 | 17, 18 | 10, 12 |
| T98G | X, Y | 10, 12 | 13 | 13 | 13, 16 | 28, 32.2 | 16 | 10, 12 |
| Hs683 | X, Y | 13, 9 | 12, 8 | 10, 9 | 12, 14 | 27, 33.2 | 14, 16 | 11, 12 |
| A172 | X, Y | 12, 9 | 11 | 12 | 12, 13 | 28, 32.2 | 14, 18 | 11, 12 |
| SF188 | X, Y | 12 | 13 | 11 | 17 | 31 | 15, 18 | 11, 14 |
| SF763 | X | 9 | 10, 12 | 10 | 16 | 27, 30 | 15 | 12 |
| SF767 | X | 11 | 11, 13 | 12, 13 | 12 | 30, 31 | 16 | 12 |
| BS153 | X | 10, 12 | 12 | 9 | 12, 17 | 28, 29 | 17 | 11, 13 |
| LN-2207 | X, Y | 10, 11 | 11, 12 | 12 | 11, 14 | 30, 31 | 14, 16 | 11, 12 |
| LN-2540GS | X, Y | 11 | 11, 13 | 11, 12 | 16 | 29, 31 | 16, 18 | 11 |
| LN-2669 | X, Y | 11, 13 | 11, 12 | 10, 13 | 16 | 30, 31.2 | 14, 15, 16 | 11, 12 |
| LN-2683GS | X | 11, 12 | 10, 8 | 12 | 16, 19 | 30, 31 | 15 | 10, 12 |
| LN-2826GS | X, Y | 10, 11 | 8 | 11, 12 | 12, 20 | 28, 31.2 | 17 | 13 |
| Cell lines | D7S820a | D8S1179 | FGA | PENTAD | PENTAE | THO1a | TPOXa | VWAa |
|---|---|---|---|---|---|---|---|---|
| LN-18 | 10, 8 | 12, 14 | 19, 23 | 11 | 10, 7 | 9 | 8 | 17, 18 |
| LN-71 | 10, 9 | 15 | 21, 22 | 10, 13 | 12, 14 | 8 | 8 | 19 |
| LN-215 | 10 | 10, 14 | 22, 25 | 13, 9 | 11, 7 | 8 | 8 | 18 |
| LN-229 | 11, 8 | 13, 14 | 23 | 10, 11 | 16, 7 | 9.3 | 8 | 16, 19 |
| LN-235 | 10, 12 | 14, 15 | 22 | 11, 12 | 14, 15 | 7, 9 | 8 | 17 |
| LN-Z308 | 10, 12 | 13, 8 | 18, 20 | 11, 9 | 10, 7 | 9.3 | 8, 9 | 15, 17 |
| LN-319 | 9 | 12 | 19, 26 | 13, 9 | 15, 17 | 9, 9.3 | 12, 8 | 15, 18 |
| LN-340 | 11, 12 | 14 | 21, 25 | 12, 15 | 5, 9 | 7, 9.3 | 8 | 17 |
| LN-382T | 11, 8 | 13, 9 | 24, 25 | 12 | 10 | 9, 9.3 | 8 | 16, 18 |
| LN-401 | 10, 9 | 10, 13 | 22, 24 | 13 | 13, 15 | 7, 8 | 8 | 14, 19 |
| LN-405 | 11, 9 | 13 | 22, 24 | 11, 8 | 17 | 8, 9.3 | 11, 8 | 15, 16 |
| LN-427 | 8, 9 | 11, 13 | 23, 24 | 8, 9 | 11 | 8, 9.3 | 11, 8 | 17 |
| LN-428 | 12, 8 | 12, 13 | 20, 25 | 13 | 14, 16 | 8, 9.3 | 11, 8 | 16 |
| LN-443 | 10 | 13, 14 | 21, 23 | 11, 12 | 13, 7 | 7, 9 | 8 | 18, 19 |
| LN-444 | 10 | 13, 14 | 21, 23 | 11, 12 | 13, 7 | 7, 9 | 8 | 18, 19 |
| LN-464 | 10, 13 | 12, 13 | 22 | 9 | 10, 14 | 9 | 12, 9 | 14, 17, 18 |
| LN-751 | 10, 12, 9 | 13, 14 | 18, 22 | 11, 13.1, 14 | 14, 15 | 6, 9 | 11, 8 | 17, 18, 20 |
| LN-827 | 12, 9 | 12 | 23 | 10, 13 | 10, 12 | 6, 9.3 | 12, 8 | 17, 19 |
| LN-992 | 9 | 12, 12.2 | 19 | 13, 9 | 15, 17 | 9, 9.3 | 12, 8 | 15, 18 |
| U87MG | 8, 9 | 10, 11 | 18, 24 | 14, 9 | 14, 7 | 9.3 | 8 | 15, 17 |
| U118MG | 9 | 14, 15 | 23 | 10, 13 | 7 | 6 | 8 | 18 |
| U138MG | 9 | 14, 15 | 18, 23 | 13, 9 | 7 | 6 | 8 | 18 |
| U178MG | 10 | 13, 14 | 22, 26 | 12, 7 | 12, 7 | 7 | 11, 8 | 18, 19 |
| U251MG | 10, 12 | 13, 15 | 21, 25 | 12 | 7 | 9.3 | 8 | 16, 18 |
| U343MG | 11, 9 | 13, 14 | 19, 20 | 10, 9 | 10, 12 | 6, 9.3 | 8, 9 | 17 |
| U373MG | 10, 12 | 13, 15 | 21, 25 | 10, 12 | 10, 7 | 9.3 | 8 | 16, 18 |
| D247MG | 13, 9 | 15 | 24, 27 | 11, 12 | 13, 18 | 6, 9 | 11, 9 | 17, 18 |
| T98G | 10, 9 | 13, 14 | 21 | 10, 11 | 16 | 7, 9.3 | 8 | 17, 20 |
| Hs683 | 11 | 12, 13 | 21.2, 22 | 13, 14 | 13, 15 | 6, 8 | 11, 8 | 18, 20 |
| A172 | 11 | 13, 14 | 20, 22 | 13, 9 | 10, 5 | 6, 9.3 | 11, 8 | 20 |
| SF188 | 10, 8 | 13, 15 | 22, 22.2 | 14 | 10, 13 | 9.3 | 11, 8 | 16, 17 |
| SF763 | 11, 12 | 13, 14 | 22 | 11, 12 | 13, 5 | 9 | 10, 11 | 16, 17 |
| SF767 | 10, 9 | 14 | 23 | 14, 9 | 12, 14 | 8, 9.3 | 10, 8 | 15, 17 |
| BS153 | 11, 9 | 13 | 21, 22 | 14, 9 | 7 | 6, 9 | 11 | 15, 18 |
| LN-2207 | 8 | 11, 13 | 22, 23 | 12, 13 | 11, 12 | 7, 9 | 8, 9 | 16, 17 |
| LN-2540GS | 10, 9 | 12, 15 | 23 | 11, 9 | 11 | 10, 8 | 8 | 15, 17 |
| LN-2669 | 8, 9 | 11, 13 | 24 | 13 | 12 | 8, 9.3 | 11, 8 | 15, 17 |
| LN-2683GS | 11 | 10, 14 | 22, 23.2 | 13, 14 | 10 | 7, 9.3 | 12, 8 | 14, 18 |
| LN-2826GS | 11 | 13, 14 | 21, 24 | 11 | 11, 5 | 6, 7 | 11, 9 | 14, 17 |
aindicates the 9 markers used in the DSMZ database.
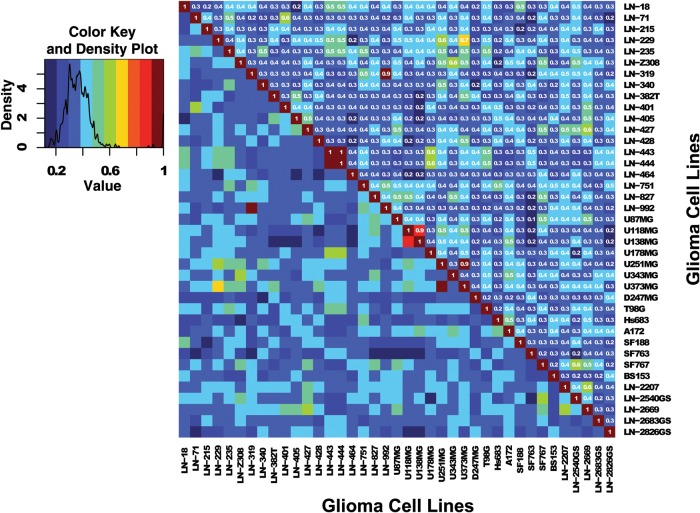
Fig. 1.
Heatmap based on the Sørensen similarity index between cell lines of the glioma cell line panel. The similarity index was computed using the 16 marker set (15 STR markers + AMEL) on 39 glioma cell lines (Glioma-CL) (Table 1). Color key and density plot are provided in the additional graphic (left). Similarity values estimated between cell lines with different origin are comprised in the range from 0.1 (dark blue) to 0.7 (yellow). High similarity values (range, 0.9–1.0) are observed for cell lines with same origin and correspond to the red and dark red squares.
From the 15 glioma cell lines established by other laboratories, the cell lines U118MG and U138MG were identified as being of the same origin, similarly to U251 and U373, as has been reported previously.5,8,21 Respective alerts are posted on the ATCC website for misidentified cell lines.
Comparison with DNA Fingerprints of 2289 Cell Lines in the DSMZ Database 9 Markers
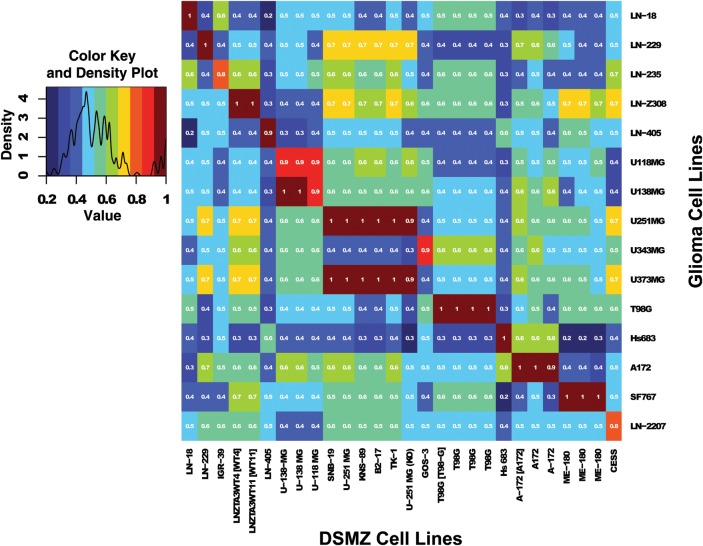
The fingerprints established for the set of 39 GBM cell lines were compared with the 9-marker fingerprint database of DSMZ and ATCC. All cell lines with a similarity score ≥0.8 to any of our characterized glioma cell lines were extracted, and respective pairwise comparisons are shown in Fig. 2. We confirmed the fingerprints of the cell lines LN-405 (score, 0.93; DSMZ# ACC189), LN-18 (score, 1; CRL-2610), and LN-229 (score, 1; CRL-2611) that the laboratory deposited with DSMZ and ATCC, respectively, or cell lines that we had obtained from ATCC originally, such as U87 (score, 1). Similarly, the in vitro genetically modified cell lines derived from LN-Z308 (LNZTA3WT4 and 11, CRL-11543 and 44) that have been deposited were identified with scores of 0.97.
Fig. 2.
Heatmap based on the Sørensen similarity index computed between the DSMZ dataset and the glioma cell line panel for markers containing similarity values superior to the cutoff of 0.8. The similarity index was computed on the 9 marker set (8 STR markers + AMEL). Colour key and density plot are provided in the additional graphic (at left). Similarity values estimated between cell lines with different origin are included in the range 0.1 (dark blue) to 0.7 (yellow). High similarity values observed (range, 0.9–1.0) for cell lines with same origin correspond to the red and dark red squares. The similarity score of 0.8 (orange) may or may not reflect similarity (see text for details). Of note, multiple cell lines with identity scores of 0.9 or 1 are extracted from the DSMZ database. Some identify redundant entries, while others reflect genetically modified cell lines of the same origin, or previously known misidentifications. However, 1 previously unknown identity was uncovered between SF767 and ME-180 with a score of 1, suggestive of cross-contamination. (See text for explications)
However, the identity score of 1 for SF767 and ME-180 (HTB-33) identifies a potential cross-contamination. ME-180 is a squamous cell carcinoma cell line of the cervix reported positive for human papillomavirus.22 No reference DNA fingerprint of SF767 was available online. We are not aware that ME-180 was ever used or even present in our laboratory.
In contrast, the U373MG identity scores of 0.9 or 1 shared with the cell lines SNB-19, U-251MG, KN-S89, B2-17, and TK-1 confirms respective alerts placed on the Web sites of the databases of ATCC, DSMZ, JCRB, or COSMIC. The similarity (score, 0.9) of GOS-3 (ACC#408) with U-343MG is in accordance with an annotation on the respective DSMZ Web site.
Cell line LN-235 exhibited a similarity score of 0.8 with the melanoma cell lines IGR-37 and IGR-39, which are both from the same patient (DSMZ# ACC 237 and 239). There was no original tumor tissue available from LN-235. However, IGR-37 and IGR39 are known to contain the classic B-raf mutation (V600E) commonly found in melanoma,23,24 which is absent in LN-235, as determined by diagnostic pyrosequencing. Of surprise, LN-2207 had a similarity score of 0.81 with the lymphoblastic cell line Cess (ATCC# TIB-190). A potential contamination could be excluded, because LN-2207 exerted a fingerprint identical to its respective original tumor tissue.
This extract based on similarity in addition illustrates the redundancy of the DSMZ database with multiple entries of cell lines that, however, may reflect different passage number/clones, as suggested by minor differences of similarity.
Evaluation of Similarity Scores for Cell Lines
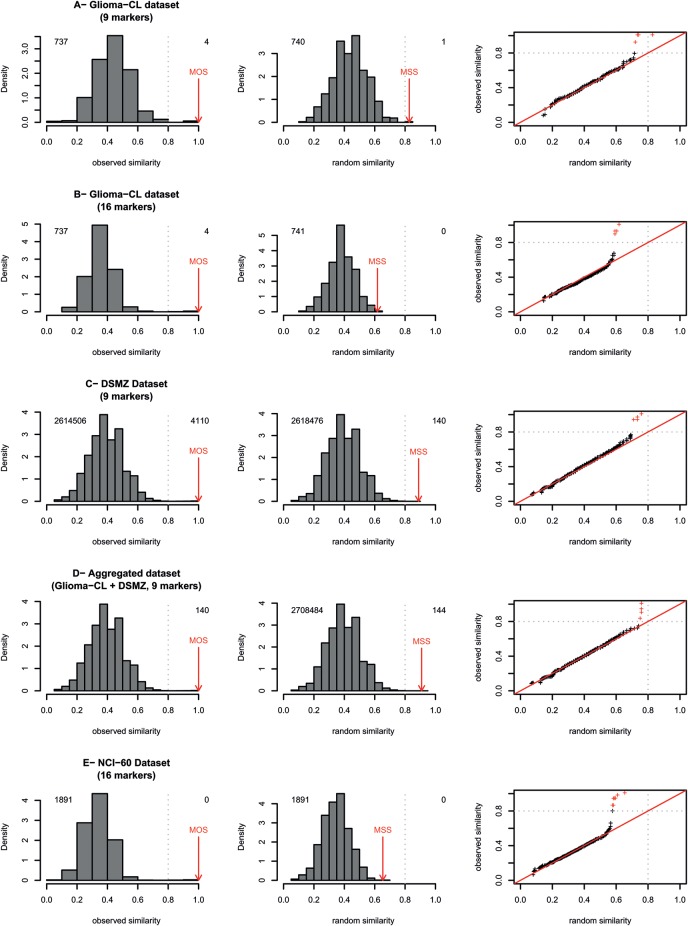
As shown above, a similarity score of 0.8, as suggested in the literature,6,7 is not sufficient to reliably discriminate between same or different origin if only a 9-marker DNA fingerprint is available. Indeed, this cutoff can be used to detect cross-contamination, but our simulations creating similar sized datasets show that this value can be observed between 2 profiles randomly rearranged. After random rearrangement of 9 markers in the glioma cell line collection, we observe that 1 similarity value was >0.8 (Fig. 3A) and 8 were >0.7. In contrast, we strictly detected no similarity values >0.7 between 2 random profiles when we consider all markers (Fig. 3B). The median MSS was ∼0.8 for the glioma cell line dataset and the NCI-60 dataset when only 9 markers were kept that are also available in the DSMZ database. We observed that the median MSS was ∼0.9 for the DSMZ dataset comprising a large number of cell lines (Fig. 3C and D and Table 2). Consequently, the cutoff of 0.8 can be used to detect potential cross-contamination, but it is not sufficient to prove or disprove same identity of 2 cell lines. In contrast, when we increased the number of markers (e.g., 16 markers) (Fig. 3B–E), we observe that it was unlikely to obtain a similarity score of 0.8 between random profiles (Table 2). Typically, when using the 16 marker glioma cell line dataset and the 16 marker NCI-60 panel, the QQ-plot representations showed that the cutoff values between observed and random distributions were ∼0.64 (Fig. 3E and Table 2).
Fig. 3.
Distributions of observed and simulated similarity values. Random similarity distributions were calculated for the 3 datasets (Glioma-CL, NCI-60, and DSMZ) using 16 markers, where available, or 9 markers based on rearranged profiles of markers randomly and independently selected from the set of genotypes in the given dataset. Quantile-quantile plot (QQ-plot) representations15 shown in the third column of the Figure provide a graphical comparison of observed versus random similarity distributions for each dataset, the red crosses represent observed values above the cutoff of 0.8. In the density plots, the number of similarity values inferior and superior to the cutoff of 0.8 is given in the left and right top of the respective panels. For each dataset, the number of similarity values is equal to (n× n− n)/2 where n corresponds to the number of cell lines contained in the collection (more details in statistical section). Red arrows identify the maximal observed similarity (MOS) and maximal simulated similarity (MSS). The grey dotted lines point to the limit of high similarity area (range, 0.8–1.0).
Table 2.
Estimated maximal simulated similarity (MSS) for the three datasets (DSMZ, Glioma-CL and NCI-60) for 9 and 16 markers
| Dataset | No. Cell lines | No. Marker | Min | Median | Mean | Max | CI 95% |
SD | MAD | |
|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||||
| Glioma-CL | 39 | 9 | 0.7143 | 0.7842 | 0.7865 | 0.9143 | 0.7333 | 0.8621 | 0.0366 | 0.0379 |
| Glioma-CL | 39 | 16 | 0.5769 | 0.6316 | 0.6362 | 0.7037 | 0.5849 | 0.6844 | 0.0262 | 0.0240 |
| NCI-60 | 62 | 9 | 0.6429 | 0.7407 | 0.7392 | 0.8462 | 0.6766 | 0.8215 | 0.0398 | 0.0422 |
| NCI-60 | 62 | 16 | 0.5882 | 0.6400 | 0.6381 | 0.7200 | 0.5903 | 0.7059 | 0.0279 | 0.0205 |
| DSMZ | 2289 | 9 | 0.8750 | 0.9032 | 0.9102 | 1.0000 | 0.8889 | 0.9616 | 0.0193 | 0.0199 |
Note: Statistic values obtained after 100 repetitions of the simulation procedure (Fig. 4). Confidence intervals at 95% (95% CIs) computed by percentile method. Abbreviations: MAD, median absolute deviation; SD, standard deviation.
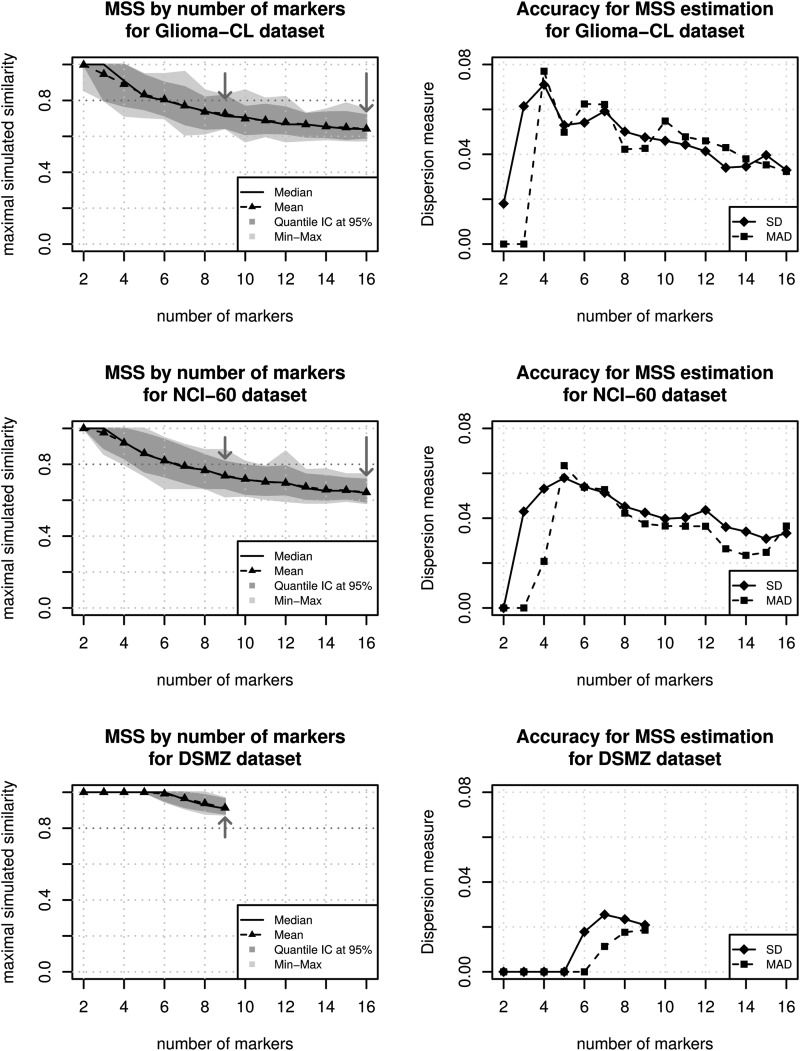
Saturation curves obtained by our second simulation procedure clearly showed the importance of the number of markers in the computation of the similarity between DNA fingerprint profiles (Fig. 4). Median and mean of MSS values were ∼0.78 for the glioma cell line dataset and ∼0.74 for the NCI-60 panel for 9 markers, and the cutoff of 0.8 is included in the confidence intervals around the median and the mean of MSS values. In other words, cross-contamination can neither be excluded nor proven at the cutoff of 0.8. In contrast, this threshold was clearly outside the confidence intervals for 16 markers, providing the power for clear distinction (Table 2, Fig. 4). Simulation results obtained for the DSMZ dataset for which only 9 markers are available show that the number and diversity of cell lines from a given collection affect the estimation of the MSS values. The DSMZ dataset contains a nonnegligible proportion of similar data. Indeed, we detected 805 cell lines with at least one similarity value equal to 1 and 1281 cell lines that exhibit at least 1 high similarity value (>0.8) in considering the 9 marker profiles (8 STR markers plus the AMEL marker). The redundancy in part originates from different spelling of the names of cell lines or database-specific added names, as is shown in Fig. 2, although slight differences may also reflect evolution by passaging in different laboratories. After the exclusion of identical and highly similar cell lines, we observed that MSS values were ∼0.8 (Supplementary material, Fig. S2) in accordance with the results observed for the NCI-60 and the glioma cell line datasets. Loss of heterozygosity is a frequent event in tumor cell lines that reduces the informativeness of the STR markers, including the AMEL marker, thereby weakening the discriminatory power of the analysis. The heterozygosity at the distinct STR markers was similar in our dataset of 39 glioma cell lines and the 2278 cell lines in the DSMZ database (0.54–079 for our dataset and 0.57–0.71 for DSMZ), whereas it was different for the AMELO marker that indicates the sex chromosomes (Supplementary material, Table S2). Heterozygosity of this marker was much more common in the glioma cell lines with 0.59, compared with those with 0.36, which may simply reflect the known higher prevalence of man affected with glioblastoma, compared with the overall patient population with cancer that is represented by cell lines.
Fig. 4.
Representation of the estimation of maximal simulated similarity (MSS) in function of the number of markers. The 3 datasets (Glioma-CL, NCI-60, and DSMZ) were used to simulate similarity scores and to compute their mean and median, as well as the 95% confidence intervals (CIs) for the median (dark grey) and the minimum-maximum intervals (light grey). The arrows indicate the position of the 9 and 16 marker profiles commonly used. For 9 markers we observed that the cutoff of 0.8, suggested by previous studies6,7 (dotted lines) is included in the confidence region defined by the percentile method. This means that at least 1 false positive value >0.8 can be expected in >5% of the datasets. In contrast, this value is outside the confidence region for the 16 markers for both datasets. This means that it is not very probable to obtain a MSS value equal or superior to 0.8 by hazard. The second series of graphics provide accuracy measures of the estimation of MSS, standard deviation (SD), and median absolute deviation (MAD), per number of markers. These figures indicate that for increasing numbers of markers used the SD and MAD are reduced, consequently the MSS values are estimated with better accuracy.
Discussion
The present study provides a 16 marker DNA fingerprint database for glioma cell lines frequently used for research. This database can be used as reference for authentication of frequently used glioma cell lines, as requested by journals and research funding agencies. The cross-comparison among and with publically available databases revealed previously unknown misidentification of 3 cell lines. For the 2 cell lines misidentified in our laboratory, the origin could be established, identifying LN-319 as a tumorigenic subline of LN-992 and establishing LN-443 as a subline of LN-444. The discovery that cell line SF767 has an identical DNA fingerprint to the squamous cell carcinoma line ME-180 will need further investigations, because no reference STR fingerprints were available. Curiously, SF767 has been described by different groups as being very different from other glioma cell lines (e.g., in terms of tumor morphology when grown in nude mice25 or in terms of patterns of E-cadherin expression).26
Furthermore, this study clearly showed that 9 marker fingerprints that are available for large number of cell lines are often insufficient to discriminate the origin of cell lines when the similarity value is close to the classical thresholds proposed in the literature (e.g., 0.8). Under these circumstances additional factors need to be considered when evaluating the similarity score of a cell line with doubts on the origin.
Number of Markers
Simulation procedures have shown that the number of markers measured has a high influence on the distribution of the similarity values and, indirectly, the value of the cutoff. In using the Sørensen score, we observed that the cutoff of 0.8 proposed by Masters et al.6,7 did not reliably discriminate between same or different origin with the 9 marker set, whereas this was much improved when considering 16 markers (Fig. 3). Our second simulation procedures confirmed that the limits of the random distribution of the similarity index decreases in function of the number of markers used (Fig. 4).
Analyses performed on the DSMZ dataset have shown the limitation of our simulation procedure when the reference database contains a high proportion of identical or highly similar profiles. The DSMZ database is based on several sources (e.g., ATCC, JCRB, and RIKEN), introducing a high proportion of duplicates (different names) or very highly similar cell lines. The set of 9 markers was clearly not sufficient to identify the difference among cell lines with efficiency and biased the estimation of MSS in over-representing some given genotypes, thereby reducing the allele diversity. Taking that finding into consideration, our simulation was not independent of the reference database that introduced an abnormal proportion of highly similarity values into the generation of random profiles. In addition, the high number of random profiles generated for the simulation associated with the DSMZ dataset may have further favored high MSS values in increasing the chance to obtain 2 similar random profiles. For this reason, we recommend careful definition of the reference database used to identify cell lines in using a priori knowledge on the nature of them and in limiting the number of duplicates.
Mutation Rate
As illustrated by Parson et al.,27 the stability of STR profiles is not the same for all markers. These authors observed that the mutation rates of markers fluctuated from 0.01% (TH01 and TPOX) through 0.28% (FGA) for cancer cell lines (i.e., K652, U937, Jurkat, and CCRF-CEM) in their study. To account for the mutation rate of a marker, a weighted similarity measure can be computed in considering the weighted sum of the partial similarity obtained for each marker14,28,29. However, the mutation rates are strongly variable among tumor cell lines. Moreover, genomes of cancer cell lines are often instable and are modified by many mechanisms, including microsatellite instability, deletions, amplifications, or rearrangements, in a tumor of origin-dependent manner. For these reasons and without a priori knowledge of the mutation rate of the tested population of cell lines, we recommend use of uniform weighting to estimate similarity between glioblastoma cell lines by default.
Threshold and Cell Origin
Definition of a threshold to determine the identity of a cell line needs to consider the number of markers, the marker stability, number of shared alleles, and number and nature of disparate alleles.27 For example, the marker for sex comprises only 2 alleles. In contrast, we count 13 distinct alleles for the marker FGA in the Glioma-CL dataset. The score used to estimate similarity between cell lines is an additional criterion to include in the definition of the threshold. In this study, we chose to use the Sørensen index, as proposed by Lynch et al.,13 to estimate the similarity among the DNA fingerprint profiles to detect the parental cell line. However, our simulation process can be generalized to apply to other similarity scores.14,30
Thresholds and similarity scores are attractive and user-friendly tools, but it is important to know their limitations. In our study, we showed that with the 9 marker STR fingerprint similarity, values close to the threshold of 0.8 are difficult to judge to exclude identity with a high probability. Additional information is required, such as presence or absence of characteristic but uncommon mutations, to decide whether the sample is different. If this is not possible, we recommend considering the number and type of necessary events to explain the difference between the 2 profiles. For example, the acquisition of a different allele is mechanistically more difficult than a mere deletion of an allele, even though they are weighed equally in the score. The definition of a reference database with a limited number of duplicates and the use of simulation procedures, as proposed in our study, can provide an efficient tool to evaluate the consistency of similarity values and thresholds for DNA fingerprint profiling studies in general.
Supplementary Material
Funding
This work was supported by the Swiss National Science Foundation (MEH, MD) and OncoSuisse (MEH, MD).
Supplementary Material
Acknowledgments
We thank Davide Sciuscio and Irene Vassallo for critical discussions throughout this project.
Conflict of interest statement. None declared.
References
- 1.Organization ATCCSD, ASN-0002 W. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10(6):441–448. doi: 10.1038/nrc2852. doi:10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tissue Culture Research Communications. 1999;18(4):329–338. [Google Scholar]
- 3.Torsvik A, Rosland GV, Svendsen A, et al. Spontaneous malignant transformation of human mesenchymal stem cells reflects cross-contamination: putting the research field on track - letter. Cancer Res. 2010;70(15):6393–6396. doi: 10.1158/0008-5472.CAN-10-1305. doi:10.1158/0008-5472.CAN-10-1305. [DOI] [PubMed] [Google Scholar]
- 4.Vogel G. To Scientists’ Dismay, Mixed-Up Cell Lines Strike Again. Science. 2010;329(5995):1004. doi: 10.1126/science.329.5995.1004. doi:10.1126/science.329.5995.1004. [DOI] [PubMed] [Google Scholar]
- 5.Lorenzi PL, Reinhold WC, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8(4):713–724. doi: 10.1158/1535-7163.MCT-08-0921. doi:10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci USA. 2001;98(14):8012–8017. doi: 10.1073/pnas.121616198. doi:10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Muzusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304, and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–338. [Google Scholar]
- 8.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. doi:10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones G, Machado J, Jr, Merlo A. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 2001;61(13):4978–4981. [PubMed] [Google Scholar]
- 10.Sciuscio D, Diserens AC, van Dommelen K, et al. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clin Cancer Res. 2011;17(2):255–266. doi: 10.1158/1078-0432.CCR-10-1931. doi:10.1158/1078-0432.CCR-10-1931. [DOI] [PubMed] [Google Scholar]
- 11.Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297–302. doi:10.2307/1932409. [Google Scholar]
- 12.Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol Skr. 1948;5:1–34. [Google Scholar]
- 13.Lynch M. The similarity index and DNA fingerprinting. Mol Biol Evol. 1990;7(5):478–484. doi: 10.1093/oxfordjournals.molbev.a040620. [DOI] [PubMed] [Google Scholar]
- 14.Legendre P, Legendre L. Numerical Ecology. Second English Edition. Amsterdam: Elsevier; 1998. [Google Scholar]
- 15.Becker RA, Chambers JM, Wilks AR. The New S Language. London: Chapman & Hall; 1988. [Google Scholar]
- 16.Davison AC, Hinkley DV. Bootstrap methods and their application. London: Cambridge University Press; 1997. [Google Scholar]
- 17.Manly BFJ. Randomization, bootstrap and Monte-Carlo methods in biology. 3rd ed. London: Chapman & Hall/CRC; 2006. [Google Scholar]
- 18.Team RDC. R: a language and environment for statistical computing. Vienna, Austria: 2011. http://cran.r-project.org/doc/FAQ/R-FAQ.html . [Google Scholar]
- 19.Lambiv WL, Vassallo I, Delorenzi M, et al. The Wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol. 2011;13(7):736–747. doi: 10.1093/neuonc/nor036. doi:10.1093/neuonc/nor036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso M, Hamelin R, Kim M, et al. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res. 2001;61(5):2124–2128. [PubMed] [Google Scholar]
- 21.Azari S, Ahmadi N, Tehrani MJ, Shokri F. Profiling and authentication of human cell lines using short tandem repeat (STR) loci: Report from the National Cell Bank of Iran. Biologicals. 2007;35(3):195–202. doi: 10.1016/j.biologicals.2006.10.001. doi:10.1016/j.biologicals.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Reuter S, Delius H, Kahn T, Hofmann B, zur Hausen H, Schwarz E. Characterization of a novel human papillomavirus DNA in the cervical carcinoma cell line ME180. J Virol. 1991;65(10):5564–5568. doi: 10.1128/jvi.65.10.5564-5568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reifenberger J, Knobbe CB, Sterzinger AA, et al. Frequent alterations of Ras signaling pathway genes in sporadic malignant melanomas. Int J Cancer. 2004;109(3):377–384. doi: 10.1002/ijc.11722. doi:10.1002/ijc.11722. [DOI] [PubMed] [Google Scholar]
- 24.Meyer P, Klaes R, Schmitt C, Boettger MB, Garbe C. Exclusion of BRAFV599E as a melanoma susceptibility mutation. Int J Cancer. 2003;106(1):78–80. doi: 10.1002/ijc.11199. doi:10.1002/ijc.11199. [DOI] [PubMed] [Google Scholar]
- 25.Ozawa T, Wang J, Hu LJ, Lamborn KR, Bollen AW, Deen DF. Characterization of human glioblastoma xenograft growth in athymic mice. In Vivo. 1998;12(4):369–374. [PubMed] [Google Scholar]
- 26.Lewis-Tuffin LJ, Rodriguez F, Giannini C, et al. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS One. 2010;5(10):e13665. doi: 10.1371/journal.pone.0013665. doi:10.1371/journal.pone.0013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parson W, Kirchebner R, Muhlmann R, et al. Cancer cell line identification by short tandem repeat profiling: power and limitations. FASEB J. 2005;19(3):434–436. doi: 10.1096/fj.04-3062fje. [DOI] [PubMed] [Google Scholar]
- 28.Estabrook GF, Rogers DJ. A general method of taxonomic description for a computed similarity measure. BioScience. 1966;16(11):789–793. doi:10.2307/1293644. [Google Scholar]
- 29.Gower JC. A general coefficient of similarity and some of its properties. Biometrics. 1971;27(4):857–871. doi:10.2307/2528823. [Google Scholar]
- 30.Duarte JM, dos Santos JB, Melo LC. Comparison of similarity coefficients based on RAPD markers in the common bean. Genet Mol Biol. 1999;22:427–432. doi:10.1590/S1415-47571999000300024. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.